Advances in Microbiology
Vol.3 No.3(2013), Article ID:33775,8 pages DOI:10.4236/aim.2013.33038
Identification and Characterization of Lactic Acid Bacteria Isolated from Fermented Rice Bran Product
Microbial Genetics, Institute of Genetic Resources, Faculty of Agriculture, Kyushu University, Fukuoka, Japan
Email: *doi@agr.kyushu-u.ac.jp
Copyright © 2013 Katsumi Doi et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received April 8, 2013; revised May 8, 2013; accepted June 8, 2013
Keywords: Rice Bran Fermentation; Lactobacillus johnsonii; 16S rRNA Gene; RAPD-PCR
ABSTRACT
To analyze the microflora in fermented rice bran product, bacterial colonies were grown under various conditions. Although cultivation temperature did not affect the number of bacterial colonies formed on agar plates, twice as many colonies formed under aerobic as under anaerobic conditions. All colonies appearing on the plates showed acid production. Based on 16S rRNA sequence analysis, nearly all of the bacteria in the fermented product were highly similar (>99%) to Lactobacillus johnsonii. In addition, several Bacillus cereus and unidentified Lactobacillus strains that grew only under anaerobic conditions at 30˚C were seen. Random Amplified Polymorphic DNA (RAPD)-PCR analysis showed the amplified patterns of all isolates to differ substantially from the reference strain L. johnsonii. We conclude that L. johnsonii-related strains predominate in fermented rice bran product, and that these bacteria produce lactic acid to decrease the pH of the fermented product. Several novel Lactobacillus strains may also occur in this environment.
1. Introduction
Rice bran is rich in nutrients, and so is utilized as a food material, worked food product and a whole food. The fermented rice bran product is widely used for pickling vegetables, as an organic fertilizer, and as animal and aquaculture feed [1-3]. Lactic acid bacteria (LAB), such as the Lactobacillus species and Tetragenococcus halophilus, or yeasts, such as Rhizopus oligosporus, Saccharomyces cerevisiae and the Aspergillus species, are the main producers of fermented rice bran product [4,5]. Interestingly, the strain mediating lactic acid fermentation of rice bran can determine some of the properties of the fermented product. For example, Lactobacillus brevis produces gamma-aminobutyric acid (GABA) with rice bran, and the fermented rice bran product shows antihypertensive properties [6,7]. Fermented rice bran may also exert inhibitory effects against pathogenic microorganisms [8]. In this way, LAB contributes to the longterm preservation of fermented rice bran products. In fact, Japanese traditional fermented rice bran mash, nukadoko, can last for as long as 100 years without spoilage [4]. In an earlier report, seven predominant operational taxonomic units closely related to known Lactobacillus species were shown to inhabit long-aged nukadoko. Slowgrowing lactobacilli stably dominated the nukadoko microbiota, with L. acidifarinae being the primary mediator of the lactic acid fermentation [1]. Because rice bran fermentation is carried out using an open fermentation system, a number of different bacteria can inhabit the bran, and their species can vary over the period of the fermentation and depending on the fermentation conditions. Consequently, accurate characterization of the microflora in fermented rice bran product and selection of functional strains is very difficult.
Characterization of a bacterial community within an environmental sample can be done using both cultivation-dependent and cultivation-independent methods [9, 10]. With either approach, the sequence of the gene encoding 16S rRNA is frequently used for molecular identification of bacterial strains. However, for closely related Lactobacillus strains, such as those belonging to the L. acidophilus and L. casei groups, it is often difficult to make a precise identification using phylogenetic analysis of the 16S rRNA nucleotide sequence [11,12]. In those cases, molecular typing using Random Amplified Polymorphic DNA (RAPD), Restriction Fragment Length Polymorphism (RFLP), Denaturing Gradient Gel Electrophoresis (DGGE) or Enterobacterial Repetitive Intergenic Consensus (ERIC)-PCR provides an effective alter native [13-16]. The RAPD-PCR method entails using short, random sequence primers (9 - 10 nucleotides) under low-stringency annealing conditions to amplify an arbitrary fragment of template DNA. Single primers anneal to near-complementary sequences anywhere within the genome, but amplification only occurs when two priming sites are sufficiently close to one another.
Here, we report the molecular identification and typing of bacteria isolated from a fermented rice bran product under aerobic and anaerobic conditions. We suggest that several novel strains were present within the fermented rice bran.
