Open Journal of Stomatology
Vol.4 No.1(2014), Article ID:42021,7 pages DOI:10.4236/ojst.2014.41005
Upregulated genes in toll-like receptor (TLR) signaling pathway in periodontitis-affected gingival tissues
![]()
Division of Periodontology, Department of Oral Biological Science, Niigata University Graduate School of Medical and Dental Sciences, Niigata, Japan
Email: *kubota@dent.niigata-u.ac.jp
Copyright © 2014 Daisuke Abe et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. In accordance of the Creative Commons Attribution License all Copyrights © 2014 are reserved for SCIRP and the owner of the intellectual property Daisuke Abe et al. All Copyright © 2014 are guarded by law and by SCIRP as a guardian.
Received 14 November 2013; revised 26 December 2013; accepted 15 January 2014
ABSTRACT
Toll-like receptor (TLR) signaling is thought to be one of the most important pathways initiating periodontitis onset. We have previously reported that the TLR signaling pathway is upregulated in periodontitis-affected gingival tissues by microarray pathway frequency analysis. The aim of the present study was to quantitatively analyze specific upregulated genes in the TLR signaling pathway, as compared to healthy controls. Healthy and periodontitis-affected gingival tissues were taken from distinct sites of 3 patients with severe chronic periodontitis. Total RNAs from 6 gingival tissue samples were used for microarray. Samples were taken from 14 chronic periodontitis patients and 14 healthy individuals for quantitative reverse transcription real-time polymerase chain reaction (qRTPCR) analysis. Data-mining analyses, such as pathway analyses, were performed and significant biological pathways in periodontitis were identified. In addition, qRT-PCR analysis was performed for 5 genes —cluster of differentiation 14 (CD14), lymphocyte antigen 96 (MD-2), interleukin-1 beta (IL-1β), interleukin 8 (IL-8), and chemokine ligand 9 (CXCL-9), which are associated with TLR signaling, in order to confirm the results of pathway analysis. qRT-PCR verified that the transcripts for 5 genes in the TLR signaling pathway were significantly upregulated (MD-2 p = 0.0082, CD14 p = 0.0322, IL-1β p = 0.0126, IL-8 p = 0.0438, CXCL-9 p = 0.0325), which was consistent with pathway analyses. We confirmed upregulated MD-2 gene expression levels and associated TLR pathway gene expression, including CD14, IL-1β, IL-8 and CXCL-9, in periodontitis-affected gingival tissues, as compared with healthy controls.
KEYWORDS
TLR Signaling Pathway; Gene Expression; Microarray; qRT-PCR
1. INTRODUCTION
Periodontitis is a chronic inflammatory disease caused by infection with bacteria, in which host cells exhibit an immune response to infection, leading to the destruction of the supporting structures of teeth [1,2]. Disease progression is also related to host-based risk factors, such as genetics, age, gender and systemic conditions, which lead to expression of multiple genes involved in complex pathways related to disease susceptibility [3]. Therefore, it is desirable to clarify the pathogenesis of periodontitis using a comprehensive approach to simultaneously identify upregulated or downregulated genes in gingival tissues at sites of active periodontal tissue destruction [4]. Microarray analyses have recently been applied to obtain comprehensive gene expression profiles of various diseases, including periodontitis [5-9]. These reports have suggested that microarray analyses can provide insight into the associations between gene expression and clinical signs. We have previously reported the comprehensive gene expression/transcriptomes in periodontitis-affected gingival tissues, with the aim to explore the pathogenesis of periodontitis, and identify the upregulated genes (ITGB-2, MMP-2, CXCL-12, CXCR-4 and Rac-2) and the downregulated genes (connexin, DSG-1, DSC-1 and nestin) in periodontitis-affected gingival tissues. These gene expression profiles suggested the stimulation of leukocyte transendothelial migration and the impairment of cell-to-cell communication in periodontitis. In a previous study, the toll-like receptor (TLR) signaling pathway was also found to be significantly upregulated in periodontitis-affected gingival tissues by pathway frequency analysis [10].
TTLR signaling is thought to be one of the most important pathways initiating periodontitis onset [11]. To date, at least 11 different TLRs have been described in humans [12]. TLRs are host cell transmembrane glycoproteins comprising an N-terminal leucine-rich repeat domain, a transmembrane region, and a C-terminal cytoplasmic signaling domain [13]. TLRs have been identified as a family of mammalian homologs of drosophila Toll, and TLR families are implicated in the recognition of conserved bacterial cell-wall component of these pathogens, particularly lipopolysaccharide (LPS), which might cause direct destruction to periodontal tissues or stimulate host cells to trigger a wide range of inflammatory responses [14]. Mammalian TLRs are expressed on innate immune cells, such as macrophages and dendritic cells, and respond to the membrane components of Gram-positive or Gram-negative bacteria. TLRs in human gingival epithelial cells and in gingival biopsies have been investigated in previous studies [15-17]. Among these, TLR2 and TLR4 function as the principal innate sensors for cell-wall components of Gram-negative bacteria in mammals, and are thought to be very important in the progress of periodontitis [18]. However, the exact mechanisms of periodontitis remain obscure, and little is known about the signaling cascade of host responses. Therefore, the present study investigating gene expression levels in TLR-signaling cascade is important. The aim of the present study was to identify the expression profiles of genes related to the TLR signaling pathway in periodontitis-affected gingival tissues.
