Advances in Biological Chemistry
Vol.4 No.5(2014), Article
ID:49049,12
pages
DOI:10.4236/abc.2014.45039
Molecular Cloning and Characterization of a Candidate ENOX Protein of Saccharomyces cerevisiae with a 25 Min Period Insensitive to Simalikalactone D Inhibition and Melatonin
Aya F. Ryuzoji1, Debby H. Parisi1, Sara Dick2, James Kim1, Dorothy M. Morré1, D. James Morré1*
1Mor-NuCo, Inc., Purdue Research Park, West Lafayette, USA
2Department of Biology, Valparaiso University, Valparaiso, USA
Email: *dj-morre@yahoo.com
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 9 June 2014; revised 25 July 2014; accepted 11 August 2014
ABSTRACT
A yeast deletion library was screened based on NADH fluorescence using a 384-well plate assay to identify a yeast isolate lacking a previously identified cell surface oxidase exhibiting an oscillatory pattern with a period length of 25 min and resistant to the ENOX1-specific inhibitor simalikalactone D (YNOX for yeast-specific ENOX = ENOX4). The cDNA was cloned from a yeast over expression library using NADH fluorescence analyzed by Fast Fourier transform and decomposition fits. The objective was to identify and sequence an ENOX homologue in Saccharomyces cerevisiae with a 25 min rather than a 24 min period length (YNOX). The finding identified YDR005C as the yeast ENOX protein with a temperature-independent 25 min period length and insensitive to inhibition by simalikalactone D. The encoded protein was expressed in bacteria and characterized. Gel slices corresponding to 55 kDa and 39 kDa His-tagged proteins exhibited 25 min oscillatory patterns not inhibited by 1 µM simalikalactone D for both NADH oxidation and reduced coenzyme Q10 oxidation as well as a protein disulfide-thiol interchange activity which alternated with the oxidative activities. Activities were phased by low-frequency electromagnetic fields but, in contrast, to yeast ENOX1, not by addition of melatonin. The assay in the presence of D2O shifted the length of the oscillatory period from 25 min to 32 min. The YDR005C deletion mutant cells lacked the ENOX4 clock output present in the wild type yeast.
Keywords:Biological Clock, ECTO-NOX (ENOX) Proteins, YNOX = ENOX4, Ultradian Oscillator, Yeast (Saccharomyces cerevisiae)

1. Introduction
All animals and plants exhibit constitutive ECTO-NOX (ENOX) proteins with common characteristics. ENOX1 proteins reside on the external plasma membrane and are distinguished from other proteins due to their ability to catalyze both protein disulfide-thiol interchange and oxidation of reduced coenzyme Q10. NAD(P)H is an alternate substrate for the natural substrate and is used in our assays [1] [2] . ENOX1 can be inhibited by extracts from the Quassia amara L. tree containing simalikalactone D [3] . ENOX2 (tNOX) activity is associated with tumors or cancers in man and can be inhibited by quinone site inhibitors with anticancer activities [4] [5] . Both ENOX1 and ENOX2 proteins are involved in the cell enlargement phase of cell growth [6] -[8] . Thus, when their activities are inhibited, the cells are unable to grow [2] -[4] [9] . The ENOX3 proteins, also known as arNOX, in man are found primarily in aged individuals [10] [11] .
ENOX proteins have several characteristics in common. Each exhibits an oscillatory activity of the protein disulfide-thiol interchange and reduced coenzyme Q and NAD(P)H oxidation having a temperature-independent period length that correlates with a potential ultradian biological clock function [1] [2] [12] -[14] . ENOX1, Accession Number 43052 located on human chromosome 13G14.11, has an oscillatory period length of 24 min. ENOX2, Accession Number AF20788 located on human chromosome XG25G26.2, has a period length of 22 min. ENOX3 proteins, Accession Number BK008759, are members of the TM-9 super-family of transmembrane proteins, each with a different chromosomal location and all exhibit a 26 min period [15] . The constitutive ECTO-NOX from a yeast has activity characteristics similar to those of other eukaryotic ENOX1 proteins [16] . However, Saccharomyces cerevisiae has an additional ENOX protein (YNOX for yeast NOX) that exhibits a 25 min oscillatory pattern for both NAD(P)H oxidase and disulfide-thiol interchange activities [17] . This activity has been designated as YNOX (for yeast-specific NOX) or ENOX4. While yeast ENOX1 activity is phased by the addition of melatonin and is inhibited by simalikalactone D, yeast ENOX4 is unresponsive to both melatonin and simalikalactone D.
The ENOX4 in yeast was over expressed by a selection process using a deletion library of both yeast ENOX1 and the ENOX4. One deletion strain had a deletion at the ENOX4 protein sequence and, thus, only showed yeast ENOX1 characteristics. The other deletion strain had a deletion at the yeast ENOX1 sequence and, thus, only showed the ENOX4 characteristics. The addition of the simalikalactone D to both deletion strains differentiated between the sequence for the yeast ENOX1 protein and that of ENOX4 [16] . Using this approach, a candidate yeast gene potentially encoding a yeast ENOX protein with a 25 min period designed as YNOX or ENOX4 was identified and characterized where activity was not inhibited by simalikalactone D and was unresponsive to melatonin.
