Open Journal of Medical Microbiology
Vol.3 No.1(2013), Article ID:28541,14 pages DOI:10.4236/ojmm.2013.31005
Hospital Delivery Room versus Outdoor Birthing Place: Differences in Airborne Microorganisms and Their Impact on the Infant
Division of Medical Microbiology, Department of Laboratory Medicine, Lund University, Lund, Sweden
Email: *tobias.olofsson@med.lu.se
Received December 26, 2012; revised January 29, 2013; accepted February 7, 2013
Keywords: Allergy; Newborns; Airborne microflora; Delivery rooms; Outdoor birthing place
ABSTRACT
The incidence of allergic airway diseases continues to increase in industrial countries while remaining much more stable in developing countries. Allergens inhaled are eventually also swallowed and evidently the gastrointestinal immune system has a role in regulating allergic responses in the pulmonary as well as the GI system. While some studies have pointed out the role of probiotic bacteria as a supplementary protection against the early development of various allergies, little attention has been paid to the composition of the airborne microflora first and continuosly inhaled by newborns and infants. This study compares the composition of two airborne microbial communities, one from hospital delivery rooms and the other from a nature reserve, evidently in use as a birthing place as early as 7500 B.C. around the air from the outdoor birthing place was marked by a far greater variation in microbial composition and a much higher representation of fungi than the air from the hospitals. The dominant bacterial species from the delivery rooms were Staphylococcus areus and Micrococcus luteus, originating from the staff and the hospital environment; the outdoor flora, however, was dominated by Pseudomonas spp. and Bacillus spp. In addition, 56% of all the bacterial isolates from the delivery rooms were most closely related to strains previously associated with clinical infections, whereas only 15% of isolates in the outdoor bacterial sample had such relationships. The role of airborne microorganisms could be important to infants with developing immune systems considering the microbial bias of hospital air presented in this study.
1. Introduction
Over the past 40 years the incidence of allergic airway disease has risen in industrialized countries and has remained stable in developing countries [1]. Several researchers have suggested that the higher incidence of asthma in industrialized countries may be attributed to environmental changes [2-4], while other studies support the possibility that lack of early microbial stimulation results in abnormal immune response to antigens later in life [5-7]. Repeated exposure of airways to antigens is thought to lead to decreased responsiveness and the development of immunologic tolerance of the antigens [8-11]. Animal studies show that inhaled aeroallergens such as microorganisms may also be swallowed [12], and antigens delivered orally may also lead to the development of antigen-specific immunologic tolerance [13-15]. In addition, oral tolerance to an allergen can block responses outside of the gastrointestinal tract (GIT), including the allergic response to that allergen in the lungs
[16-18]. Hence, the immune systems of the lungs and the GIT are connected and dependent on each other. Both the lungs and the GIT of infants are sterile, but after delivery the microbiota of the GIT is acquired from the environment [19]. The GIT of an infant is initially colonized by the mother’s vaginal, perinal, and skin flora. Major factors affecting the nature of the early microbial populations are vaginal delivery versus cesarian delivery, antibiotic use in the mother, and bottle feeding versus breastfeeding [2,20,21]. Several studies have shown that the GIT of infants are first inhabited by facultative anaerobes such as Enterobacteriaceae, Enterococcus, Streptococcus, and Staphylococcus [22-24]. Later on, but still in the first week of life, Bifidobacterium, Bacteroides, and Clostridium are also established [25].
Several studies demonstrate evidence of different microbiota among lower income and middle income individuals from industrial versus developing countries [26- 31]. Estonia has a markedly lower incidence of allergic disease than Sweden. It was demonstrated that the microbiota of children from poor Estonian societies had a different composition from those of Swedish children. The analyses revealed that the microbiota of the Swedish children was associated with increased aerobic microbes and decreased anaerobic microbes, especially Lactobacillus, in fecal samples [26]. In another study [27], it was discovered that infants with developed allergies showed fewer Bifidobacterium and Enterococcus but more Clostridium spp.
Hospitals harbour microorganisms, many of which are multiply resistant to antibiotics and cause hospital-acquired infections. Thorough and continuous cleaning can significantly disturb the normal niches of microorganisms found on patients and staff and in the hospital environment [32-35]. Many hospital-acquired infections are due to well-known pathogens that seem to survive easily in the harsh hospital environment. Well-known examples are Clostridium difficile, linked with antibiotic-associated colitis [36,37], and Bacillus cereus, with infections recorded in maternity, surgical, and intensive care units [38, 39]. Additionally, bacterial infective genera such as multiresistant Klebsiella [40], Enterobacter as a source of septicaemia [41], Escherichia in operating theaters [42], Serratia in an intensive therapy unit [43], and Pseudomonas in a special care baby unit outbreak [44], have been found in hospitals. Furthermore, Acinetobacterradioresistens was found in an increasing number of hospital-acquired infections and outbreaks [45], and A. baumani was the cause of an epidemiological outbreak [46]. Better known organisms from hospital environments are methicillin-resistant Staphylococcus aureus (MRSA) [47], and vancomycin-resistant Enterococcus (VRE) [48]. Finally the hospital-acquired infection organisms Legionella spp. and Aspergillus spp. are known to survive the hospital environment [49,50].
In a study [51] of the bacterial flora on the hands of 119 nurses working in two neonatal intensive care units and found Acinetobacterbaumani, A. iwoffi, Enterobacteragglomerans, E. cloace, Klebsiellaoxytoca, K. pneumonia, Pseudomonas aeroginosa, P. flourescens, Staphylococcus aureus, S. warneri, and S. epidermis, with the last two most dominant. In another study [52], sampling the bacterial aerosol in various areas of a dental clinic Staphylococcus epidermis was most frequently found, closely followed by Micrcoccus spp. and Corynebacteria spp., and Staphylococcus aureus, Pseudomonas spp., and fungi were also found.
To our knowledge no previous studies have compared the airborne microbiota inhaled by newborns after delivery in hospitals with the microbiota of outdoor air in more natural settings. In this study we wanted to discover the microbial composition of the hospital air in some Swedish delivery rooms because the airborne microorganisms first inhaled by newborns might have an impact on their developing immune systems. As a control we used the air of a possible outdoor birthing place in a Swedish primeval forest, with supposedly conserved fauna and flora and records of human activities circa 7500 B.C. [53].
2. Materials and Methods
2.1. Samples Collection
Once each month for one year, the airborne microorganisms from Solviken in the Kullaberg Nature Reserve (Skåne, Sweden) were sampled. On each occasion the sampling was performed with a Trypton Soy Broth agar (TSB, Oxoid Ltd., Basingstoke, Hampshire, England) on a surface plate 14.0 cm in diameter, for a total of 12 plates. The plates were circled manually in the air, without lids, sweeping to 0.5 m above the ground for 1 min to collect both airborne and ground surface microorgansims. Sampling was also performed in eight hospital delivery rooms in one single day during the summer. In each of eight delivery rooms in the University Hospital of Malmö (Malmö, Sweden) one uncovered TSB surface agar plate, 14 cm in diameter was placed by a nurse on a shelf and left for 1 h. Some rooms were empty at the time but most rooms were occupied with mothers giving birth. Plates from both Kullaberg and the delivery rooms were incubated aerobically at 22˚C for 5d. One of every morphologically different colony of bacteria, yeasts, and molds were selected and re-cultivated for purity as isolates. The recultivation was performed on TSB agar surface plates, incubated aerobically at 22˚C for 5d.