2. MATERIALS AND METHODS
2.1. Subjects
This study was approved by the regional ethics committee of the Faculty of Dentistry, Niigata University, and all subjects provided written informed consent prior to participation. A total of 28 subjects were recruited from patients attending Niigata University Medical & Dental Hospital, Niigata, Japan. Fourteen patients with generalized severe chronic periodontitis and who had received conventional periodontal treatment more than 4 - 8 weeks before the study were selected (Periodontitis group, Group P). Another 14 individuals with good clinical periodontal health and no history of periodontal disease, impacted teeth or severe dental caries were also enrolled (Healthy group, Group H). The clinical and demographic characteristics of subjects are shown in Tables 1 and 2.
2.2. Collection Of Gingival Tissue Samples
Two distinct gingival samples were collected for microarray analyses as described previously (9). Fourteen pe-
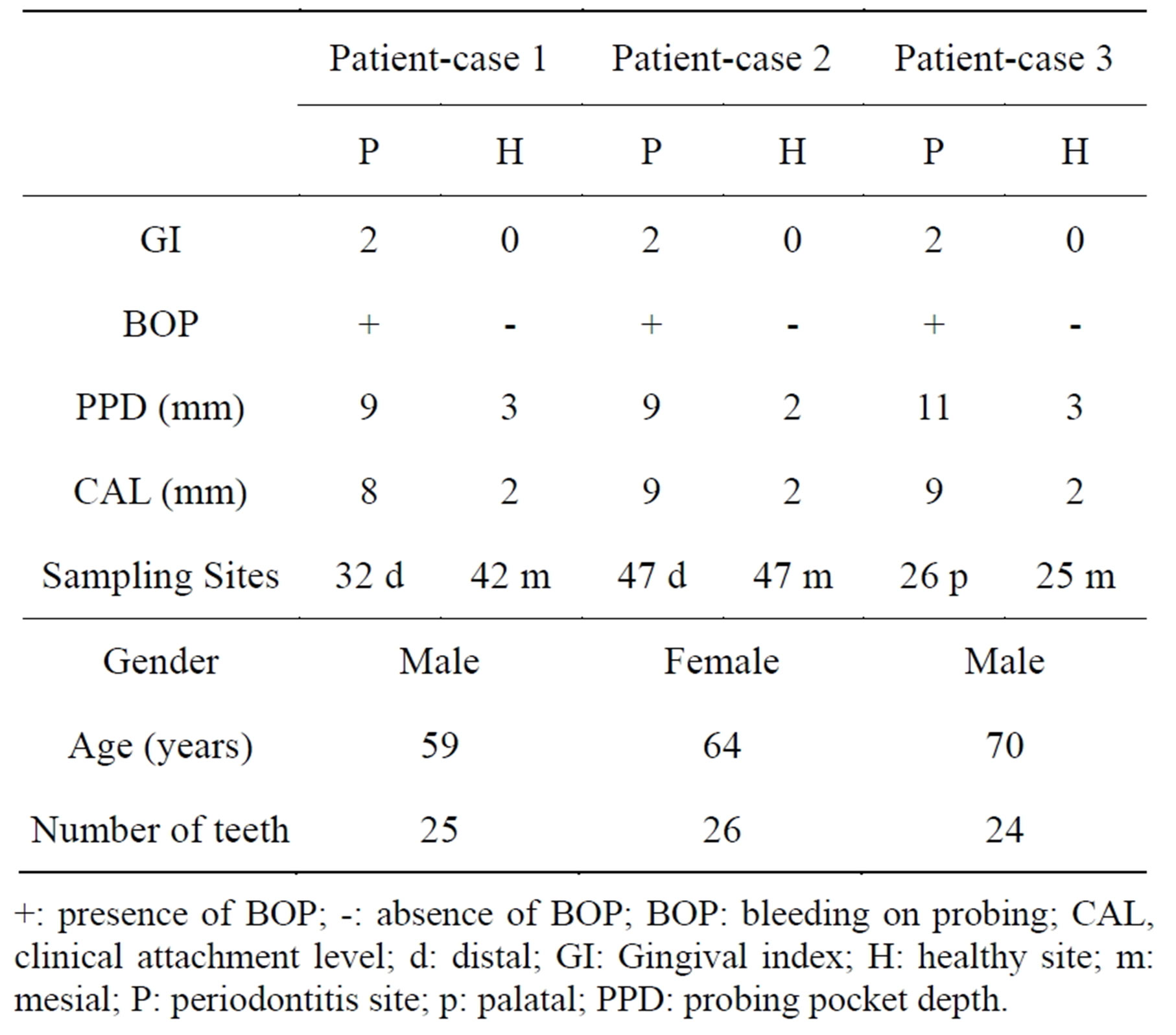
Table 1. General characteristics and clinical periodontal variables of study participants for microarray.
+: presence of BOP; -: absence of BOP; BOP: bleeding on probing; CAL, clinical attachment level; d: distal; GI: Gingival index; H: healthy site; m: mesial; P: periodontitis site; p: palatal; PPD: probing pocket depth.