2. Methods
Candidates for ENOX1 and ENOX4 were selected from a deletion library of nonessential yeast genes (strain BY4741 courtesy of Mark Hall, Purdue University) which showed patterns with 5 to 7 - 8 maxima. The deletion candidates used for ENOX4 were affected by the presence of ENOX1 inhibitor simalikalactone D. Of these ENOX4 candidates, those that showed a 25 min oscillatory pattern of NADH oxidation and/or reduced coenzyme Q10 assays were selected. The expression library screen was strain MORF courtesy of Tony Hazbun, Purdue University.
2.1. Saccharomyces Growth and Preparation
Yeasts were grown at room temperature with shaking in rich media (YEPD) for 1 - 2 days until saturation. The yeast strains were maintained on YEPD agar plates and stored at 4˚C. Yeast cells were inactivated by heating for 1 h at 70˚C prior to assay.
2.2. Purification of ENOX4
The yeast whole cell pellet with overexpression of His-tagged ENOX4 isolated from the overexpression library (Yeast ORF library, Open Biosystems),was resuspended in 20 mM Tris-HCl, pH 8.0 with 0.5 mM benzamidine, 0.5 mM PMSF and 1 mM 6-aminohexanoic acid. Cells were lysed by three passages through a French pressure cell at 20,000 psi. The resultant pellet was extracted sequentially. The supernatant following centrifugation at 10,000 rpm for 15 min at 40˚C was saved. The resultant pellet was then extracted with 20 mM Tris-HCl, pH 8.0, containing 1% Triton X-100, 20 mM Tris-HCl, pH 8.0, with 0.3% sarcosine and in 20 mM Tris-HCl, pH 8.0 with 0.4% SDS. The combined supernatants were analyzed by SDS-PAGE and western blot and silver stain to determine the location of ENOX4. Additionally the supernatant was applied to a nickel nitrilotriacetic acid (Ni-NTA) column to bind to the histidine tag and eluted with imidazole to further purify the protein. The identity of the ENOX4 protein was confirmed by sequencing.
2.3. SDS-PAGE Gel Slicing
The SDS-PAGE gels were sliced every 0.5 cm and the slices were eluted with 50 mM Tris-MES, pH 7.0 overnight in 4˚C. The gel slice elutes were assayed for ENOX activity based on NADH oxidation. Proteins were determined by the bicinchoninic acid (BCA) method with bovine serum albumin as a standard [18] .
2.4. SDS-Page and Western Blot
Yeast samples were resolved on 10% SDS polyacrylamide gels were transferred to a nitrocellulose membrane at 90 V for 1 h. A 5% solution of non-fat milk powder was used for blocking and the probe used for the western blot was a 1:2500 anti-histidine antibody (Genscript, Cat. No. A001865).
2.5. Measurements of Enzymatic Activities
Measurements of enzymatic activities were as described previously [16] . The oxidation of NADH was measured from the disappearance of NADH at 340 nm. An extinction coefficient of 6.22 mM−1·cm−1 was used to determine specific activity. The oxidation of reduced coenzyme Q10 (CoQ10H2) was determined from the disappearance of reduced CoQ at both 290 nm and 410 nm. An extinction coefficient of 0.805 mM−1·cm−1 was used to calculate the rate of Q10H2 oxidation. The increase in absorbance due to the cleavage of DTDP was monitored at 340 nm. The specific activity of the cleavage reaction was calculated using a millimolar absorption coefficient of 6.21.
2.6. NADH Fluorescence
As YEPD medium is fluorescent, heat-inactivated yeast cells were washed with PBS by centrifuging the cells in the benchtop mini centrifuge (VWR, Pennsylvania) for 1 - 2 min, the supernatant was discarded and the pellet was resuspended in PBS. This suspension was used for measurements of NADH fluorescence.
The inactivated yeast suspension in PBS was added to wells of a black 96-well plate at a dilution of 1:10 in a volume of 20 µl. The PBS for the experiment was supplemented with 2% glucose to support enzyme activity. The plate was loaded into a Fluoroskan fluorescent plate reader and the samples were measured once every min for 2 - 8 h, with excitation at 355 nm and emission at 460 nm. Data were analyzed by Fast Fourier Transform (Minitab 15) and decomposition analysis (Minitab 15) [19] .
3. Results
The yeast deletion library of non-essential genes was screened by NADH fluorescence and analyzed with decomposition fits. A total of 4846 strains were screened. NADH fluorescence of each strain was analyzed by fitting the data to a 24 min period and inspecting the resulting graphs for loss of complexity compared to the wild-type strain. The wild type exhibited at least 7 or more maxima in the decomposition fits (Figure 1(A)).