2. Materials and Methods
2.1. Chemical Analysis of Fermented Rice Bran Product
Fermented rice bran product was obtained from Create Co. Ltd. (Tosu, Saga, Japan). The materials used for fermentation were 10% (w/w) rice bran, 1% (w/w) maltose, 1% (w/w) leaf mold and spring water. Fermentation was carried out for 20 days at 37˚C. The fermented sample was then kept at 4˚C for as short a time as possible before use. The pH of the fermented products was measured in 1-g samples using a φ 310 pH meter (Beckman Coulter, Inc., Brea, CA, USA). The total protein content of the products was determined using a Protein Rapid Assay kit (Wako Pure Chemical Industries Ltd., Osaka, Japan). When calculating sample protein concentrations, bovine serum albumin (Sigma-Aldrich Co. LLC., St. Louis, MO, USA) was used to construct the standard curve.
2.2. Isolation of LAB under Various Conditions
To isolate LAB, fermented samples were suspended in 0.1 M sodium phosphate buffer (pH 7) to a final concentration of 10% (wt/vol) through vigorous vortexing followed by centrifugation at 2000 × g at 4˚C for 5 min. The supernatant containing the bacterial suspension was transferred to a clean tube, after which 100-μl aliquots of the bacterial suspension was spread on GYP-CaCO3 agar [17] containing 0.5% CaCO3, 10 ppm cycloheximide and 10 ppm sodium azide in serial dilutions from 10−1 to 10−8. Cultivation was then carried out for 36 h at 30˚C or 37˚C under anaerobic condition in an Anaero Packor Anaero Pack Rectangular Jar (Mitsubishi Gas Chemical Company, Inc., Tokyo, Japan), or in a static aerobic culture.
Twenty-seven colonies randomly selected from each of the four culture conditions were picked up and purified twice by isolation on MRS agar (Becton, Dickinson and Company, Franklin Lakes, NJ, USA). The sugar fermentation patterns of the isolates were then determined over a period of 72 h using an API 50 CHL kit (bioMe´rieux, Marcy l’Etoile, France) according to the manufacturer’s instructions. The purified strains were stored in 25% glycerol solution at −20˚C and later cultured in MRS broth for 36 h at 30˚C or 37˚C before use for genome DNA extraction.
2.3. 16S rRNA Gene Sequence Analysis
Bacterial cells grown in MRS broth under each of the four conditions were used for genomic DNA extraction and purification. DNA was extracted using an Illustra Bacteria Genomic Prep Mini Spin Kit (GE Healthcare, Uppsala, Sweden) following the manufacturer’s instructtions. The 16S rRNA gene was amplified using polymerase chain reaction (PCR) with ExTaq Polymerase (TaKaRa Bio, Shiga, Japan) and primers 27f (5’-AGAGTTTGATCCTGGCTCAG-3’) and 1492r (5’-ACGGCTACCTTGTTACGACTT-3’). The PCR protocol entailed initial denaturation at 95˚C for 5 min, followed by 30 cycles of denaturation for 1.5 min at 95˚C, annealing for 1.5 min at 55˚C and extension for 1.5 min at 72˚C, and a final extension for 10 min at 72˚C. The PCR products were subjected to electrophoresis in 2.0% agarose gels in 1X TAE (100 V for 50 min), and were then recovered using a Fast Gene Gel/PCR Extraction Kit (NIPPON Genetics Co. Ltd., Tokyo, Japan). The purified DNAs were then inserted into pTA2 vector using a TArget clone-plus-kit (TOYOBO, Osaka, Japan), after which Escherichia coli DH5α were transformed with the recombinant plasmid, and the transformants were spread onto LB agar plates containing IPTG (40 μg/ml) and X-Gal (150 μg/ml). Isolation of plasmids from the transformants was carried out using Xprep Plasmid DNA Mini Kit (AS ONE Corporation, Osaka, Japan).
Nucleotide sequencing was performed using a Big Dye Terminator v3.1 Cycle Sequencing Kit (Life Technologies, Carlsbad, CA, USA) with M13 Primer M3 (5’- GTAAAACGACGGCCAGT-3’) or M13 Primer RV (5’- CAGGAAACAGCTATGAC-3’). Sequences were determined using an Applied Biosysytems Gene Analyzer 3130xl (Life Technologies). A homology search for the 16S rRNA sequences was carried out with the Nucleotide Sequence Data Library using the BLAST program. Phylogenetic trees were then constructed with Clustal W ver. 2.1 (http://clustalw.ddbj.nig.ac.jp/) using the neighborjoining (NJ) method. Distance matrices for the aligned sequences were calculated using the two-parameter method of Kimura [18]. E. coli was used as an out group organism.