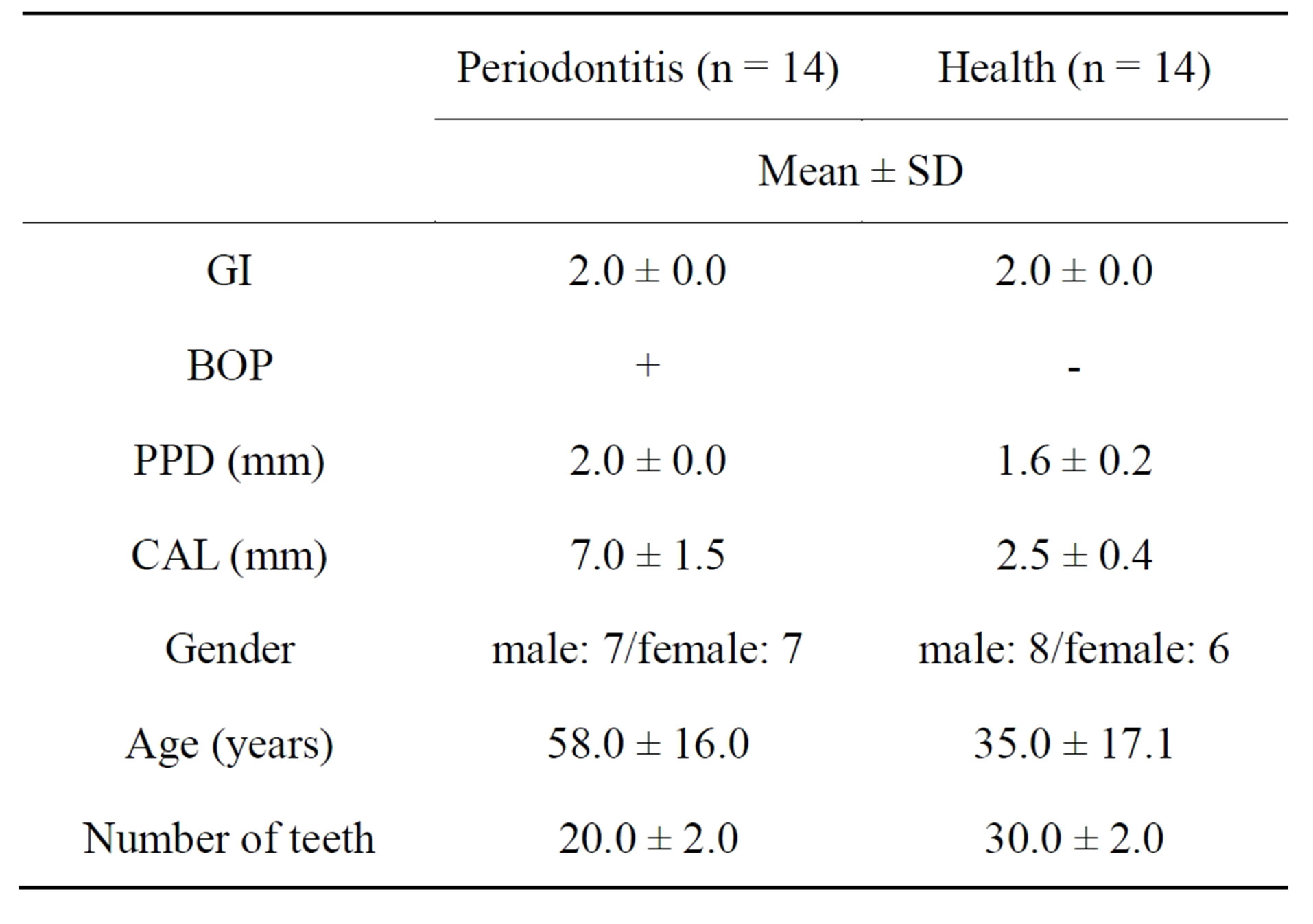
Table 2. General characteristics and clinical periodontal variables of the study participants (qRT-PCR).
riodontitis-affected gingival samples were taken from Group P subjects and fourteen healthy gingival tissue samples were harvested from Group H subjects. Diseased sites showed bleeding on probing and a gingival index of ³1, and had a probing pocket depth and clinical attachment loss of ³5 mm. Healthy sites had PPD of £2 mm with neither clinical attachment loss nor gingival inflammation. Periodontitis-affected and healthy gingival (connective and epithelial) tissue samples were respectively obtained during periodontal flap surgery and tooth extraction, as described previously [19].
2.3. RNA Extraction
Immediately after sampling, gingival tissues were immersed in 500 μl of RNA stabilization reagent (RNA later; Qiagen, Valencia, CA), removed and kept overnight at 4˚C. Samples were homogenized thoroughly using tissue homogenizer (Polytron homogenizer; Kinematica, Lucerne, Switzerland) and total RNA was isolated using the RNAiso® reagent (TaKaRa Bio Inc., Otsu, Japan). Quality control and quantitation of total RNA were determined using the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA) before microarray experiments.