Each decomposition fit was inspected and the maxima were counted. Thirty-two candidate genes with the requisite number of 5 maxima when deleted yielded one strain YDR005C with protein properties consistent with its identity as the ENOX4 protein of Saccharomyces cerevisiae (Figure 1(B)).
The uninduced yeast lysate and the induced yeast lysate both showed similar oscillatory patterns with the
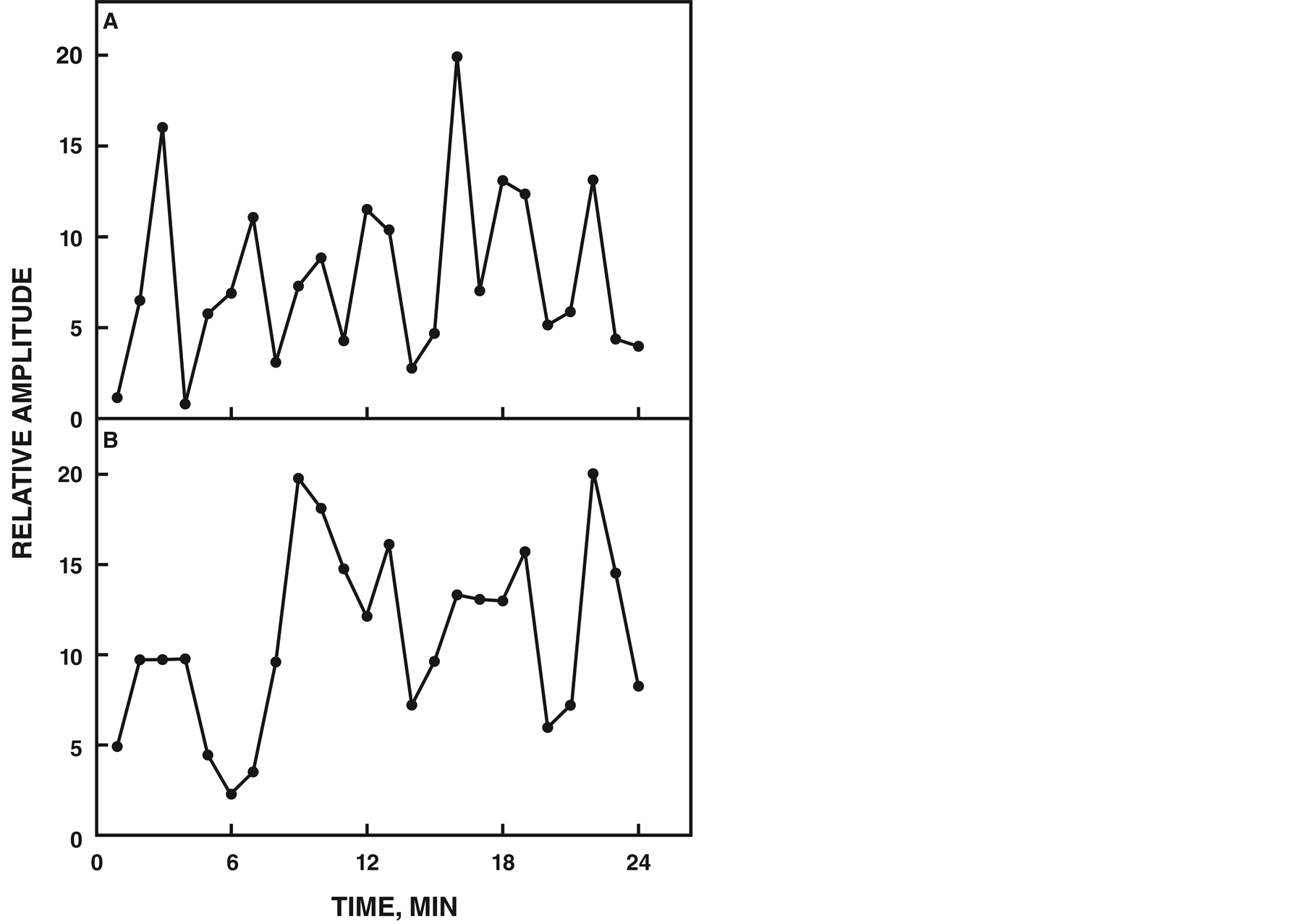
Figure 1. NADH fluorescence decomposition fits. A. Wild type yeast strain showing a pattern of 7 maxima. B. YDR005C showing a pattern of 5 maxima.
induced lysate having stronger amplitude for both NADH (Figure 2) and ubiquinol (Figure 3) oxidase activities. The pattern of oscillation of the maxima was similar to that of the constitutive ENOX1 of other species in that the pattern was asymmetric with the first two maxima separated by approximately 6 min. The remaining maxima were separated by an average of approximately 4.75 min instead of the usual 4.5 min which combined for a total of 25 min per oscillatory YDR005C cycle.