2.4. RAPD-PCR Analysis
RAPD-PCR was carried out using an Arbitrary Primer Set (Nippongene, Tokyo, Japan) and Gene Taq FP polymerase (Nippongene). The PCR protocol entailed initial denaturation for 3 min at 95˚C, followed by 30 cycles of denaturation for 15 s at 95˚C, annealing for 2 min at 30˚C and extension for 1 min at 72˚C, and a final extension for 5 min at 72˚C. The PCR products were analyzed using 2.0% agarose gel electrophoresis (1X TAE at 100 V for 50 min), then photographed. Numerical analysis of the protein pattern was performed using the GelCompar system (version 4.0; Applied Maths, Sint-Martens-Latem, Belgium), which normalizes the fragment pattern data for band intensity and relative band position with respect to nucleotide chain length. For construction of the dendrogram, similarity between pairs was expressed using the pair-group method with arithmetic averages (UPGMA). All strains were analyzed at least three times to ensure the reproducibility of the fingerprinting pattern.
3. Results
3.1. Chemical Properties and Microflora of Fermented Rice Bran
The average pH and total protein content of the fermented rice bran were 3.08 ± 0.08 and 461 μg/ml, respectively. After incubation for 3 days on agar plates, numerous colonies with a clear zone around their edges were seen (Figure 1). Nearly all of the colonies were ash gray or beige in color and were smooth. Around 106 cfu/ ml formed clearzones in the samples tested (Table 1). Acid-producing colonies appeared with equal frequency at 30˚C and 37˚C. On the other hand, twice as many colonies grew under aerobic conditions as grew under anaerobic conditions.
3.2. Classification of Isolated LAB Based on Sugar Utilization and 16S rRNA Gene Sequencing Analysis
Total DNAs were isolated from 27 colonies grown under each condition, after which the 16S rRNA gene sequences were determined. From the subsequent phylogenetic analysis, 25 isolated strains were categorized as genus Lactobacillus (Figure 2), with isolates showing more than 99% similarity to L. johnsonii. However, the 16S rRNA gene sequences for three strains, C30An7, C30An8 and C30An22, showed less than 95% similarity to those of any deposited bacterial strains. In addition, the 16S rRNA gene sequences of two strains, C30An16 and C30Ae21, were identical to that of Bacillus cereus. These nucleotide sequences were deposited in the DDBJ database under accession numbers AB809566 to AB- 809592.
The physiological characterization of the three as yet undeposited strains (C30An7, C30An8 and C30An22), as well as two other randomly selected strains (C37An3 and C37An9), was accomplished using an API kit (Table 2). All isolated strains grew on and fermented amygdalin, Dcellobiose, salicin and D-sucrose. Acid was not produced from adonitol, arabinose, erythritol, esculin, lactose, mannitol, cellobiose, melezitose, melibiose, raffi nose, rhamnose, sorbitol or xylose. The fermentation of Nacetyl glucosamine, D-fructose, D-galactose, D-glucose, glycerol, D-maltose, D-mannose, D-melibiose, D-raffinose, D-ribose and trehalose was strain-dependent.
3.3. RAPD-PCR Analysis
The 27 isolated strains were each genotyped using RAPD-PCR with 25 primers, and reproducible RAPDPCR patterns were obtained with two of the primers, AP-A-2 (5’-TGGATTGGTC-3’) and AP-A-23 (5’- GATCTGACTG-3’). Based on UPGMA cluster analysis of the RAPD-PCR profiles, the strains were divided into six clusters (i.e., 1, 4, 5, 6, 7 and 8) or differentiated strains within others (clusters 2 and 3). The results ob-
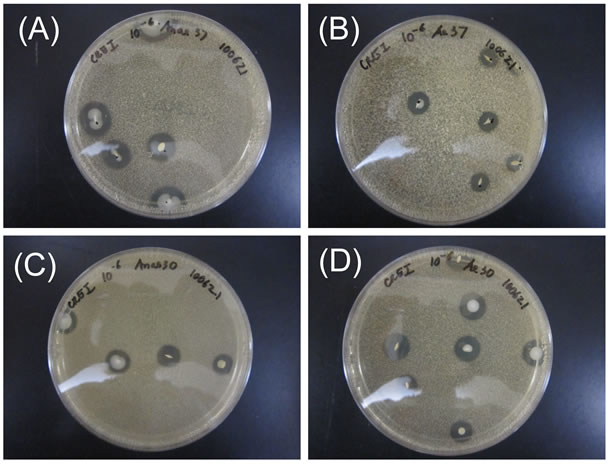
Figure 1. Morphologies of colonies formed from rice bran fermentation. Colonies were grown under four conditions: (A) anaerobic at 37˚C; (B) aerobic at 37˚C; (C) anaerobic at 30˚C; and (D) aerobic at 30˚C. Colonies that produced acid (causing a clear zone) were observed and picked up for further experimentation.