2.4. Microarray Analyses
Microarray analyses and pathway frequency analyses were performed as described previously (9). Briefly, three micrograms of total RNA was reverse-transcribed using a GeneChip® T7-Oligo(dt) Promoter Primer kit (Affymetrix, Santa Clara, CA) and a complementary DNA Synthesis kit (M-MLV version; TaKaRa Bio). Synthesis of biotinylated complementary RNA was performed using a GeneChip® IVT Labeling kit (Affymetrix) for in vitro transcription. After fragmentation, 10 μg of complementary RNA was hybridized at 45˚C for 16 h on a GeneChip® Human Genome U133 Plus 2.0 Array® (Affymetrix). GeneChips were washed and stained in a GeneChip® Fluidics Station 450 (Affymetrix). Fluorescence intensities for chips were examined on a GeneChip Scanner 3000 7G (Affymetrix) and were calculated using Microarray Suite version 5.0 (MAS5.0) with Affymetrix default settings and global scaling, as a normalization method for the complete database comprising six measurements of expression (three in periodontitis and three in a healthy state) of 38,500 genes. Data were filtered to ensure both statistical and biological significance. Values were normalized against the median signal values for each array. Genes that showed a statistically significant higher level of expression when compared with the control group were selected using a paired t-test (p < 0.05). Healthy and periodontitis sites were compared, and genes found to exhibit at least two-fold changes in their gene expression levels between periodontitis and control sites were selected [20]. Data sets presented in the study have been deposited in the Gene expression Omnibus (GEO, National Center for Biotechnology Information) database (accession no.: GSE 23586). The functional assignment in the Kyoto Encyclopedia of Gene and Genomes pathway (KEGG) is the process of linking a set of genes in the genome with a network of interacting molecules in the cell, such as a pathway or complex representing a higher-order biological function [21]. To analyze the biological profiles included in each group, KEGG pathway analyses were performed as described previously [22].
2.5. Post-Hoc Confirmation with qRT-PCR
qRT-PCR primers and probes for cluster of differentiation 14 (CD14; Hs02621496_sl), lymphocyte antigen 96
(MD-2; Hs00209770_ml), interleukin-1 beta (IL-1β; Hs01555413-ml), interleukin-8 (IL-8; Hs01567913-gl), chemokine ligand 12 (CXCL-12; Hs00970536_ml) were purchased from Applied Biosystems (Foster City, CA). Quantitative real-time RT-PCR to confirm the expression of genes belonging to the pathways was performed using a sequence detection system (ABI PRISM 7900HT Sequence Detection System; Applied Biosystems). Briefly, a reaction solution composed of premixed PCR buffer (TaqMan Universal PCR Master Mix, Applied Biosystems), forward and reverse primers (final concentration, 300 nM each), specific probe (TaqMan probe, Applied Biosystems; final concentration, 200 nM), and a complementary DNA(cDNA) mixture (25 ng) were mixed to a total volume of 25 μl. Conditions for qRT-PCR were as follows: preheating at 50˚C for 2 min and at 95˚C for 10 min, followed by 40 cycles of shuttle heating at 95˚C for 15 s and at 60˚C for 1 min. All PCRs were run in duplicate. Sequence detection software (ABI Prism SDS version 2.0, Applied Biosystems) was used to analyze the standards and to carry out quantification. The relative quantity of each mRNA was normalized against the relative quantity of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) determined using specific endogenous control primers (Predeveloped TaqMan Assay Reagent Endogenous Control, Applied Biosystems). The comparative threshold cycle (Ct) (i.e., the number of PCR cycles necessary to obtain the threshold signal of fluorescence) method was used to quantify the amplified transcripts. Differences in transcript levels between healthy and periodontitis patients were analyzed by Mann-Whitney Utest (p < 0.05).
3. RESULTS
3.1. Microarray Analyses
Pathway analysis identified significantly and differentially expressed biological pathways in periodontitis-affected gingival tissues. Fifteen pathways, including leukocyte transendothelial migration, cytokine-cytokine receptor interaction, complement and coagulation cascades, hematopoietic cell lineage, Alzheimer’s disease, Wnt signaling and TLR signaling pathways, were significantly upregulated, showing at least two-fold changes in expression (9). We decided to focus our attention on the TLR signaling pathway (Figure 1), because of its statistical significance (P-value), known functional relevance and location in periodontal tissues.