2.2. PCR of Bacterial Isolates
One colony from the purified isolates was placed in 2.0 ml Eppendorf tubes together with 0.25 ml sterile water and 10 - 15 glass beads (2.0 mm). Cells were disintegrated by shaking for 45 min in an Eppendorf mixer 5432 (Eppendorf, Hamburg, Germany). After centrifugation (20,200 × g for 5 min), 1 μl of the supernatant was used in the following PCR reaction.
Amplification was conducted with primers designed to anneal to conserved regions of bacterial 16S rRNA genes. The forward primer ENV1 (5’-AGA GTT TGA TII TGG CTC AG-3’) corresponded to positions 8 - 27 in Escherichia coli 16S rRNA, and the reverse primer ENV2 (5’-CGG ITA CCT TGT TAC GAC TT-3’) corresponded to positions 1511-1492 [54]. The PCR reaction contained 5 μl 10× PCR buffer (100 mMTris-HCl, 15 mM MgCl2, 500 mMKCl, pH 8.3), 200 μmol∙1−1 of each deoxyribonucleotide triphosphate, 2.5 U of Taq DNA polymerase (Roche Diagnostics, Mannheim, Germany), 10 pmol of each primer and 1 - 10 μl template in a total volume of 50 μl. Amplification was performed with a T-personal (WhatmanBiometra, Goettingen, Germany) as follows: 30 cycles at 95˚C for 15 s, 48˚C for 30 s, and 72˚C for 90 s followed by an elongation step at 72˚C for 10 min. The PCR product was stored at –20˚C for sequencing.
2.3. Sequencing and Identification of Isolate DNA
PCR products were sequenced by a sequencing company (MWG Operon Eurofins, Ebersberg, Germany) with universal primer ENV1. These partial 16S rDNA sequences were searched against GenBank (National Centre for Biotechnology Information, Rockville Pike, Bethesda, MD) using the Advanced BLAST similarity search option [55], accessible from the homepage of the National Centre for Biotechnology Information (http://www.ncbi.nlm.nih.gov/). For comparison, sequences were also searched against another software, The Ribosomal Database Project II [56], accessible from the homepage (http://rdp.cme.msu.edu/).The partial sequences were mostly around 550 - 900 base pairs (ranging from the 8th bp). A bacterial isolate was defined as a specific phylotype when it diverged by 5 bases or more from the closest related isolate. The identification of mold isolates was performed to the morphological genus level. The identification of yeasts was performed visually by noting differences in colony appearances, and by microscope, but never to genus or species level.
3. Results
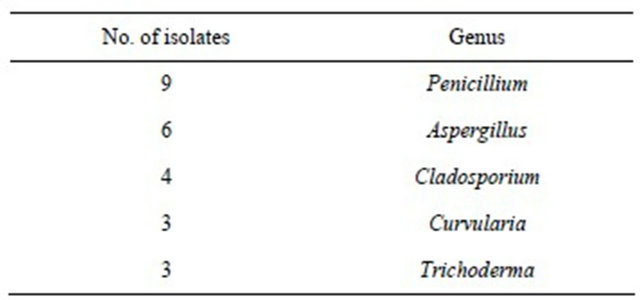
A total of 355 bacterial sequences were identified and all showed a similarity of >91% to type strains in the RDP. In addition 28 colonies of yeast were morphologically identified and 105 colonies of mold were identified to the level of genus. A total of 242 microorganisms (142 bacterial sequences, 75 mold colonies, and 25 yeast colonies) originated in the Kullaberg Nature Reserve, and 246 microorganisms (213 bacterial sequences, 30 mold colonies, and 3 yeast colonies) originated in the delivery rooms. The bacterial diversity was larger in the samples from Kullaberg, constituting 88 different phylotypes among 41 different genera and 76 different species of the 142 isolated bacteria (Table 1). From the delivery rooms 213 bacteria were isolated, representing 80 different phylotypes among 35 different genera and 68 different species (Table 2). A number of possibly novel bacterial species with <97% sequence similarity to the nearest type strain were found from both Kullaberg (n 14) and the delivery rooms (n 10). Bacterial isolates constituted 142 (59%) of the isolated microorganisms from Kullaberg and 213 (87%) from the delivery rooms. Kullaberg had 75 (31%) mold isolates representing 12 genera (Table 3) compared with 30 (12%) representing 5 genera from the hospital (Table 4). Finally, yeasts were found to be more dominant in the samples from Kullaberg, with 25 isolates (10%), than in the hospital samples, with 3 isolates (1.4%). The dominant bacterial genera from Kullaberg were Pseudomonas (25 isolates; 18%), Bacillus (16 isolates; 11%), Rhodococcus (14 isolates; 10%), Arthrobacter (10 isolates; 7%), Micrococcus (9 isolates; 6%), Microbacterium (6 isolates; 4%), Curtobacterium (6 isolates; 4%), Sporosarcina (6 isolates; 4%), and Paenibacillus 4 isolates; 3%) (Figure 1). The dominant bacterial genus from the delivery rooms were markedly different with 47 isolates of Staphylococcus (22%), 36 isolates of Micrococcus (17%), 17 isolates of Arthrobacter (8%), 14 isolates of Acinetobacter (7%), 13 isolates of Microbacterium (6%), 13 isolates of Bacillus (6%), 9 isolates of Paenibacillus (4%), 8 isolates of Kocuria (4%), and7 isolates of Rhodococcus (3%) (Figure 2). The percentage of isolated bacterial species previously found in clinical samples were much higher in the delivery room samples (51%) (Table 1) than in the samples from the nature reserve Kullaberg (18%) (Table 2).
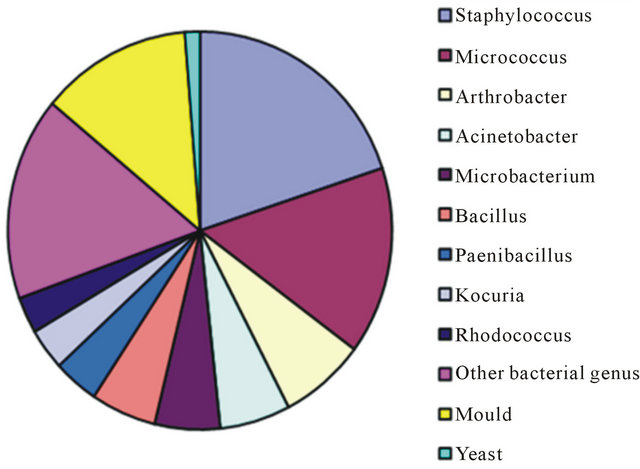
Figure 1. The dominant airborne microorganism flora from the Kullaberg Nature Reserve, Sweden.
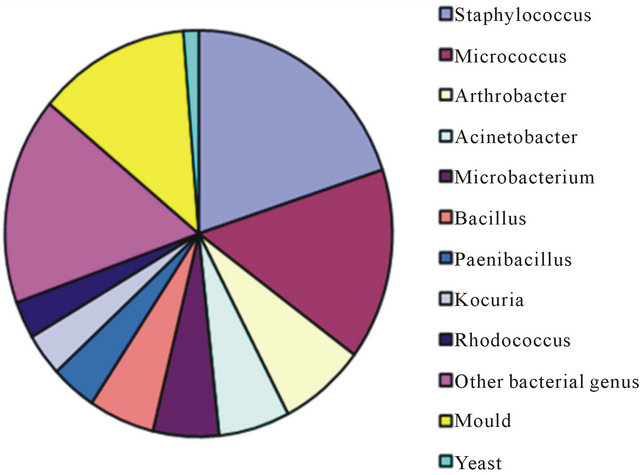
Figure 2. The dominant airborne microorganism flora from five hospital delivery rooms.