Table 1. Numbers of colonies formed under the indicated conditions.
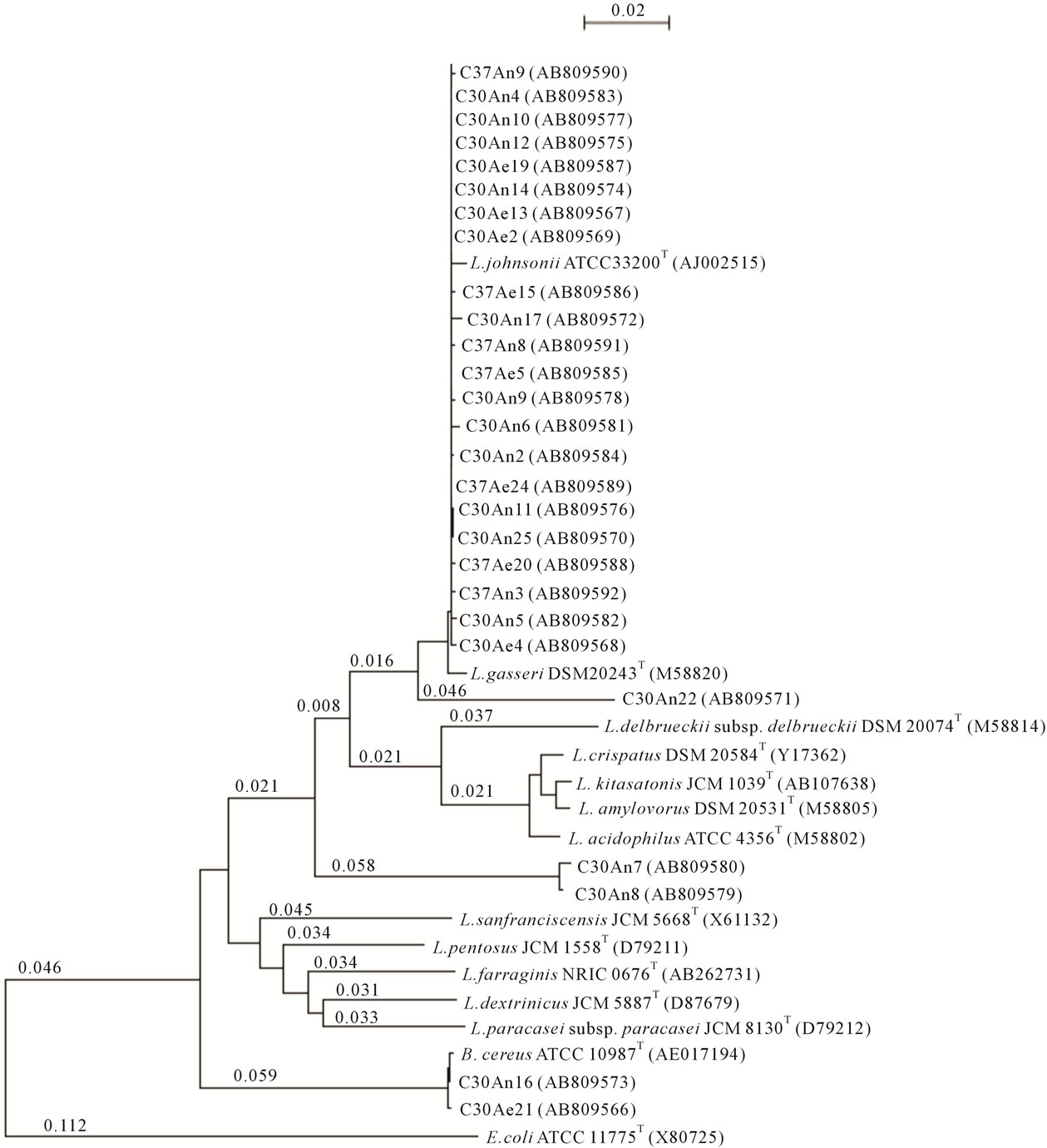
Figure 2. Phylogenetic tree based on the 16S rRNA gene sequences of the isolates determined using the neighbor-joining method. C30An, C30Ae, C37An and C37Ae indicate strains isolated under anaerobic conditions at 30˚C, aerobic conditions at 30˚C, anaerobic conditions at 37˚C, and aerobic conditions at 37˚C, respectively.
tained with primer AP-A-2 are shown in Figure 3. Group definitions were based on reproducible differences in one or more bands. Strains within a group had at least 90% similarity. The results obtained with primer AP-A-23 resembled those obtained with primer AP-A-2 (Data not shown).