3.2. Quantitative Real-Time RT-PCR
In order to validate pathway analysis, five genes in the TLR signaling pathway (CD-14, MD-2, IL-1β, IL-8 and CXCL-9) that showed a high change rate in each path-
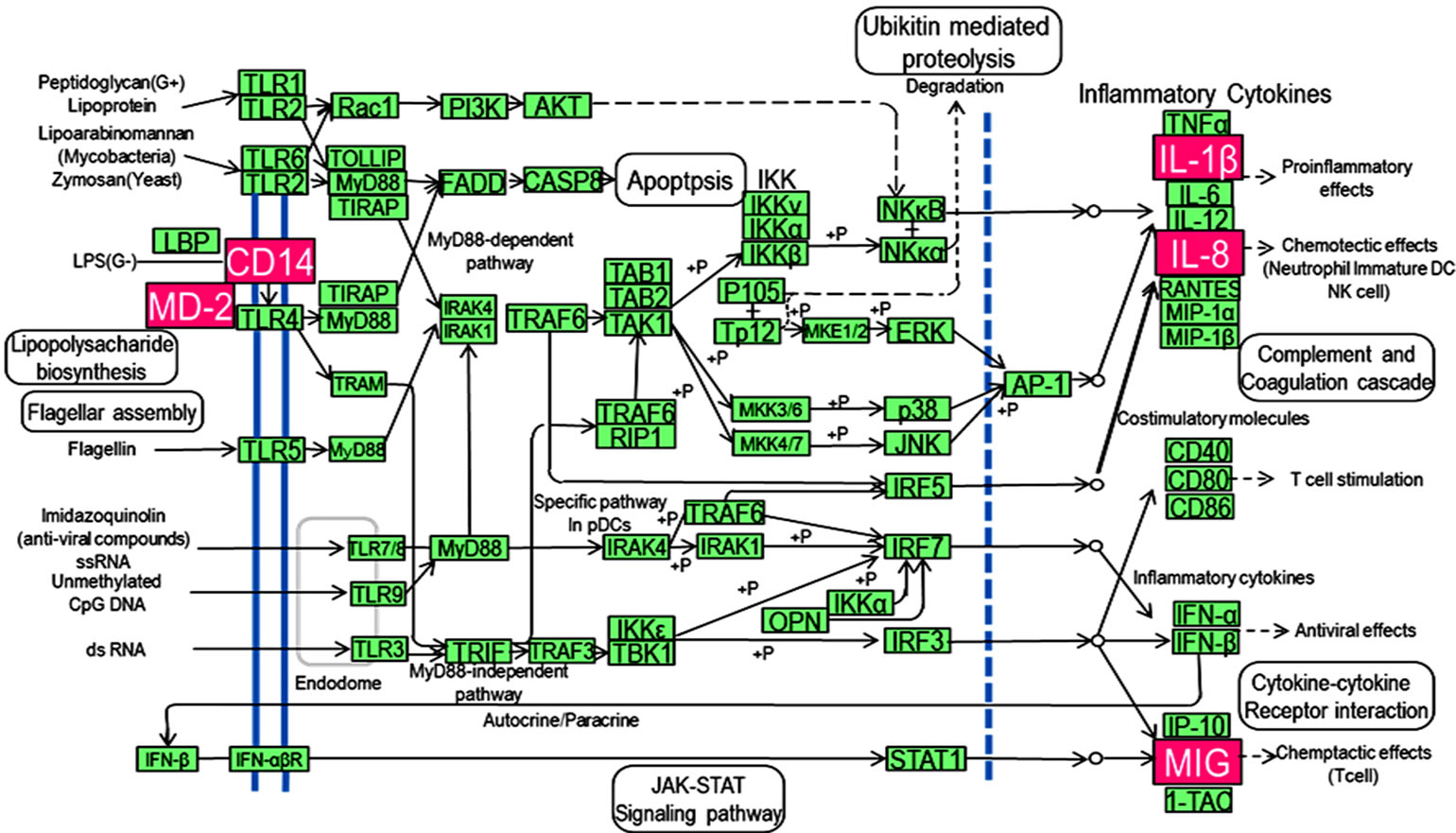
Figure 1. Kyoto Encyclopedia of Gene and Genomes (KEGG) pathway map of toll-like signaling pathway. Genes shown with pink backgrounds are expressed at higher levels in periodontitis-affected gingival tissues when compared with healthy tissues, as determined by pathway analysis.
way were selected and analyzed by quantitative real-time RT-PCR. All five genes were significantly upregulated (Figure 2). These results were consistent with those of pathway analysis.
4. DISCUSSION
In this study, we focused on the TLR signaling pathway, as well as its constituent genes. The data obtained from qRT-PCR were consistent with the results from microarray experiments. We identified several upregulated (CD- 14, MD-2, IL-1β, IL-8 and CXCL-9) gene expression profiles in periodontitis-affected gingival tissues, as compared to healthy controls. The TLR signaling pathway is reported to be important in various diseases, including diabetes [23] and cancers [24], in addition to periodontitis, as shown in the present study. We first confirmed upregulated MD-2 gene expression levels and TLR pathway-associated gene expression, including CD14, IL-1β, IL-8 and CXCL-9, in periodontitis-affected gingival tissues, as compared with healthy controls. MD-2 is a 20- to 30-kDa glycoprotein that was originally discovered based on sequence homology with MD-1, a protein found associated with a B cell homologue of TLR4, RP105 [25]. TLR4 signaling complex is MD-2 which binds to the extracellular domain of TLR4 and causes their surface expression levels to increase where it facilitates LPSmediated NF-κB activation and IL-8 production [26] In addition to bacterial recognition and activation of inflammatory cascades, MD-2 has recently been shown to be an opsonin and an acute-phase protein after infection by

Figure 2. Relative gene transcript levels against glyceraldehyde 3-phosphate dehydrogenase (GAPDH) for lymphocyte antigen 96(MD-2), cluster of differentiation 14(CD14), interleukin-1 beta (IL-1β), interleukin 8 (IL-8), chemokine ligand 9(CXCL-9), which belong to the toll-like signaling pathway. Statistical analysis was performed by Mann-Whitney U test (p < 0.05). Y-axis: mRNA levels relative to GAPDH. Box and whisker plots: bottoms and tops of the boxes represent the lower and upper quartiles, respectively. The line near the middle of the box is the median. The ends of the vertical lines, or “whiskers”, indicate the minimum and maximum data values.