Table 1. Thebacterial flora of air from the Kullaberg Nature Reserve. Identity of 16S rRNA gene sequences generated from isolates selected for morphological differences after cultivation. Isolates are presented with the clinically recorded type strains to which they are most closely related.




aAmong the number of isolates found closely related to a specific type strain, the number of phylotypes (if more than one) are shown in brackets; bType strain numbers are followed by GenBank accession numbers.
Of 213 bacterial isolates from the delivery rooms, 120 (56%) were most closely related to type strains previously associated with clinical samples. From the Kullaberg Nature Reserve, of 142 bacterial isolates, only 22 (15% of the isolates) were most closely related to type strains previously associated with clinical samples.
4. Discussion
We hypothesised that the air from the delivery rooms would have less variation in microorganisms than that from the nature reserve and would also have a higher incidence of microorganisms with clinical origin. There are several reasons to believe that Kullaberg, the first piece of land to emerge in Sweden after the last ice age [57], hosts a greater diversity of microorganisms than hospital delivery rooms. First, the soil of the forest of Solviken in the Kullaberg Nature Reservehas been uncultivated since the first leafed trees immigrated from the south around 7500 B.C. [57]. Second, 70% of all the plant species recorded in Sweden can be found in Kullaberg. Third, according to an inventroy of plant species in Kullaberg, the particular locale of Solviken supported the most plant species of 70 locales investigated [58]. Finally, fauna and flora undisturbed by human settlement or agriculture will in time create a unique environment [57].
The total number of microbial isolates was about the same from each of the two different locales investigated, but the percentages of mold and yeast were higher and the overall diversity of microorganisms was greater in the air of the nature reserve than in the delivery rooms. To our knowledge this has not been reported before, even though it seems reasonable considering the higher biological activity in the nature reserve. A general picture indicates a higher frequency and diversity of molds and yeasts in the air from the Nature reserve compared to the hospital. Both environments had a high incidence of molds, although molds were found twice as frequently in air from the nature reserve. Molds were identified to only the genus level and the results showed 12 different genera in the samples from Kullaberg compared to 5 different genera from the hospital. Yeasts were clearly represented in the samples from the nature reserve but almost absent in the delivery rooms. Yeasts were not identified to species or genus level but differentiated morphologically through observation of their colonies on plates and cells under the microscope. Nevertheless, 25 different types of yeast colonies were found in the samples from Kullaberg compared to only 3 different colonies from the hospital.
Species level identification of the bacterial flora needs to be clarified. In the present study, partial sequences above 550 bases were evaluated which was in agreement with other studies [59,60]. The relationships of the isolates to described taxa in the database are given in Tables 1 and 2. It has been suggested that a similarity in 16S rDNA of >97% indicates closely related species. However, in one study [59], it was shown that there was only 50% chance for two 16S rDNA to belong to the same species if the similarity was >99.8 %. Thus, the descriptions in Tables 1 and 2 are not definite; the taxa defined are not necessarily true species but should be called phylotypes [61], or molecular species [60].
The bacterial flora of the delivery rooms was largely consistent with previous reports of hospital environments, e.g. Staphylococcus, which made up a major part of the hand flora of housekeeping staff and neonatal intensive care unit nurses and was also abundant in aerosols [51; 52]. Two of its species, Staphylococcus epidermidis and S. capitis, were found to be two of the first colonizers of an infant GIT [24]. Micrococcus spp. occurred frequently in the hospital air in this study, as previously described in
Table 2. Thebacterial flora of air from hospital delivery rooms. Identity of 16S rRNA gene sequences generated from isolates selected for morphological differences after cultivation. Isolates are presented with the clinically recorded type strains to which they are most closely related.
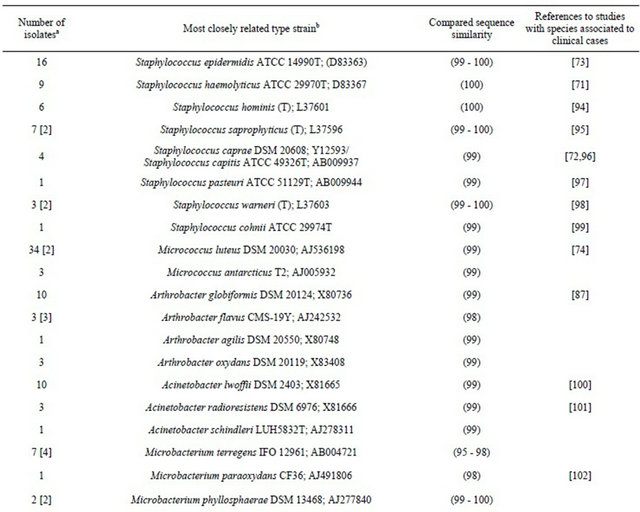
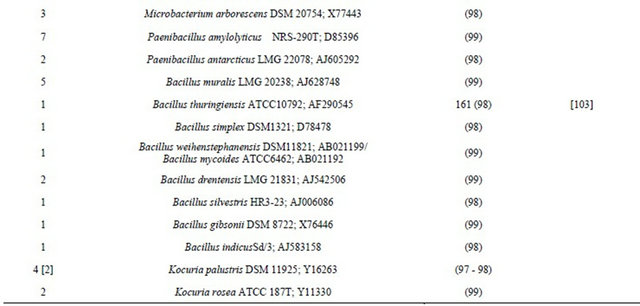

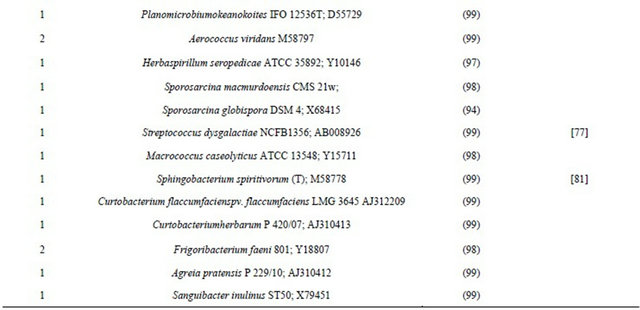
aAmong the number of isolates found closely related to a specific type strain, the number of phylotypes (if more than one) are shown in brackets. bType strain numbers are followed by GenBank accession numbers.
Table 3. Themold flora in air from the Kullaberg Nature Reserve. Identification to the genus level was performed using a morphology identification scheme generated from isolates selected for morphological differences after cultivation.