A separate pattern was observed for DNAs from the B. cereus strains (cluster 6).The profiles of the other isolates, which were identified as Lactobacillus species based on their16S rRNA gene sequences, were quite different from the L. gasseri type strain. Clusters 1, 2 and 3 were divided into subgroups of the L. johnsonii type strain. The RAPD-PCR profiles for strains C30An8 and C30An22 showed weak similarity (<95% in 16S rRNA

Table 2. Sugar fermentation patterns determined using the API 50CH test.
sequence) to L. johnsonii and belonged to the same cluster (cluster 4). By contrast, the profile of strain C30An7 was similar to isolates in cluster 8.
4. Discussion
Rice bran is usually fermented in an open system at room temperature. Consequently, there is rapid growth of LAB and reduction of pH, which inhibits growth of bacteria that could cause spoilage. Other functions of the rice bran fermentation are to add flavor and nutritional value to food, to eliminate allergens [19] and to act as a fertileizer. After prolonged growth on rice bran agar plates, frequently isolated bacteria and fungi include Candida albicans, Enterococcus faecium, E. coli, Klebsiella pneumonia and Pseudomonas aeruginosa [8]. Although the fermented rice bran produced by Create Co. Ltd. has the ability to stimulate plant growth (data not shown), the bacterial community in the product has never been investigated. For that reason, we analyzed the bacterial community in fermented rice bran product cultured under different conditions, focusing in particular on the LAB community. Twice as many acid-producing colonies formed on GYP-CaCO3 agar plates under aerobic conditions as under anaerobic conditions. On the other hand, the incubation temperature (30˚C and 37˚C) had no effect on bacterial growth. This is consistent with the earlier finding that growth of L. curvatus LTH 1174 differed only slightly between 20˚C and 35˚C, though growth was markedly reduced at 38˚C [20]. In addition, the growth of L. curvatus L442 at 25˚C was similar to that observed at 30˚C [21], and growth temperature also had little effect on L. casei strains [22].
From the phylogenetic analysis of the 16S rRNA gene sequence, we determined that L. johnsonii was the predominant strain in the fermented rice bran product tested in the present study; other bacteria were rarely detected [23]. L. johnsonii strains are known to be widely distributedin the human body and in fermented and probiotic foods [24], but there have been no reports of its isolation from fermented rice bran. Molecular analysis of longaged nukadoko, a traditional Japanese fermented rice bran bed used for pickling vegetables, showed that Lactobacillus species, including L. namurensis and L. ace-
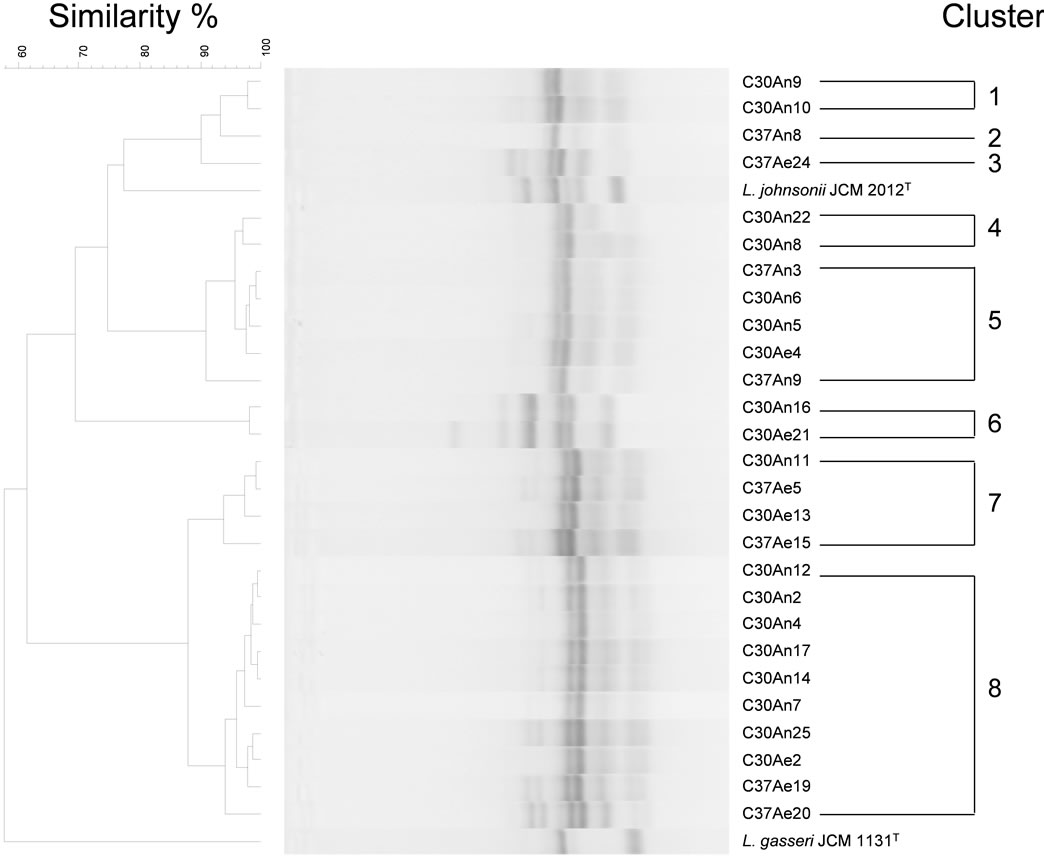