several types of pathogens [27]. Thus, the locally enhanced MD-2 gene expression may facilitate LPS-mediated NF-κB activation as a result of production of cytokines and chemokines. CD14, a 55-kDa cell-surface molecule, plays an important role in signal transduction in LPS-responsive cells, such as monocytes/macrophages, neutrophils and human gingival fibroblasts [28,29]. LPS strongly activates monocytes via the CD14 receptor to secrete TNF, and neutrophils have also been shown to respond to LPS through CD14 receptor signal transudation [30]. This protein acts as an essential component of the CD14-TLR4/MD-2 receptor complex for microbial cell wall component recognition (such as lipopolysaccharide) during infection and is a critical factor for cell surface expression of TLR4 [31,32]. Thus, CD14 protein acts as receptor for the detection of bacterial lipopolysaccharide (LPS) and binding to TLR4 and MD-2, which stimulates immune responses such as the production of interleukin or chemokines as a result of periodontal tissue destruction.
IL-8 is a member of the CXC chemokine family of cytokines and is one of the major mediators of inflammation, which contributes to the sequestration of neutrophils at tissue injury sites and specifically directs polymorphonuclear cell migration [33]. Infiltration by mononuclear cells is a major event in chronic inflammation such as that found in periodontitis and atherosclerosis. Levels of IL-8 are reported to show increases in gingival crevicular fluid (GCF) [34]. IL-8 can also promote the recruitment of tissue stem cells to injury sites [35]. A previous study indicated that LPS is able to induce the expression of IL-8 in dental pulp stem cells (DPSCs) [36]. We speculate that LPS induces the expression of IL-8, and this process is upregulated in vivo in periodontitisaffected gingival tissues, as well as in the dental pulp stem cells. Our study also indicated that IL-8 plays a role in mediating the immune response of the periodontal tissue.
IL-1β is a 17.5-kDa cytokine protein that is a member of the IL-1 cytokine family. This cytokine is produced by activated macrophages as a proprotein [37], which is proteolytically processed into its active form by caspase 1. This cytokine is an important mediator of the inflammatory response, and is involved in a variety of cellular activities, including cell proliferation, differentiation and apoptosis [38]. Proinflammatory cytokines such as IL-1β are known to be synthesized and lead to the activation of the NF-κB in oral gingivitis and periodontitis [39].
Chemokine (C-X-C motif) ligand 9 (CXCL9) is a small cytokine belonging to the CXC chemokine family that is also known as monokine induced by gamma interferon (MIG). CXCL9 is a T-cell chemoattractant, which is induced by IFN-γ [40]. Thus, pathogen recognition by TLRs provokes rapid activation of innate immunity by inducing the production of proinflammatory cytokines and upregulation of costimulatory molecules. Further investigation is necessary to determine the associations between periodontitis and TLR signaling.
5. CONCLUSION
We identified upregulated genes (MD-2, CD14, IL-1β, IL-8 and CXCL-9) in periodontitis-affected gingival tissues that may be related to the TLR signaling pathway and to the pathogenesis of periodontitis.
ACKNOWLEDGEMENTS
This work was supported by Grants-in-aid for Scientific Research (No. 21592622 to T. Kubota and No. 23890062 to D. Abe) from the Japan Ministry of Education, Culture, Sports, Science and Technology of Japan, and Niigata University Research Project Grant of 2009 and 2011 (to T. Kubota).
REFERENCES
- Haffajee, A.D. and Socransky, S.S. (1994) Microbial etiological agents of destructive periodontal disease. Periodontology, 5, 78-111.
- Moore, W.E. and Moore, L.V. (1994) The bacteria of periodontal diseases. Periodontology, 5, 66-77. http://dx.doi.org/10.1111/j.1600-0757.1994.tb00019.x
- Kinane, D.F., Peterson, M. and Stathopoulou, P.G. (2006) Environmental and other modifying factors of the periodontal diseases. Periodontology, 40, 107-119. http://dx.doi.org/10.1111/j.1600-0757.2005.00136.x
- Kim, D.M., Ramoni, M.F., Nevins, M. and Fiorellini, J.P. (2006) The gene expression profile in refractory periodontitis patients. Journal of Periodontology, 77, 1043- 1050. http://dx.doi.org/10.1902/jop.2006.050254
- Chung, C.H., Bernard, P.S. and Perou, C.M. (2002) Molecular portraits and the family tree of cancer. Nature Genetics, 32, 533-540. http://dx.doi.org/10.1038/ng1038
- Thornton, S., Sowders, D., Aronow, B., Witte, D.P., Brunner, H.I., Giannini, E.H. and Hirsch, R. (2002) DNA microarray analysis reveals novel gene expression profiles in collagen-induced arthritis. Clinical Immunology, 105, 155-168. http://dx.doi.org/10.1006/clim.2002.5227