YDR005C encodes a 395 aa protein of molecular weight 44,730 and isoelectric point pH 5.15 (Figure 4). It contained a S219LGPFG putative adenine binding motif and putative copper binding motifs Y246PDH and Y335LYas well as a potential protein disulfide interchange motif C23KIVFSC. There were essentially no homologies between the amino acid sequence of the YDR005C and the yeast ENOX1 protein YM117W with the 24 min period length [16] .
The YDR005C gene product, when over expressed and analyzed, did exhibit properties associated with the ENOX proteins of other species but with a period length of 25 min rather than 24 min. Sliced and pulverized SDS-PAGE gels from which the protein was eluted overnight in assay buffer revealed enzymatic activity in the regions of the gel occupied by two molecular weights of 55 and 39 kDa (Figure 5). NADH oxidase activity was localized in slices 5 and 7, corresponding to molecular weight ranges of 49 - 60 kDa and 36 - 41 kDa. These slices contained the protein based on western blot localization of the His-tag at molecular weights of 55 kDa and 39 kDa (Figure 5, inset).
The gel slice elute containing the 55 kDa His-tagged YDR005C was assayed to determine the NADH oxidase activity and the pattern of oscillatory maxima (Figure 6). The assay showed a similar symmetric pattern of five maxima with a period length of 25 min and the failure of 1 µM simalikalactone D to inhibit the activity.
When hydroquinones were employed as a natural membrane located substrate using the gel slice elute corresponding to the 55 kDa His-tagged YDR005C, a 25 min period of 5 maxima was seen either as an increase in absorbance at 410 nmor as a decrease in absorbance at 290 nm (Figure 7). The addition of 1 µM simalikalactone D
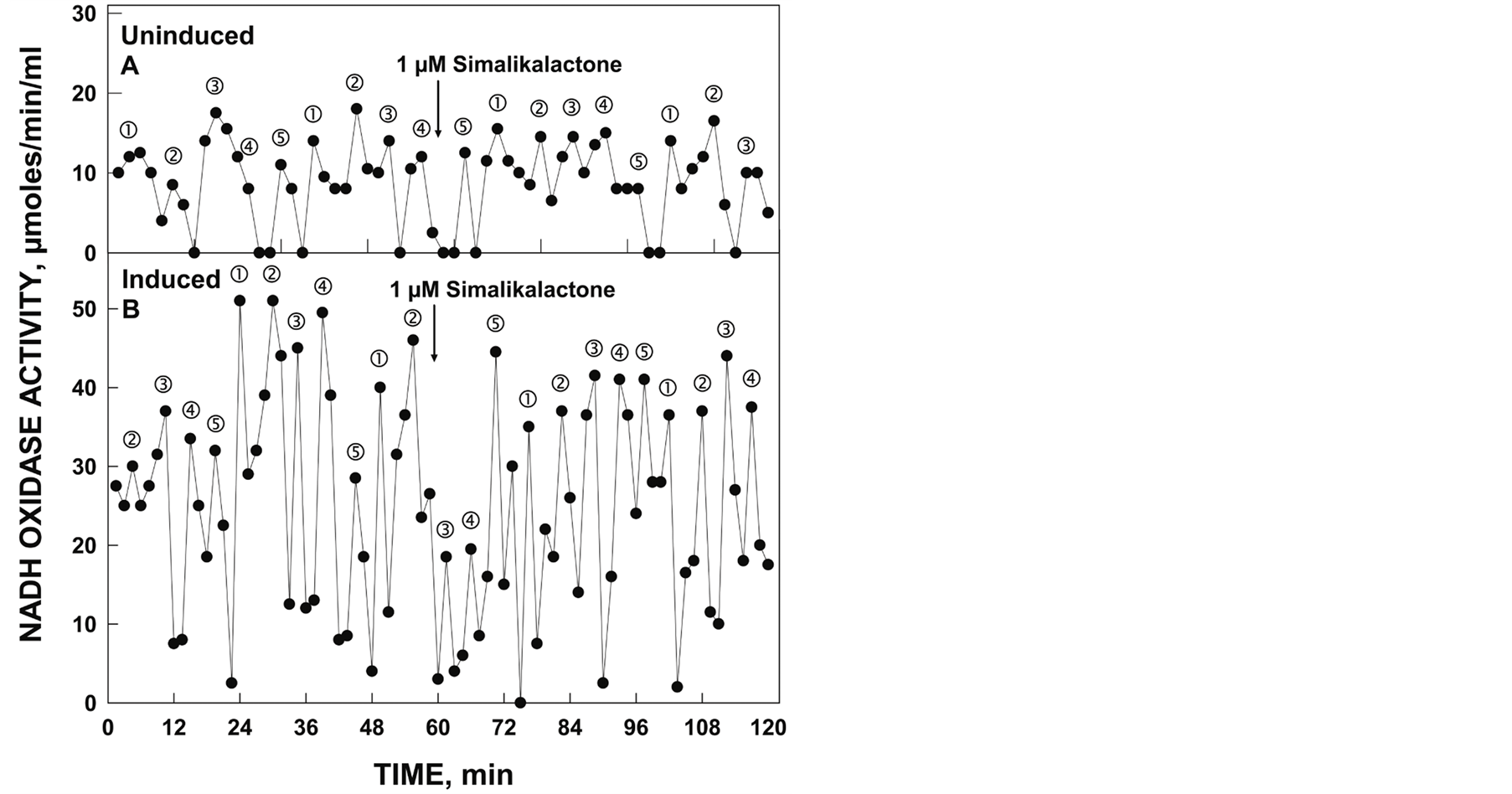
Figure 2. Oxidation of NADH by YDR005C yeast lysate comparing uninduced (A) and induced (B) yeast. Uninduced yeast was grown in the presence of glucose and the induced yeast was grown in the absence of glucose. Maxima labeled and ‚ were separated from each other by 6 min whereas maxima labeled ƒ, „ and … are separated from each other and from maxima and ‚ by 4.75 min to produce a 25 min [6 + 4 × (4.75) min]. Activity was not inhibited by the addition of 1 µM simalikalactone D (arrow).