Figure 3. Comparison of the RAPD-PCR profiles of the 27 strains studied and the deduced dendrogram obtained using UPGMA. The RAPD-PCR pattern was obtained using the primer AP-A-2 (5’-TGGATTGGTC-3’). The scale bar indicates the correlation values (Pearson’s coefficient, ×100).
totolerans, predominate [4]. In addition, L. plantarum has been isolated from fermented rice bran for its synbiotic effect [25]. Our phylogenetic analysis indicates that most strains isolated from fermented rice bran products are highly similar to L. johnsonii, and this cluster was readily distinguishable from L. gasseri, which belongs to the L. acidophilus group. The strains closely related to L. johnsonii occurred independently of the growth temperature and oxygen partial pressure. However, the 16S rRNA gene sequence of three strains, C30An7, C30An8 and C30An22 showed, respectively, 87.9%, 90.0% and 93.2% similarity to L. johnsonii and 87.5, 89.4% and 92.8% similarity to L. gasseri. Given that the threshold value for species recognition based on 16S rRNA homology is 97% [26], these three strains may be novel Lactobacillus species. Moreover, all three strains grew under the same conditions (30˚C, anaerobic culture), making it likely that those conditions are especially suitable for growth of the novel strains in the fermented rice bran product.
The RAPD-PCR patterns of the isolated LAB strains were recognizable as those of type strains of L. johnsonii and L. gasseri, whereas the patterns of the three presumably novel strains were equated with other isolated LAB strains. Thus the isolated LABs did not show a high degree of diversity. It may be that the culture methods used in this study limited the growth of other LAB. If so, use of RAPD-PCR could enable characterization of the diversity of LAB in fermented products. Isolation using agar plates, 16S rRNA gene sequence determination and gene typing of the isolated LAB strains are only the first steps in this research. With further analysis, we aim to comprehensively characterize the biodiversity of LAB in fermented rice bran product. For example, we will assess the culture-independent diversity of microbial populations in rice bran during fermentation using Denaturing Gradient Gel Electrophoresis (DGGE). The three novel strains identified could be characterized using ERICPCR analysis, recA gene sequence analysis and DNADNA hybridization with L. acidophilus group. We will report these results elsewhere as soon as possible.
5. Acknowledgements
We are grateful to Dr. Maki Kitahara (Microbe Division, Japan Collection of Microorganisms, RIKEN BioResource Center) for helpful discussion and for bacterial identification using GelCompar. This research was supported by the Small and Medium Enterprise Agency of Japan (No. T218410006) and Japanese Grant Aid for Human Resource Development Scholarship. We thank Mrs. Y. Toyomasu and C. Ishikawa (Create Co. Ltd.) for providing fermented rice bran products.