- Colangelo, V., Schurr, J., Ball, M.J., Pelaez, R.P., Bazan, N.G. and Lukiw, W.J. (2002) Gene expression profiling of 12633 genes in Alzheimer hippocampal CA1: Transcription and neurotrophic factor down-regulation and upregulation of apoptotic and pro-inflammatory signaling. Journal of Neuroscience Research, 70, 462-473. http://dx.doi.org/10.1002/jnr.10351
- Beikler, T., Peters, U., Prior, K., Eisenacher, M. and Flemmig, T.F. (2008) Gene expression in periodontal tissues following treatment. BMC Med Genomics (serial online), 1, 30. http://dx.doi.org/10.1186/1755-8794-1-30
- Abe, D., Kubota, T., Morozumi, T., Shimizu, T., Nakasone, N., Itagaki, M. and Yoshie, H. (2011) Altered gene expression in leukocyte transendothelial migration and cell communication pathways in periodontitis-affected gingival tissues. Journal of Periodontal Research, 46, 345- 353. http://dx.doi.org/10.1111/j.1600-0765.2011.01349.x
- Sun, Y., Shu, R., Li, C.L. and Zhang, M.Z. (2010) Gramnegative periodontal bacteria induce the activation of tolllike receptors 2 and 4, and cytokine production in human periodontal ligament cells. Journal of Periodontology, 81, 1488-1496. http://dx.doi.org/10.1902/jop.2010.100004
- Jin, M.S., Kim, S.E., Heo, J.Y., Lee, M.E., Kim, H.M., Paik, S.G., Lee, H. and Lee, J.O. (2007) Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a triacylatedlipopeptide. Cell, 130, 1071-1082. http://dx.doi.org/10.1016/j.cell.2007.09.008
- Janeway Jr., C.A. and Medzhitov, R. (2002) Innate immune recognition. Annual Review of Immunology, 20, 197-216. http://dx.doi.org/10.1146/annurev.immunol.20.083001.084359
- Kinane, D.F., Shiba, H., Stathopoulou, P.G., Zhao, H., Lappin, D.F., Singh, A., Eskan, M.A., Beckers, S., Waigel, S., Alpert, B. and Knudsen, T.B. (2006) Gingival epithelial cells heterozygous for Toll-like receptor 4 polymorphisms Asp299Gly and Thr399ile are hyporesponsive to Porphyromonas gingivalis. Genes and Immunity, 7, 190- 200. http://dx.doi.org/10.1038/sj.gene.6364282
- Beklen, A., Hukkanen, M., Richardson, R. and Konttinen, Y.T. (2008) Immunohistochemical localization of tolllike receptors1-10 in periodontitis. Oral Microbiology and Immunology, 23, 425-431. http://dx.doi.org/10.1111/j.1399-302X.2008.00448.x
- Beklen, A., Sorsa, T. and Konttinen, Y.T. (2009) Tolllike receptors 2 and 5 in human gingival epithelial cells co-operate with T-cell cytokine interleukin-17. Oral Microbiology and Immunology, 24, 38-42. http://dx.doi.org/10.1111/j.1399-302X.2008.00473.x
- Akira, S., Takeda, K. and Kaisho, T. (2001) Toll-like receptors: Critical proteins linking innate and acquired immunity. Nature Immunology, 2, 675-680. http://dx.doi.org/10.1038/90609
- Kumar, H., Kawai, T. and Akira, S. (2009) Toll-like receptors and innate immunity. Biochemical and Biophysical Research Communications, 388, 621-625. http://dx.doi.org/10.1016/j.bbrc.2009.08.062
- Sun, Y., Shu, R., Li, C.L. and Zhang, M.Z. (2009) Gramnegative periodontal bacteria induce the activation of tolllike receptors 2 and 4, and cytokine production in human periodontal ligament cells. Journal of Periodontology, 81, 1488-1496. http://dx.doi.org/10.1902/jop.2010.100004
- Sun, Y., Guo, Q.M., Liu, D.L., Zhang, M.Z. and Shu, R. (2010) In vivo expression of Toll-like receptor 2, toll-like receptor 4, CSF2 and LY64 in Chinese chronic periodontitis patients. Oral Disease, 16, 343-350. http://dx.doi.org/10.1111/j.1601-0825.2009.01630.x
- Kinane, D.F., Peterson, M. and Stathopoulou, P.G. (2006) Environmental and other modifying factors of the periodontal diseases. Periodontology, 40, 107-119. http://dx.doi.org/10.1111/j.1600-0757.2005.00136.x
- Koshi, R., Sugano, N., Orii, H., Fukuda, T. and Ito, K. (2007) Microarray analysis of nicotine-induced changes in gene expression in a macrophage-like human cell line. Journal of Periodontal Research, 42, 518-526. http://dx.doi.org/10.1111/j.1600-0765.2007.00976.x
- Kanehisa, M. and Goto, S. (2000) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Research, 28, 27-30. http://dx.doi.org/10.1093/nar/28.1.27
- Handfield, M., Mans, J.J., Zheng, G., Lopez, M.C., Mao, S., Progulske-Fox, A., Narasimhan, G., Baker, H.V., Lamont, R.J. (2005) Distinct transcriptional profiles characterize oral epithelium-microbiota interactions. Cell Microbial, 7, 811-823. http://dx.doi.org/10.1111/j.1462-5822.2005.00513.x
- Meyers, A.J., Shah, R.R., Gottlieb, P.A. and Zipris, D. (2010) Altered toll-like receptor signaling pathways in human type 1 diabetes. Journal of Molecular Medicine, 12, 1221-1231. http://dx.doi.org/10.1007/s00109-010-0666-6
- Miura, Y., Shimazu, R., Miyake, K., Akashi, S., Ogata, H., Yamashita, Y., Narisawa, Y. and Kimoto, M. (1998) RP105 is associated with MD-1 and transmits an activation signal in human B cells. Blood, 92, 2815-2822.