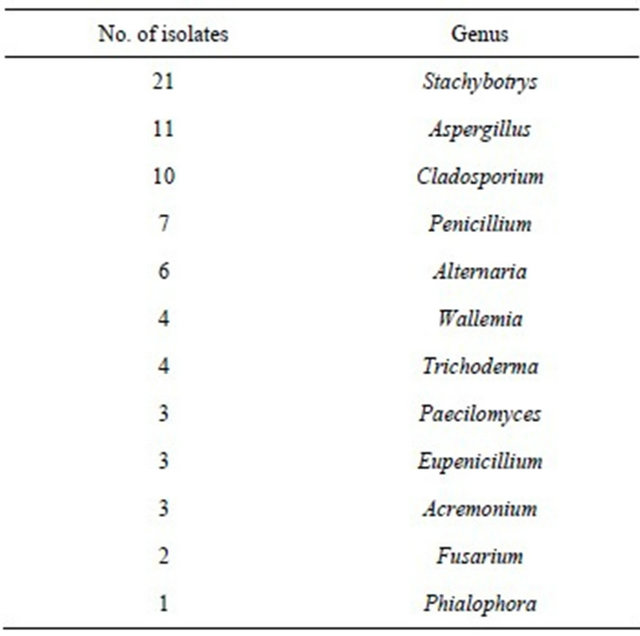
Table 4. Themold flora in air from hospital delivery rooms. Identification to the genus level was performed using a morphology identification scheme generated from isolates selected for morphological differences is after cultivation.
aerosols [52]. The dominant bacterial flora from the air of the nature reserve was different from those of the hospital delivery rooms in several respects. First, air from the nature reserve contained almost no Staphylococcus spp., much fewer Micrococcus luteus, and very few Acinetobacter spp. Instead it was dominated by more Pseudomonas spp. and much more Bacillus spp. than in the hospital. Interestingly, a study of the bioaerosol distributed in a general hospital in Korea [62], showed an almost identical result in the dominant airborne bacteria and fungi found in the delivery rooms as in our present study.
A relatively high portion of bacteria found in the delivery rooms was known to be associated with clinical infections (56% of the bacterial isolates Table 2). Early administration of Lactobacillus rhamnosus was shown to prevent small children at risk (one or more first-degree relatives with allergic disease) from developing eczema, an early manifestation of subsequent allergy [63,64], and the role of intestinal microbiota in modulating the immunological system of children and as an important factor in the prevention of various allergic diseases has been discussed [65-69]. For example, it was shown that early probiotic therapy reduced the frequency of allergies and repeated infections and this effect extended into adulthood [70] airways repeatedly exposed to antigens may lose responsiveness and may develop immunologic tolerance to the antigens [8-11]. Hence, if some bacteria have been shown to modulate the immune system reaction to allergens in the GIT, it can be speculated that the composition of the first and continuously inhaled airborne microorganisms may also influence the developing immune system and thereby potential allergic diseases. The microfloral diversity was only slightly higher at the nature reserve than in the delivery rooms; however, the sampling time in the nature reserve was only one minute versus one hour in the delivery rooms. The numbers and diversity of microorganisms in inhaled air from the nature reserve are therefore expected to be much higher than in air from delivery rooms over the same length of time, and the fewer and less varied microorganisms in hospital air might affect the developing immune system of a newborn.
What is less speculative is the fact that infants are at much greater risk of inhaling dangerous bacteria in the hospital during their first minutes, hours, and days than they would be if they were born in an open environment such as the nature reserve. Examples of such bacteria, or closely related bacteria, dominant in the passive air samples of the delivery rooms over merely one hour include Staphylococcus spp., which cause septicemia in term and preterm infants[71,72]; S. epidermidis, which causes infections in infants [73]; Micrococcus luteus, which causes recurrent bacteremia [74]; Acinetobacter johnsonii, which causes bloodstream infections [75]; Pseudomonas luteola, whichcauses both cutaneous abscess and bacteremia [76]; Streptococcus dysgalactiae subsp. equisimilis, which causes vertebral osteomyelitis (spinal infection) [77]; Corynebacterium pseudodiphtheriticum, which is reported as a respiratory tract pathogen [78]; Corynebacterium diphtheriae, which causes respiratory diphtheria [79]; Kytococcuss chroeteri, which leads to fatal bacteremic pneumonia [80]; and Sphingobacterium spiritivorum, which causes extrinsic allergic alveolitis (hypersensitivity pneumonitis) [81]. The microbes acquired from the mother at the time of birth through vaginal delivery rather than cesarean section seems to be an important start to establishing a proper indigenous microbial community [21,82-84]. Further studies should be conducted to evaluate the role of newborn exposure to a higher variety of airborne microorganisms and its influence on the development of the immune system and future allergic prevalence.
5. Acknowledgements
This study was financed by the Swedish research foundation Gyllenstiernska Krapperupsstiftelsen.
REFERENCES
- D. M. Mannino, D. M. Homa, C. A. Pertowski, A. Ashizawa, L. L. Nixon, C. A. Johnson, L. B. Ball, E. Jack and D. S. Kang, “Surveillance for Asthma—United States, 1960-1995,” MMWR CDC SurveillSumm, Vol. 47, No. 1, 1998, pp. 1-27.
- P. G. Burney, C. Luczynska, S. Chinn and D. Jarvis, “The European Community Respiratory Health Survey,” European Respiratory Journal, Vol. 7, No. 5, 1994, pp. 954- 960. doi:10.1183/09031936.94.07050954
- M. I. Asher, U. Keil, H. R. Anderson, R. Beasley, J. Crane, F. Martinez, E. A. Mitchell, N. Pearce, B. Sibbald, A. W. Stewart, et al., “International Study of Asthma and Allergies in Childhood (ISAAC): Rationale and Methods,” European Respiratory Journal, Vol. 8, No. 3, 1995, pp. 483-491. doi:10.1183/09031936.95.08030483
- R. Beasley, J. Crane, C. K. Lai and N. Pearce, “Prevalence and Etiology of Asthma,” Journal of Allergy and Clinical Immunology, Vol. 105, No. 2, 2000, pp. S466- S472.
- D. P. Strachan, “Hay Fever, Hygiene, and Household Size,” British Medical Journal, Vol. 299, No. 6710, 1989, pp. 1259-1260. doi:10.1136/bmj.299.6710.1259
- M. Wills-Karp, J. Santeliz and C. L. Karp, “The Germless Theory of Allergic Disease: Revisiting the Hygiene Hypothesis,” Nature Reviews Immunology, Vol. 1, No. 1, 2001, pp. 69-75. doi:10.1038/35095579
- D. T. Umetsu, J. J. McIntire, O. Akbari, C. Macaubas and R. H. DeKruyff, “Asthma: An Epidemic of Dysregulated Immunity,” Nature Immunology, Vol. 3, No. 8, 2002, pp. 715-720. doi:10.1038/ni0802-715
- G. Hall, C. G. Houghton, J. U. Rahbek, J. R. Lamb and E. R. Jarman, “Suppression of Allergen Reactive Th2 Mediated Responses and Pulmonary Eosinophilia by Intranasal Administration of an Immunodominant Peptide Is Linked to IL-10 Production,” Vaccine, Vol. 21, No. 5-6, 2003, pp. 549-561. doi:10.1016/S0264-410X(02)00394-8
- K. Takabayashi, L. Libet, D. Chisholm, J. Zubeldia and A. A. Horner, “Intranasal Immunotherapy Is More Effective than Intradermal Immunotherapy for the Induction of Airway Allergen Tolerance in Th2-Sensitized Mice,” Journal of Immunology, Vol. 170, No. 7, 2003, pp. 3898-3905.