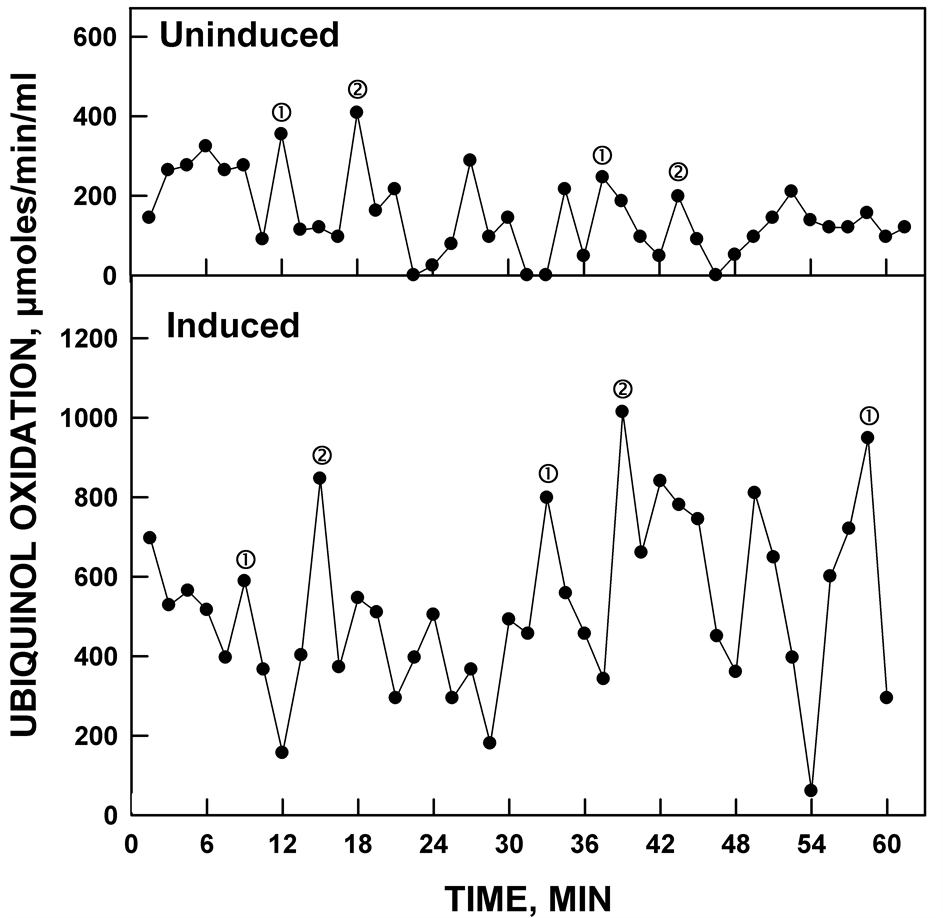
Figure 3. Ubiquinol oxidation activity (average of ubiquinol disappearance at A290 and at A410) of yeast lysates comparing uninduced and induced yeast. Uninduced yeast was grown in the absence of glucose and the induced yeast was grown in the presence of glucose. Both show similar oscillatory patterns with the induced lysate having the stronger ubiquinol oxidation activity.

Figure 4. Sequence of YDR005C. A putative NADH-binding sequence is bolded, potential protein disulfide-thiol interchange sites are denoted by dashed underlines and putative copper-binding sites are underlined.