REFERENCES
- J. Nakayama, H. Hoshiko, M. Fukuda, H. Tanaka, N. Sakamoto, S. Tanaka, K. Ohue, K. Sakai and K. Sonomoto, “Molecular Monitoring of Bacterial Community Structure in Long-Aged Nukadoko: Pickling Ded of Fermented Rice Bran Dominated by Slow-Growing Lactobacilli,” Journal of Bioscience and Bioengineering, Vol. 104, No. 6, 2007, pp. 481-489. doi:10.1263/jbb.104.481
- C. G. Schmidt and E. B. Furlong, “Effect of Particle Size and Ammonium Sulfate Concentration on Rice Bran Fermentation with the Fungus Rhizopus oryzae,” Bioresource Technology, Vol. 123, 2012, pp. 36-41. doi:10.1016/j.biortech.2012.07.081
- J. H. Koh, K. W. Yu and H. J. Suh, “Biological Activities of Saccharomyces cerevisiaeand Fermented Rice Bran as Feed Additives,” Letters in Applied Microbiology, Vol. 35, No. 1, 2002, pp. 47-51. doi:10.1046/j.1472-765X.2002.01131.x
- N. Sakamoto, S. Tanaka, K. Sonomoto and J. Nakayama, “16S rRNA Pyrosequencing-Based Investigation of the Bacterial Community in Nukadoko, a Pickling Bed of Fermented Rice Bran,” International Journal of Food Microbiology, Vol. 144, No. 3, 2011, pp. 352-359. doi:10.1016/j.ijfoodmicro.2010.10.017
- O. O. Oduguwa, M. O. Edema and A. O. Ayenia, “Physico-Chemical and Microbiological Analyses of Fermented Corn Cob, Rice Bran and Cowpea Husk for Use in Composite Rabbit Feed,” Bioresource Technology, Vol. 99, No. 6, 2008, pp. 1816-1820. doi:10.1016/j.biortech.2007.03.036
- S. Yokoyama, J. Hiramatsu and K. Hayakawa, “Production of Gamma-Aminobutyric Acid from Alcohol Distillery Lees by Lactobacillus brevis IFO-12005,” Journal of Bioscience and Bioengineering, Vol. 93, No. 1, 2002, pp. 95-97. doi:10.1016/S1389-1723(02)80061-5
- K. Hiwatashi, A. Narisawa, M. Hokari and K. Toeda, “Antihypertensive Effect of Honey-Based Beverage Containing Fermented Rice Bran in Spontaneously Hypertensive Rats,” Journal of The Japanese Society for Food Science and Technology, Vol. 57, No. 1, 2010, pp. 40-43. doi:10.3136/nskkk.57.40
- I. Yamada, “Studies on Growth and Formation of Capsule of Bacterial Culture by Rice Bran,” Vol. 3, Kyushu University, Kyushu, 2004.
- D. G. Burns, H. M. Camakaris, P. H. Janssen and M. L. Dyall-Smith, “Combined Use of Cultivation-Dependent and Cultivation-Independent Methods Indicates that Members of Most Haloarchaeal Groups in an Australian Crystallizer Pond Are Cultivable,” Applied and Environmental Microbiology, Vol. 70, No. 9, 2004, pp. 5258-5265. doi:10.1128/AEM.70.9.5258-5265.2004
- R. Temmerman, G. Huys and J. Swings, “Identification of Lactic Acid Bacteria: Culture-Dependent and Culture-Independent Methods,” Trends in Food Science and Technology, Vol. 15, No. 7-8, 2004, pp. 348-359. doi:10.1016/j.tifs.2003.12.007
- L. A. Sarmiento-Rubiano, B. Berger, D. Moine, M. Zúñiga, G. Pérez-Martínez and M. J. Yebra, “Characterization of a Novel Lactobacillus Species Closely Related to Lactobacillus johnsonii Using a Combination of Molecular and Comparative Genomics Methods,” BMC Genomics, Vol. 11, 2010, p. 504. doi:10.1186/1471-2164-11-504
- H. Sato, M. Torimura, M. Kitahara, M. Ohkuma, Y. Hotta and H. Tamura, “Characterization of the Lactobacillus casei group Based on the Profiling of Ribosomal Proteins Coded in S10-spc-alpha Operons as Observed by MALDITOF MS,” Systematic and Applied Microbiology, Vol. 35, No. 7, 2012, pp. 447-454. doi:10.1016/j.syapm.2012.08.008
- K. Doi, Y. Nishizaki, Y. Fujino, T. Ohshima, S. Ohmomo and S. Ogata, “Pediococcus lolii sp. nov., Isolated from Ryegrass Silage,” International Journal of Systematic and Evolutionary Microbiology, Vol. 59, No. 5, 2009, pp. 1007-1010. doi:10.1099/ijs.0.005793-0
- A. Michinaka and T. Fujii, “Efficient and Direct Identification of Fructose Fermenting and Non-Fermenting Bacteria from Calf Gut Microbiota Using Stable Isotope Probing and Modified T-RFLP,” Journal of General and Applied Microbiology, Vol. 58, No. 4, 2012, pp. 297-307. doi:10.2323/jgam.58.297
- K. Gori, M. Ryssel, N. Arneborg and L. Jespersen, “Isolation and Identification of the Microbiota of Danish Farmhouse and Industrially Produced Surface-Ripened Cheeses,” Microbial Ecology, Vol. 65, No. 3, 2013, pp. 602- 615. doi:10.1007/s00248-012-0138-3
- Y. Fujino, R. Kawatsu, F. Inagaki, A. Umeda, T. Yokoyama, Y. Okaue, S. Iwai, S. Ogata, T. Ohshima and K. Doi, “Thermus thermophilus TMY Isolated from Silica Scale Taken from a Geothermal Power Plant,” Journal of Applied Microbiology, Vol. 104, No. 1, 2008, pp. 70-78.