- O. Akbari, P. Stock, R. H. DeKruyff and D. T. Umetsu, “Role of Regulatory T Cells in Allergy and Asthma,” Current Opinion in Immunology, Vol. 15, No. 6, 2003, pp. 627-633. doi:10.1016/j.coi.2003.09.012
- C. A. Herrick and K. Bottomly, “To Respond or Not to Respond: T cells in Allergic Asthma,” Nature Reviews Immunology, Vol. 3, No. 5, 2003, pp. 405-412. doi:10.1038/nri1084
- T. E. Pickett, M. F. Pasetti, J. E. Galen, M. B. Sztein and M. M. Levine, “In Vivo Characterization of the Murine Intranasal Model for Assessing the Immunogenicity of Attenuated Salmonella enterica Serovar Typhi Strains as Live Mucosal Vaccines and as Live Vectors,” Infection and Immunity, Vol. 68, No. 1, 2000, pp. 205-213. doi:10.1128/IAI.68.1.205-213.2000
- O. Alpan, “Oral Tolerance and Gut-Oriented Immune Response to Dietary Proteins,” Current Allergy and Asthma Reports, Vol. 1, No. 6, 2001, pp. 572-577. doi:10.1007/s11882-001-0067-6
- N. K. M. Smith, A. D. Eaton, L. M. Finlayson and P. Garside, “Oral Tolerance,” American Journal of Respiratory and Critical Care Medicine, Vol. 162, No. 4, 2000, pp. S175-S178.
- H. L. Weiner, “Oral Tolerance: Immune Mechanisms and the Generation of Th3-Type TGF-Beta-Secreting Regulatory Cells,” Microbes and Infection, Vol. 3, No. 11, 2001, pp. 947-954. doi:10.1016/S1286-4579(01)01456-3
- M. Russo, S. Jancar, A. L. Pereira de Siqueira, J. Mengel, E. Gomes, S. M. Ficker and A. M. Caetano de Faria, “Prevention of Lung Eosinophilic Inflammation by Oral Tolerance,” Immunology Letters, Vol. 61, No. 1, 1998, pp. 15-23. doi:10.1016/S0165-2478(97)00155-7
- M. Russo, M. A. Nahori, J. Lefort, E. Gomes, A. de Castro Keller, D. Rodriguez, O. G. Ribeiro, S. Adriouch, V. Gallois, A. M. de Faria and B. B. Vargaftig, “Suppression of Asthma-Like Responses in Different Mouse Strains by Oral Tolerance,” American Journal of Respiratory Cell and Molecular Biology, Vol. 24, No. 5, 2001, pp. 518- 526.
- Y. Chung, J. Cho, Y. S. Chang, S. H. Cho and C. Y. Kang, “Preventive and Therapeutic Effects of Oral Tolerance in a Murine Model of Asthma,” Immunobiology, Vol. 206, No. 4, 2002, pp. 408-423. doi:10.1078/0171-2985-00190
- L. Dethlefsen, M. McFall-Ngai and D. A. Relman, “An Ecological and Evolutionary Perspective on Human-Microbe Mutualism and Disease,” Nature, Vol. 449, No. 7164, 2007, pp. 811-818. doi:10.1038/nature06245
- G. W. Tannock, “Normal Microflora: An Introduction to Microbes Inhabiting the Human Body,” Chapman & Hall, London, 1995.
- M. G. Dominguez-Bello, M. J. Blaser, R. E. Ley and R. Knight, “Development of the Human Gastrointestinal Microbiota and Insights from High-Throughput Sequencing,” Gastroenterology, Vol. 140, No. 6, 2011, pp. 1713- 1719. doi:10.1053/j.gastro.2011.02.011
- P. L. Stark and A. Lee, “The Microbial Ecology of the Large Bowel of Breast-Fed and Formula-Fed Infants during the First Year of Life,” Journal of Medical Microbiology, Vol. 15, No. 2, 1982, pp. 189-203. doi:10.1099/00222615-15-2-189
- S. Fanaro, R. Chierici, P. Guerrini and V. Vigi, “Intestinal Microflora in Early Infancy: Composition and Development,” Acta Paediatrica Supplement, Vol. 91, No. 441, 2003, pp. 48-55.
- C. F. Favier, E. E. Vaughan, W. M. De Vos and A. D. Akkermans, “Molecular Monitoring of Succession of Bacterial Communities in Human Neonates,” Applied and Environmental Microbiology, Vol. 68, No. 1, 2002, pp. 219-226. doi:10.1128/AEM.68.1.219-226.2002
- R. I. Mackie, A. Sghir and H. R. Gaskins, “Developmental Microbial Ecology of the Neonatal Gastrointestinal Tract,” American Journal of Clinical Nutrition, Vol. 69, No. 5, 1999, pp. 1035S-1045S.
- B. Bjorksten, P. Naaber, E. Sepp and M. Mikelsaar, “The Intestinal Microflora in Allergic Estonian and Swedish 2-Year-Old Children,” Clinical & Experimental Allergy, Vol. 29, No. 3, 1999, pp. 342-346. doi:10.1046/j.1365-2222.1999.00560.x
- B. Bjorksten, E. Sepp, K. Julge, T. Voor and M. Mikelsaar, “Allergy Development and the Intestinal Microflora during the First Year of Life,” Journal of Allergy and Clinical Immunology, Vol. 108, No. 4, 2001, pp. 516-520. doi:10.1067/mai.2001.118130
- M. F. Bottcher, E. K. Nordin, A. Sandin, T. Midtvedt and B. Bjorksten, “Microflora-Associated Characteristics in Faeces from Allergic and Nonallergic Infants,” Clinical & Experimental Allergy, Vol. 30, No. 11, 2000, pp. 1590- 1596. doi:10.1046/j.1365-2222.2000.00982.x
- M. Kalliomaki, P. Kirjavainen, E. Eerola, P. Kero, S. Salminen and E. Isolauri, “Distinct Patterns of Neonatal Gut Microflora in Infants in Whom Atopy Was and Was Not Developing,” Journal of Allergy and Clinical Immunology, Vol. 107, No. 1, 2001, pp. 129-134. doi:10.1067/mai.2001.111237
- P. V. Kirjavainen, E. Apostolou, T. Arvola, S. J. Salminen, G. R. Gibson and E. Isolauri, “Characterizing the Composition of Intestinal Microflora as a Prospective Treatment Target in Infant Allergic Disease,” FEMS Immunology & Medical Microbiology, Vol. 32, No. 1, 2001, pp. 1-7. doi:10.1111/j.1574-695X.2001.tb00526.x
- P. V. Kirjavainen, T. Arvola, S. J. Salminen and E. Isolauri, “Aberrant Composition of Gut Microbiota of Allergic Infants: A Target of Bifidobacterial Therapy at Weaning?” Gut, Vol. 51, No. 1, 2002, pp. 51-55. doi:10.1136/gut.51.1.51
- I. M. Maurer, “Hospital Hygiene,” Wright PSG, Bristol, 1985.
- B. J. Collins, “The Hospital Environment: How Clean Should a Hospital Be?” Journal of Hospital Infection, Vol. 11, No. A, 1988, pp. 53-56.
- H. Gaze, “Dirt in Hospitals. Sweeping Change?” Nursing Times, Vol. 86, No. 24, 1990, pp. 27-28.
- M. Thomson and P. Hempshall, “Dirt Alert,” Nursing Times, Vol. 94, No. 28, 1998, pp. 63-64.