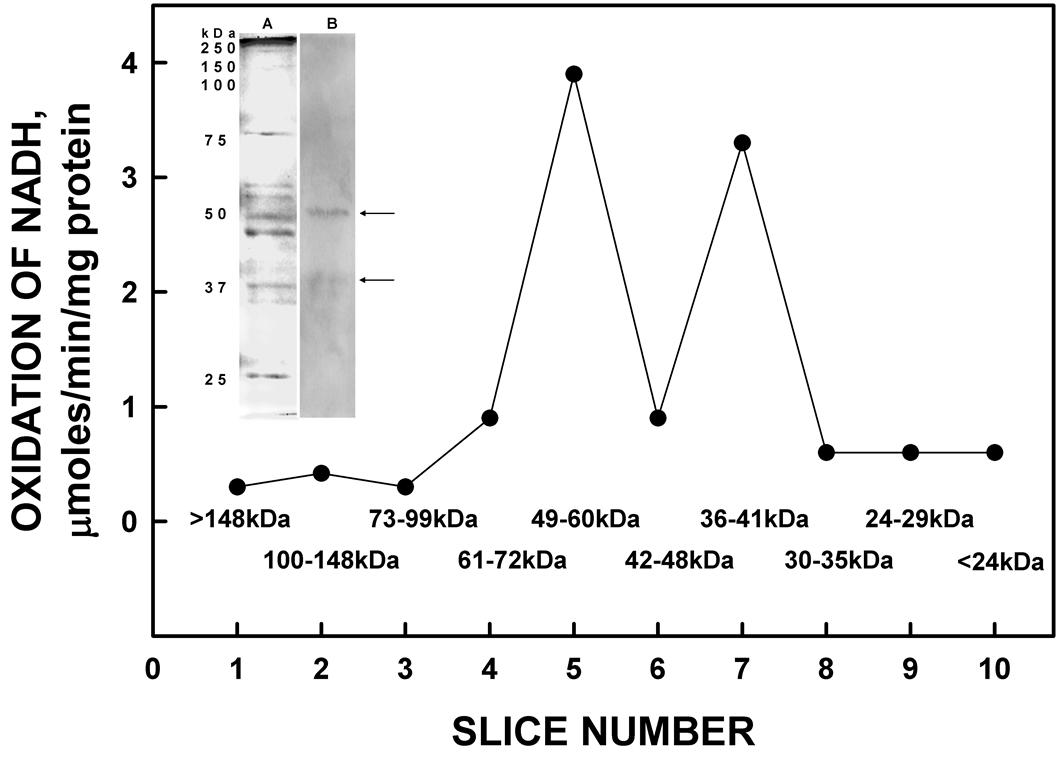
Figure 5. Resolution of recombinant His-tagged ENOX1 by SDSPAGE. Eluted activity was restricted to the high molecular weight region of the gel consistent with a molecular weight of 55 kDa with the His tag. The inset shows silver staining (A) and western blot (B) analysis of His-tagged ENOX4 protein [Slice 1: >148 kDa; Slice 2: 100 - 148 kDa; Slice 3: 73 - 99 kDa; Slice 4: 61 - 72 kDa; Slice 5: 49 - 60 kDa; Slice 6: 42 - 48 kDa; Slice 7: 36 - 41 kDa; Slice 8: 30 - 35 kDa; Slice 9: 24 - 29 kDa; Slice 10: <42 kDa]. The arrows indicate positions of YDR005C proteins.
was without effect on hydroquinone oxidation catalyzed by YDR005C.
The protein disulfide-thiol interchange activity of the gel slice elute containing the desired 55 kDa His-tagged YDR005C was determined by cleavage of dithiodipyridine substrate (Figure 8). The five maxima oscillatory pattern was obtained similar to the NADH oxidase activity. However, in contrast to NADH oxidation where maxima and ‚ often appear to dominate (e.g., Figure 6), the maxima separated by 4.75 min, labeled ƒ, „, and …, were of greater amplitude than maxima labeled and ‚ separated by 6 min. The YDR005C-catalyzed cleavage of the dithiodipyridine substrate was not inhibited by 1 µM simalikalactone D (Figure 8, dotted lines and open circles).
Unlike the constitutive ENOX1, the activity of YDR005C was not phased by addition of 10 mM melatonin. With ENOX1 proteins, a new maximum of NADH oxidation corresponding to maximum of the periodicity pattern was observed 24 min following melatonin addition. With YDR005C, melatonin addition was without effect (Figure 9).