- M. Kozaki, T. Uchimura and S. Okada, “Experimental Manual of Lactic Acid Bacteria,” Asakurasyoten, Tokyo, 1992.
- M. Kimura, “A Simple Method for Estimating Evolutionary Rates of Base Substitutions through Comparative Studies of Nucleotide Sequences,” Journal of Molecular Evolution, Vol. 16, No. 2, 1980, pp. 111-120. doi:10.1007/BF01731581
- D. Kim and G. D. Han, “High Hydrostatic Pressure Treatment Combined with Enzymes Increases the Extractability and Bioactivity of Fermented Rice Bran,” Innovative Food Science and Emerging Technologies, Vol. 16, 2012, pp. 191-197. doi:10.1016/j.ifset.2012.05.014
- W. Messens, J. Verluyten, F. Leroy and Luc De Vuyst, “Modelling Growth and Bacteriocin Production by Lactobacillus curvatus LTH 1174 in Response to Temperature and pH Values Used for European Sausage Fermentation Processes,” International Journal of Food Microbiology, Vol. 81, No. 1, 2003, pp. 41-52. doi:10.1016/S0168-1605(02)00168-X
- M. Mataragas, J. Metaxopoulos, M. Galiotou and E. H. Drosinos, “Influence of pH and Temperature on Growth and Bacteriocin Production by Leuconostoc mesenteroides L124 and Lactobacillus curvatus L442,” Meat Science, Vol. 64, No. 3, 2003, pp. 265-271. doi:10.1016/S0309-1740(02)00188-2
- W. S. Tan, M. F. Budinich, R. Ward, J. R. Broadbent and J. L. Steele, “Optimal Growth of Lactobacillus casei in a Cheddar Cheese Ripening Model System Requires Exogenous Fatty Acids,” Journal of Dairy Science, Vol. 95, No. 4, 2012, pp. 1680-1689.
- T. Fujisawa, Y. Benno, T. Yaeshima, and T. Mitsuoka, “Taxonomic Study of the Lactobacillus acidophilus Group, with Recognition of Lactobacillus gallinarum sp. nov. and Lactobacillus johnsonii sp. nov. and Synonymy of Lactobacillus acidophilus Group A3 (Johnson et al., 1980) with the Type Strain of Lactobacillus amylovorus (Nakamura 1981),” International Journal of Systematic and Evolutionary Microbiology, Vol. 42, No. 3, 1992, pp. 487-491.
- W. P. Hammes and C. Hertel, “The Genera Lactobacillus and Carnobacterium,” In: M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer and E. Stackebrandt, Eds., The Prokaryotes, Vol. 4, 3rd Edition, Springer, New York, pp. 320-403.
- E. Zubaidah, M. Nurcholis, S. N. Wulan and A. Kusuma, “Comparative Study on Synbiotic Effect of Fermented Rice Bran by Probiotic Lactic Acid Bacteria Lactobacillus casei and Newly Isolated Lactobacillus plantarum B2 in Wistar Rats,” APCBEE Procedia, Vol. 2, 2012, pp. 170-177. doi:10.1016/j.apcbee.2012.06.031
- P. Vandamme, B. Pot, M. Gillis, P. de Vos, K. Kersters, and J. Swings, “Polyphasic Taxonomy, a Consensus Approach to Bacterial Systematics,” Microbiological Reviews. Vol. 60, No. 2, 1996, pp. 407-438.
NOTES
*Corresponding author.