- R. Fekety, K. H. Kim, D. Brown, D. H. Batts, M. Cudmore and J. Silva Jr., “Epidemiology of Antibiotic-Associated Colitis; Isolation of Clostridium Difficile from the Hospital Environment,” The American Journal of Medicine, Vol. 70, No. 4, 1981, pp. 906-908. doi:10.1016/0002-9343(81)90553-2
- W. A. Rutala, M. F. Gergen and D. J. Weber, “Inactivation of Clostridium Difficile Spores by Disinfectants,” Infection Control and Hospital Epidemiology, Vol. 14, No. 1, 1993, pp. 36-39. doi:10.1086/646628
- B. R. Birch, B. S. Perera, W. A. Hyde, V. Ruehorn, L. A. Ganguli, J. M. Kramer and P. C. Turnbull, “Bacillus Cereus Cross-Infection in a Maternity-Unit,” Journal of Hospital Infection, Vol. 2, No. 4, 1981, pp. 349-354. doi:10.1016/0195-6701(81)90067-0
- D. Barrie, P. N. Hoffman, J. A. Wilson and J. M. Kramer, “Contamination of Hospital Linen by Bacillus cereus,” Epidemiology and Infection, Vol. 113, No. 2, 1994, pp. 297-306. doi:10.1017/S0950268800051724
- H. M. Aucken, M. E. Kaufmann and T. L. Pitt., “The Spread of Multi-Resistent Epidemic Klebsiella Strain in London and beyond,” 4th International Conference of the Hospital Infection Society, Edinburgh International Conference Centre, 1998.
- M. W. Casewell, J. E. Cooper and M. Webster, “Enteral Feeds Contaminated with Enterobacter cloacae as a Cause of Septicaemia,” British Medical Journal, Vol. 282, No. 6268, 1981, p. 973. doi:10.1136/bmj.282.6268.973
- A. A. Forder, “Buckets and Mops in Operating-Theatres,” Lancet, Vol. 1, No. 7815, 1973, p. 1325. doi:10.1016/S0140-6736(73)91349-4
- J. L. Whitby, J. N. Blair and A. Rampling, “Cross-Infection with Serratia marcescens in an Intensive-Therapy Unit,” Lancet, Vol. 2, No. 7768, 1972, pp. 127-129. doi:10.1016/S0140-6736(72)91612-1
- S. Heard, S. Lawrence, B. Holmes and M. Costas, “A Pseudo-Outbreak of Pseudomonas on a Special Care Baby Unit,” Journal of Hospital Infection, Vol. 16, No. 1, 1990, pp. 59-65. doi:10.1016/0195-6701(90)90049-T
- E. K. Musa, N. Desai and M. W. Casewell, “The Survival of Acinetobacter calcoaceticus Inoculated on Fingertips and on Formica,” Journal of Hospital Infection, Vol. 15, No. 3, 1990, pp. 219-227. doi:10.1016/0195-6701(90)90029-N
- P. Pina, P. Guezenec, S. Grosbuis, L. Guyot, J. C. Ghnassia and P. Y. Allouch, “An Acinetobacter baumanii Outbreak at the Versailles Hospital Center,” Biology and Pathology (Paris), Vol. 46, No. 6, 1998, pp. 385-394.
- R. A. Cox, C. Conquest, C. Mallaghan and R. R. Marples, “A Major Outbreak of Methicillin-Resistant Staphylococcus aureus Caused by a New Phage-Type (EMRSA- 16),” Journal of Hospital Infection, Vol. 29, No. 2, 1995, pp. 87-106. doi:10.1016/0195-6701(95)90191-4
- T. R. Frieden, S. S. Munsiff, D. E. Low, B. M. Willey, G. Williams, Y. Faur, W. Eisner, S. Warren and B. Kreiswirth, “Emergence of Vancomycin-Resistant Enterococci in New York City,” Lancet, Vol. 342, No. 8863, 1993, pp. 76-79. doi:10.1016/0140-6736(93)91285-T
- L. A. Mermel, S. L. Josephson, C. H. Giorgio, J. Dempsey and S. Parenteau, “Association of Legionnaires’ Disease with Construction: Contamination of Potable Water?” Infection Control and Hospital Epidemiology, Vol. 16, No. 2, 1995, pp. 76-81. doi:10.1086/647060
- V. G. Loo, C. Bertrand, C. Dixon, D. Vitye, B. DeSalis, A. P. McLean, A. Brox and H. G. Robson, “Control of Construction-Associated Nosocomial Aspergillosis in an Antiquated Hematology Unit,” Infection Control and Hospital Epidemiology, Vol. 17, No. 6, 1996, pp. 360-364. doi:10.1086/647317
- A. E. Aiello, J. Cimiotti, P. Della-Latta and E. L. Larson, “A Comparison of the Bacteria Found on the Hands of ‘Homemakers’ and Neonatal Intensive Care Unit Nurses,” Journal of Hospital Infection, Vol. 54, No. 4, 2003, pp. 310-315. doi:10.1016/S0195-6701(03)00146-4
- A. Al Maghlouth, Y. Al Yousef and N. Al Bagieh, “Qualitative and Quantitative Analysis of Bacterial Aerosols,” Journal of Contemporary Dental Practice, Vol. 5, No. 4, 2004, pp. 91-100.
- T. Nilsson. “Från Istid till Badsässong,” 2003. http://www.skane-online.se/kullaberg/franis.htm
- J. Brosius, M. L. Palmer, P. J. Kennedy and H. F. Noller, “Complete Nucleotide Sequence of a 16S Ribosomal RNA gene from Escherichia coli,” Proceedings of the National Academy of Sciences of USA, Vol. 75, No. 10, 1978, pp. 4801-4805.doi:10.1073/pnas.75.10.4801
- S. F. Altschul, T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller and D. J. Lipman, “Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs,” Nucleic Acids Research, Vol. 25, No. 17, 1997, pp. 3389-3402. doi:10.1093/nar/25.17.3389
- J. R. Cole, B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. McGarrell, G. M. Garrity and J. M. Tiedje, “The Ribosomal Database Project (RDP-II): Sequences and Tools for High-Throughput rRNA Analysis,” Nucleic Acids Research, Vol. 33, 2005, pp. D294-D296.
- L. Påhlsson, “The Trees That Bear Witness to Ancient Times,” Helsingborg Daily, Helsingborg, 2003.
- J. Kraft, “Inventory of Plant Flora in Brunnby Parish with Kullaberg,” The Botanical Society of Lund, Lund, 1982.
- J. Keswani and W. B. Whitman, “Relationship of 16S rRNA Sequence Similarity to DNA Hybridization in Prokaryotes,” International Journal of Systematic and Evolutionary Microbiology, Vol. 51, No. 2, 2001, pp. 667-678.
- [61] A. Suau, R. Bonnet, M. Sutren, J. J. Godon, G. R. Gibson, M. D. Collins and J. Dore, “Direct Analysis of Genes Encoding 16S rRNA from Complex Communities Reveals Many Novel Molecular Species within the Human Gut,” Applied and Environmental Microbiology, Vol. 65, No. 11, 1999, pp. 4799-4807.