When assayed in D2O, the period length of YDR005C was increased from 25 min to 32 min (Figure 10). An increase of 25% in period length when assayed in heavy water is a general characteristic of all ENOX proteins. In addition, the period of purified yeast YDR005C based on rate of NADH oxidation was shifted by exposure of
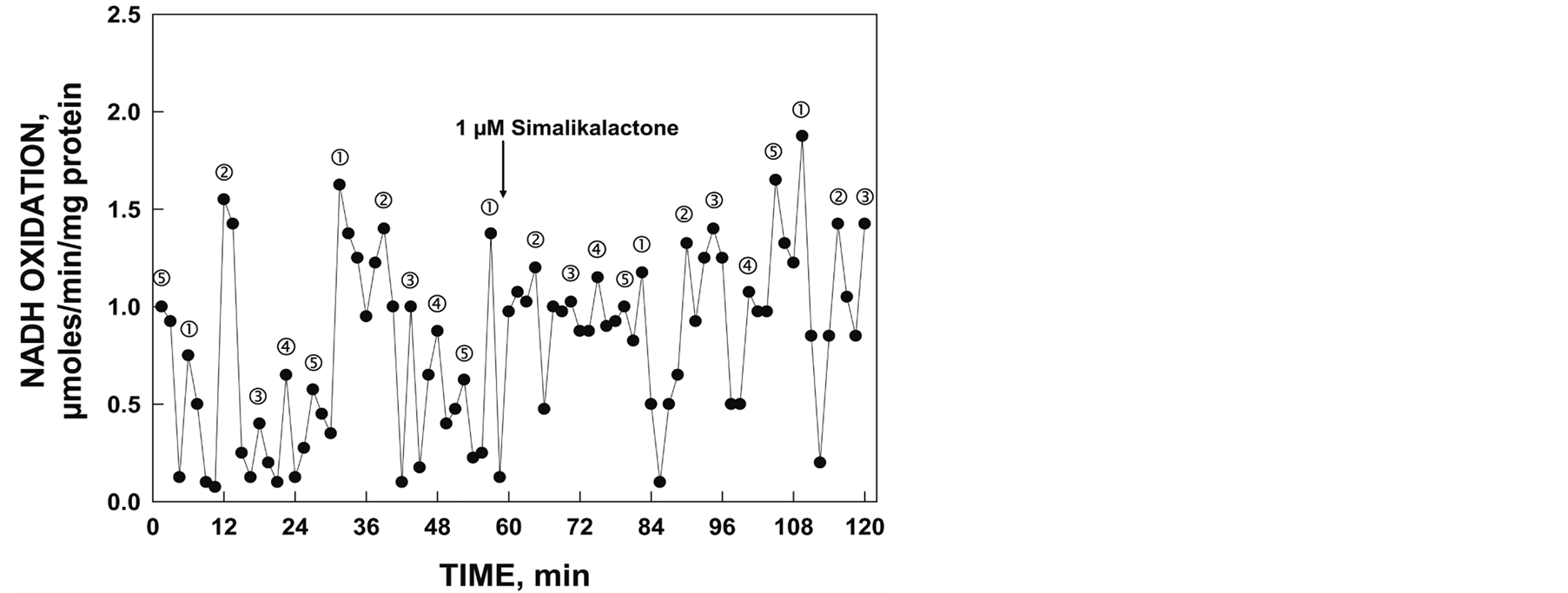
Figure 6. NADH oxidase activity of gel slice eluate corresponding to position of 55 kDa His-tagged YDR005C and response to 1 mM simalikalactone D (arrow) determined spectrophotometrically. Illustrated is the asymmetric oscillatory pattern of five maxima with a period length of 25 min.

Figure 7. Oxidation of reduced coenzyme Q by gel slice eluate corresponding to the position of the 55 kDa His-tagged YDR005C measured either by an increase in A410. (A) or by a decrease in A290 (B). As with NADH oxidation of Figure 4, the activity oscillated with prominent maxima separated by 6 min and ‚ followed by 3 additional maxima separated by 4.75 min to generate the 25 min period. Addition of 1 µM simalikalactone D (arrow) did not inhibit the activity.
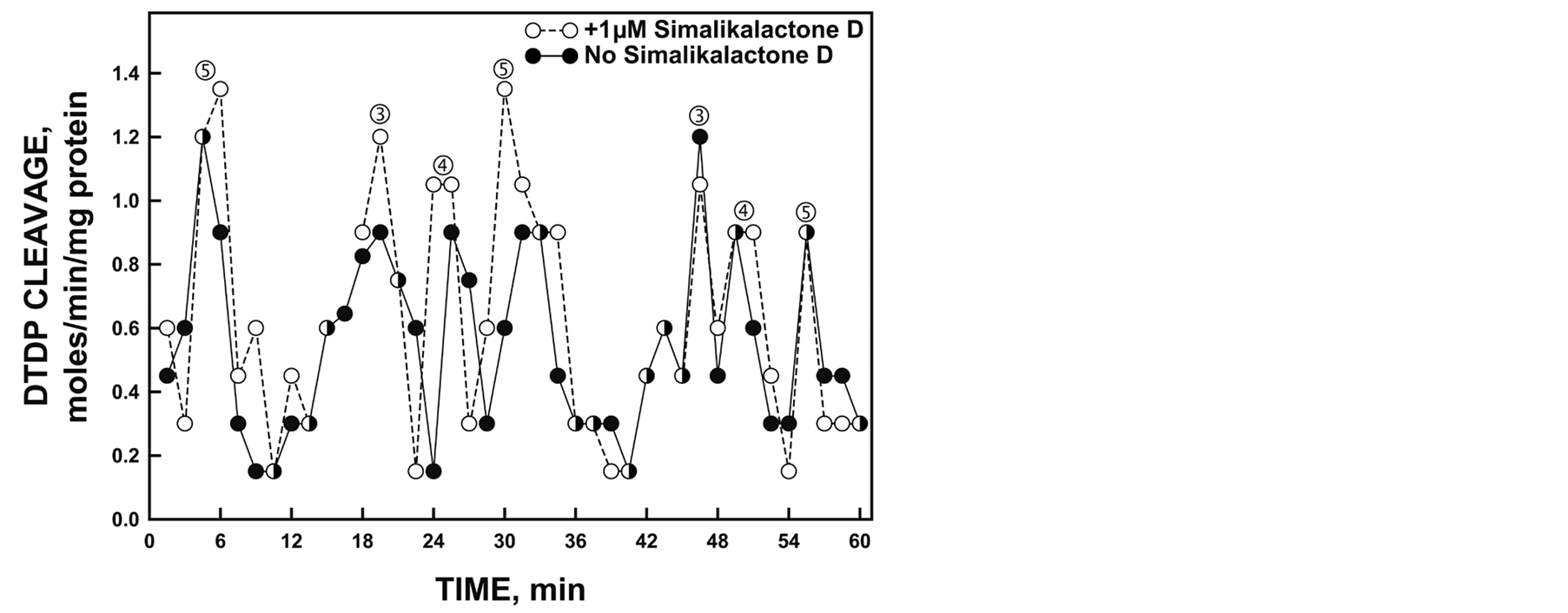
Figure 8. Protein disulfide-thiol interchange activity of gel slice eluate corresponding to the position of the 55 kDa His-tagged YDR005C determined simultaneously using paired spectrophotometers by cleavage of a dithiodipyridine (DTDP) substrate in the absence (solid