- Sauter, K.S., Brcic, M., Franchini, M. and Jungi, T.W. (2007) Stable transduction of bovine TLR4 and bovine MD-2 into LPS-nonresponsive cells and soluble CD14 promote the ability to respond to LPS. Veterinary Immunology and Immunopathology, 118, 92-104. http://dx.doi.org/10.1016/j.vetimm.2007.04.017
- Tissières, P. and Pugin, J. (2009) The role of MD-2 in the opsonophagocytosis of Gram-negative bacteria. Current Opinion in Infectious Diseases, 22, 286-291. http://dx.doi.org/10.1097/QCO.0b013e32832ae2fc
- Cheng, Y.X., Qi, X.Y., Huang, J.L., Hu, M., Zhou, L.M., Li, B.S. and Xu, X.X. (2012) Toll-like receptor 4 signaling promotes the immunosuppressive cytokine production of human cervical cancer. European Journal of Gynaecological Oncology, 33, 291-294.
- Haziot, A., Rong, G.W., Bazil, V., Silver, J. and Goyert, S.M. (1994) Recombinant soluble CD14 inhibits LPS-induced TNF-a production by cells in whole blood. Journal of Immunology, 152, 5868-5876.
- Watanabe, A., Takeshita, A., Kitano, S. and Hanazawa, S. (1996) CD14-mediated signal pathway of porphyromonas gingivalis LPS in human gingival fibroblasts. Infection and Immunity, 64, 4488-4494.
- Haziot, A., Tsuberi, B.Z. and Goyert, S.M. (1993) Neutrophil CD14: Biochemical properties and role in the secretion of TNF-α in response to LPS. Journal of Immunology, 150, 5556-5565.
- Visintin, A., Halmen, K.A., Khan, N., Monks, B.G., Golenbock, D.T. and Lien, E. (2006) MD-2 expression is not required for cell surface targeting of Toll-like receptor 4 (TLR4). Journal of Leukocyte Biology, 80, 1584-1592. http://dx.doi.org/10.1189/jlb.0606388
- Wagner, J.G. and Roth, R.A. (2000) Neutrophil migration mechanisms, with an emphasis on the pulmonary vasculature. Pharmacological Reviews, 52, 349-374.
- Sakai, A., Ohshima, M., Sugano, N., Otsuka, K. and Ito, K. (2006) Profiling the cytokines in gingival crevicular fluid using a cytokine antibody array. Journal of Periodontology, 77, 856-864. http://dx.doi.org/10.1902/jop.2006.050340
- Kim, D.S., Kim, J.H., Lee, J.K., Choi, S.J., Kim, J.S., Jeun, S.S., Oh, W., Yang, Y.S. and Chang, J.W. (2009) Overexpression of CXC chemokine receptors is required for the superior gliomatracking property of umbilical cord blood-derived mesenchymal stem cells. Stem Cells and Development, 18, 511-519. http://dx.doi.org/10.1089/scd.2008.0050
- Chang, J., Zhang, C., Tani-Ishii, N., Shi, S. and Wang, C.Y. (2005) NFκB activation in human dental pulp stem cells by TNF and LPS. Journal of Dental Research, 84, 994-998. http://dx.doi.org/10.1177/154405910508401105
- Matsuki, Y., Yamamoto, T. and Hara, K. (1993) Localization of interleukin-1 (IL-1) mRNA-expressing macrophages in human inflamed gingiva and IL-1 activity in gingival crevicular fluid. Journal of Periodontal Research, 1, 35-42. http://dx.doi.org/10.1111/j.1600-0765.1993.tb01048.x
- Fettelschoss, A., Kistowska, M., LeibundGut-Landmann, S., Beer, H.D., Johansen, P., Senti, G., Contassot, E., Bachmann, M.F., French, L.E., Oxenius, A. and Kündig, T.M. (2011) Inflammasome activation and IL-1β target IL-1α for secretion as opposed to surface expression. Proceedings of the National Academy of Sciences of the United States of Ameria, 108, 18055-18060. http://dx.doi.org/10.1073/pnas.1109176108
- Steinberg, T., Dannewitz, B., Tomakidi, P., Hoheisel, J.D., Mu¨ssig, E., Kohl, A. and Nees, M. (2006) Analysis of interleukin-1β-modulated mRNA gene transcription in human gingival keratinocytes by epithelia-specific cDNA microarrays. Journal of Periodontal Research, 41, 426- 446. http://dx.doi.org/10.1111/j.1600-0765.2006.00884.x
- Proost, P., Verpoest, S., Van de Borne, K., Schutyser, E., Struyf S, Put W, Ronsse I, Grillet B, Opdenakker, G. and Van Damme, J. (2004) Synergistic induction of CXCL9 and CXCL11 by toll-like receptor ligands and interferon-γ in fibroblasts correlates with elevated levels of CXCR3 ligands in septic arthritis synovial fluids. Journal of Leukocyte Biology, 75, 777-784. http://dx.doi.org/10.1189/jlb.1003524
NOTES
*Corresponding author.