- [62] I. Kroes, P. W. Lepp and D. A. Relman, “Bacterial Diversity within the Human Subgingival Crevice,” Proceedings of the National Academy of Sciences of USA, Vol. 96, No. 25, 1999, pp. 14547-14552. doi:10.1073/pnas.96.25.14547
- [63] K. Y. Kim, Y. S. Kim and D. Kim, “Distribution Characteristics of Airborne Bacteria and Fungi in the General Hospitals of Korea,” Industrial Health, Vol. 48, No. 2, 2010, pp. 236-243. doi:10.2486/indhealth.48.236
- [64] M. Kalliomaki, S. Salminen, H. Arvilommi, P. Kero, P. Koskinen and E. Isolauri, “Probiotics in Primary Prevention of atopic Disease: A Randomised Placebo-Controlled Trial,” Lancet, Vol. 357, No. 9262, 2001, pp. 1076-1079. doi:10.1016/S0140-6736(00)04259-8
- [65] M. Kalliomaki, S. Salminen, T. Poussa, H. Arvilommi and E. Isolauri, “Probiotics and Prevention of Atopic Disease: 4-Year Follow-Up of a Randomised Placebo-Controlled Trial,” Lancet, Vol. 361, No. 9372, 2003, pp. 1869-1871. doi:10.1016/S0140-6736(03)13490-3
- [66] H. Majamaa and E. Isolauri, “Probiotics: A Novel Approach in the Management of Food Allergy,” Journal of Allergy and Clinical Immunology, Vol. 99, No. 2, 1997, pp. 179-185. doi:10.1016/S0091-6749(97)70093-9
- [67] E. Isolauri, T. Arvola, Y. Sutas, E. Moilanen and S. Salminen, “Probiotics in the Management of Atopic Eczema,” Clinical & Experimental Allergy, Vol. 30, No. 11, 2000, pp. 1604-1610. doi:10.1046/j.1365-2222.2000.00943.x
- [68] K. Laiho, U. Hoppu, A. C. Ouwehand, S. Salminen and E. Isolauri, “Probiotics: On-Going Research on Atopic Individuals,” British Journal of Nutrition, Vol. 88, No. 1, 2002, pp. S19-S27. doi:10.1079/BJN2002626
- [69] V. Rosenfeldt, E. Benfeldt, S. D. Nielsen, K. F. Michaelsen, D. L. Jeppesen, N. H. Valerius and A. Paerregaard, “Effect of Probiotic Lactobacillus Strains in Children with Atopic Dermatitis,” Journal of Allergy and Clinical Immunology, Vol. 111, No. 2, 2003, pp. 389-395. doi:10.1067/mai.2003.389
- [70] M. Viljanen, E. Savilahti, T. Haahtela, K. JuntunenBackman, R. Korpela, T. Poussa, T. Tuure and M. Kuitunen, “Probiotics in the Treatment of Atopic Eczema/ Dermatitis Syndrome in Infants: A Double-Blind PlaceboControlled Trial,” Allergy, Vol. 60, No. 4, 2005, pp. 494- 500. doi:10.1111/j.1398-9995.2004.00514.x
- [71] R. Lodinova-Zadnikova, B. Cukrowska and H. Tlaskalova-Hogenova, “Oral Administration of Probiotic Escherichia coli after Birth Reduces Frequency of Allergies and Repeated Infections Later in Life (after 10 and 20 Years),” International Archives of Allergy and Immunology, Vol. 131, No. 3, 2003, pp. 209-211. doi:10.1159/000071488
- [72] A. Jain, J. Agarwal and S. Bansal, “Prevalence of Methicillin-Resistant, Coagulase-Negative Staphylococci in Neonatal Intensive Care Units: Findings from a Tertiary Care Hospital in India,” Journal of Medical Microbiology, Vol. 53, No. 9, 2004, pp. 941-944.
- [73] P. C. Ng, V. C. Chow, C. H. Lee, J. M. Ling, H. L. Wong and R. C. Chan, “Persistent Staphylococcus Capitis Septicemia in a Preterm Infant,” The Pediatric Infectious Disease Journal, Vol. 25, No. 7, 2006, pp. 652-654. doi:10.1097/01.inf.0000225785.32137.d3
- [74] V. Milisavljevic, F. Wu, J. Cimmotti, J. Haas, P. DellaLatta, E. Larson and L. Saiman, “Genetic Relatedness of Staphylococcus Epidermidis from Infected Infants and Staff in the Neonatal Intensive Care Unit,” Am J Infect Control, Vol. 33, No. 6, 2005, pp. 341-347. doi:10.1016/j.ajic.2005.02.003
- [75] C. von Eiff, N. Kuhn, M. Herrmann, S. Weber and G. Peters, “Micrococcus luteus as a Cause of Recurrent Bacteremia,” American Journal of Infection Control, Vol. 15, No. 8, 1996, pp. 711-713. doi:10.1097/00006454-199608000-00019
- [76] H. Seifert, A. Strate, A. Schulze and G. Pulverer, “Vascular Catheter-Related Bloodstream Infection Due to Acinetobacter johnsonii (Formerly Acinetobacter calcoaceticus var. lwoffi): Report of 13 Cases,” Clinical Infectious Diseases, Vol. 17, No. 4, 1993, pp. 632-636. doi:10.1093/clinids/17.4.632
- [77] M. Dalamaga, K. Karmaniolas, C. Chavelas, S. Liatis, H. Matekovits and I. Migdalis, “Pseudomonas luteola Cutaneous Abscess and Bacteraemia in a Previously Healthy Man,” Scandinavian Journal of Infectious Diseases, Vol. 36, No. 6-7, 2004, pp. 495-497. doi:10.1080/00365540310016196
- [78] A. Kumar, J. Sandoe and N. Kumar, “Three Cases of Vertebral Osteomyelitis Caused by Streptococcus dysgalactiae subsp. Equisimilis,” Journal of Medical Microbiology, Vol. 54, No. 11, 2005, pp. 1103-1105.
- [79] K. Ahmed, K. Kawakami, K. Watanabe, H. Mitsushima, T. Nagatake and K. Matsumoto, “Corynebacterium pseudodiphtheriticum: A Respiratory Tract Pathogen,” Clinical Infectious Diseases, Vol. 20, No. 1, 1995, pp. 41-46. doi:10.1093/clinids/20.1.41
- [80] C. Ohuabunwo, J. Perevoscikovs, A. Griskevica, P. Gargiullo, A. Brilla, L. Viksna, S. Glismann, M. Wharton and C. Vitek, “Respiratory Diphtheria among Highly Vaccinated Military Trainees in Latvia: Improved Protection from DT Compared with Td Booster Vaccination,” Scandinavian Journal of Infectious Diseases, Vol. 37, No. 11-12, 2005, pp. 813-820. doi:10.1080/00365540500262658
- [81] I. Mohammedi, C. Berchiche, K. Becker, K. Belkhouja, D. Robert, C. von Eiff and J. Etienne, “Fatal Kytococcusschroeteribacteremic pneumonia,” Journal of Infection, Vol. 51, No. 2, 2005, pp. E11-E13. doi:10.1016/j.jinf.2004.06.013
- [82] P. Kampfer, S. Engelhart, M. Rolke and J. Sennekamp, “Extrinsic Allergic Alveolitis (Hypersensitivity Pneumonitis) Caused by Sphingobacterium spiritivorum from the Water Reservoir of a Steam Iron,” Journal of Clinical Microbiology, Vol. 43, No. 9, 2005, pp. 4908-4910. doi:10.1128/JCM.43.9.4908-4910.2005
- [83] C. Palmer, E. M. Bik, D. B. DiGiulio, D. A. Relman and P. O. Brown, “Development of the Human Infant Intestinal Microbiota,” PLoS Biology, Vol. 5, No. 7, 2007, p. e177. doi:10.1371/journal.pbio.0050177
- [84] R. E. Ley, M. Hamady, C. Lozupone, P. J. Turnbaugh, R. R. Ramey, J. S. Bircher, M. L. Schlegel, T. A. Tucker, M. D. Schrenzel, R. Knight and J. I. Gordon, “Evolution of Mammals and Their Gut Microbes,” Science, Vol. 320, No. 5883, 2008, pp. 1647-1651. doi:10.1126/science.1155725
- [85] J. Walter and R. Ley, “The Human Gut Microbiome: Ecology and Recent Evolutionary Changes,” Annual Review of Microbiology, Vol. 65, 2011, pp. 411-429. doi:10.1146/annurev-micro-090110-102830
- [86] E. Castagnola, F. Fioredda, M. A. Barretta, L. Pescetto, A. Garaventa, E. Lanino, C. Micalizzi, R. Giacchino and G. Dini, “Bacillus sphaericusbacteraemia in Children with Cancer: Case Reports and Literature Review,” Journal of Hospital Infection, Vol. 48, No. 2, 2001, pp. 142-145. doi:10.1053/jhin.2001.0995
- [87] O. H. Cuello, M. J. Caorlin, V. E. Reviglio, L. Carvajal, C. P. Juarez, E. Palacio de Guerra and J. D. Luna, “Rhodococcusgloberulus keratitis after Laser in Situ Keratomileusis,” Journal of Cataract & Refractive Surgery, Vol. 28, No. 12, 2002, pp. 2235-2237. doi:10.1016/S0886-3350(01)01347-5
- [88] J. Milanowski, J. Dutkiewicz, H. Potoczna, L. Kus and B. Urbanowicz, “Allergic Alveolitis among Agricultural Workers in Eastern Poland: A Study of Twenty Cases,” Annals of Agricultural and Environmental Medicine, Vol. 5, No. 1, 1998, pp. 31-43.
- [89] F. Altuntas, O. Yildiz, B. Eser, K. Gundogan, B. Sumerkan and M. Cetin, “Catheter-Related Bacteremia Due to Kocuriarosea in a Patient Undergoing Peripheral Blood Stem Cell Transplantation,” BMC Infectious Diseases, Vol. 4, No. 1, 2004, p. 62. doi:10.1186/1471-2334-4-62
- [90] P. Bemer-Melchior, A. Haloun, P. Riegel and H. B. Drugeon, “Bacteremia Due to Dietziamaris in an Immunocompromised Patient,” Clinical Infectious Diseases, Vol. 29, No. 5, 1999, pp. 1338-1340. doi:10.1086/313490
- [91] H. Levenga, P. Donnelly, N. Blijlevens, P. Verweij, H. Shirango and B. de Pauw, “Fatal Hemorrhagic Pneumonia Caused by Infection Due to Kytococcus sedentarius— A Pathogen or Passenger?” Annals of Hematology, Vol. 83, No. 7, 2004, pp. 447-449. doi:10.1007/s00277-003-0831-x
- [92] C. G. Loftus, G. C. Harewood, F. R. Cockerill and J. A. Murray, “Clinical Features of Patients with Novel Yersinia Species,” Digestive Diseases and Sciences, Vol. 47, No. 12, 2002, pp. 2805-2810. doi:10.1023/A:1021081911456
- [93] B. La Scola, R. J. Birtles, M. N. Mallet and D. Raoult, “Massiliatimonae gen. nov., sp. nov., Isolated from Blood of an Immunocompromised Patient with Cerebellar Lesions,” Journal of Clinical Microbiology, Vol. 36, No. 10, 1998, pp. 2847-2852.
- [94] X. Y. Han and R. A. Andrade, “Brevundimonas diminuta Infections and Its Resistance to Fluoroquinolones,” Journal of Antimicrobial Chemotherapy, Vol. 55, No. 6, 2005, pp. 853-859. doi:10.1093/jac/dki139
- [95] W. E. Kloos, C. G. George, J. S. Olgiate, L. Van Pelt, M. L. McKinnon, B. L. Zimmer, E. Muller, M. P. Weinstein and S. Mirrett, “Staphylococcus hominis subsp. novobiosepticus subsp. nov., a Novel Trehaloseand N-AcetylD-Glucosamine-Negative, Novobiocinand MultipleAntibiotic-Resistant Subspecies Isolated from Human Blood Cultures,” International Journal of Systematic and Evolutionary Microbiology, Vol. 48, No. 3, 1998, pp. 799-812. doi:10.1099/00207713-48-3-799
- [96] R. Raz, R. Colodner and C. M. Kunin, “Who Are You— Staphylococcus saprophyticus?” Clinical Infectious Diseases, Vol. 40, No. 6, 2005, pp. 896-898. doi:10.1086/428353
- [97] S. M. Wang, C. C. Liu, H. W. Tseng, Y. J. Yang, C. H. Lin, A. H. Huang and Y. H. Wu, “Staphylococcus capitis Bacteremia of Very Low Birth Weight Premature Infants at Neonatal Intensive Care Units: Clinical Significance and Antimicrobial Susceptibility,” Journal of Microbiology, Immunology and Infection, Vol. 32, No. 1, 1999, pp. 26-32.
- [98] O. Chesneau, A. Morvan, F. Grimont, H. Labischinski and N. el Solh, “Staphylococcus pasteuri sp. nov., Isolated from Human, Animal, and Food Specimens,” International Journal of Systematic and Evolutionary Microbiology, Vol. 43, No. 2, 1993, pp. 237-244. doi:10.1099/00207713-43-2-237
- [99] N. Announ, J. P. Mattei, S. Jaoua, F. Fenollar, H. Sati, C. Chagnaud, J. Roudier and S. Guis, “Multifocal Discitis Caused by Staphylococcus warneri,” Joint Bone Spine, Vol. 71, No. 3, 2004, pp. 240-242. doi:10.1016/S1297-319X(03)00126-X
- [100] M. Alvarez Posadilla, P. Linares Torres, C. Bailador Andres, P. Suarez Alvarez and J. L. OlcozGoni, “Bacteriemia Caused by Staphylococcus cohnii Associated with Acute Cholecystitis,” Annals of Internal Medicine, Vol. 23, No. 1, 2006, pp. 51-52.
- [101] H. Seifert, R. Baginski, A. Schulze and G. Pulverer, “The Distribution of Acinetobacter Species in Clinical Culture Materials,” Zentralblatt für Bakteriologie, Vol. 279, No. 4, 1993, pp. 544-552. doi:10.1016/S0934-8840(11)80427-5
- [102] A. Jawad, A. M. Snelling, J. Heritage and P. M. Hawkey, “Exceptional Desiccation Tolerance of Acinetobacter radioresistens,” Journal of Hospital Infection, Vol. 39, No. 3, 1998, pp. 235-240. doi:10.1016/S0195-6701(98)90263-8
- [103] K. Laffineur, V. Avesani, G. Cornu, J. Charlier, M. Janssens, G. Wauters and M. Delmee, “Bacteremia Due to a Novel Microbacterium Species in a Patient with Leukemia and Description of Microbacteriumparaoxydans sp. nov,” Journal of Clinical Microbiology, Vol. 41, No. 5, 2003, pp. 2242-2246. doi:10.1128/JCM.41.5.2242-2246.2003
- [104] S. G. Jackson, R. B. Goodbrand, R. Ahmed and S. Kasatiya, “Bacillus cereus and Bacillus thuringiensis Isolated in a Gastroenteritis Outbreak Investigation,” Letters in Applied Microbiology, Vol. 21, No. 2, 1995, pp. 103-105. doi:10.1111/j.1472-765X.1995.tb01017.x
- [105] J. P. Casalta, P. E. Fournier, G. Habib, A. Riberi and D. Raoult, “Prosthetic Valve Endocarditis Caused by Pseudomonas luteola,” BMC Infectious Disease, Vol. 5, 2005, p. 82. doi:10.1186/1471-2334-5-82
NOTES
*Corresponding author.