lines and symbols) and presence (dotted lines and open symbols) of 1 mM simalikalactone D. The spacing of labeled maxima is the same as that of Figure 5. However, in contrast, maxima labeled ƒ, „ and … separated by 4.75 min are generally of greater amplitude than maxima and ‚ separated by 6 min consistent with an alternation of the interchange and oxidative (NADH and ubiquinol) activities.
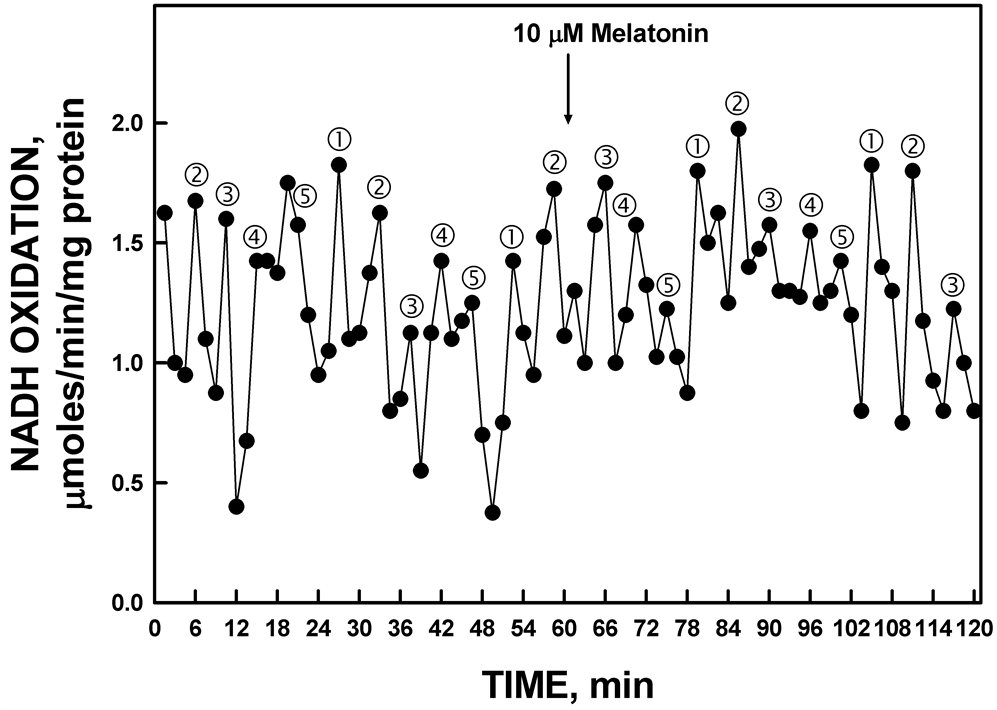
Figure 9. The activity period of NADH oxidation by recombinant YDR005C was unaffected by the addition of melatonin.
the assay cuvette to a low-frequency electromagnetic field (50 µT for 20 sec) (Figure 11).
4. Discussion
As reported previously [16] , the yeast Saccharomyces cerevisiae exhibits an ENOX1 activity with a period length of 24 min similar to that of other eucaryotes [15] . However, S. cerevisiae has a second ENOX1-like activity with a period length of 25 min [16] . The latter is different from the yeast ENOX1 and ENOX1 proteins in general not only from the longer period length but also by being resistant to the specific ENOX1 inhibitor simalikalactone D and failure of the 25 min rhythm to be phased by melatonin. In this report, the ENOX with a period length of 25 min, previously referred to as YNOX for yeast NOX and now designated as ENOX4, is identified as YDR-005C based on screening of a yeast deletion library and cloning from a yeast overexpression library.
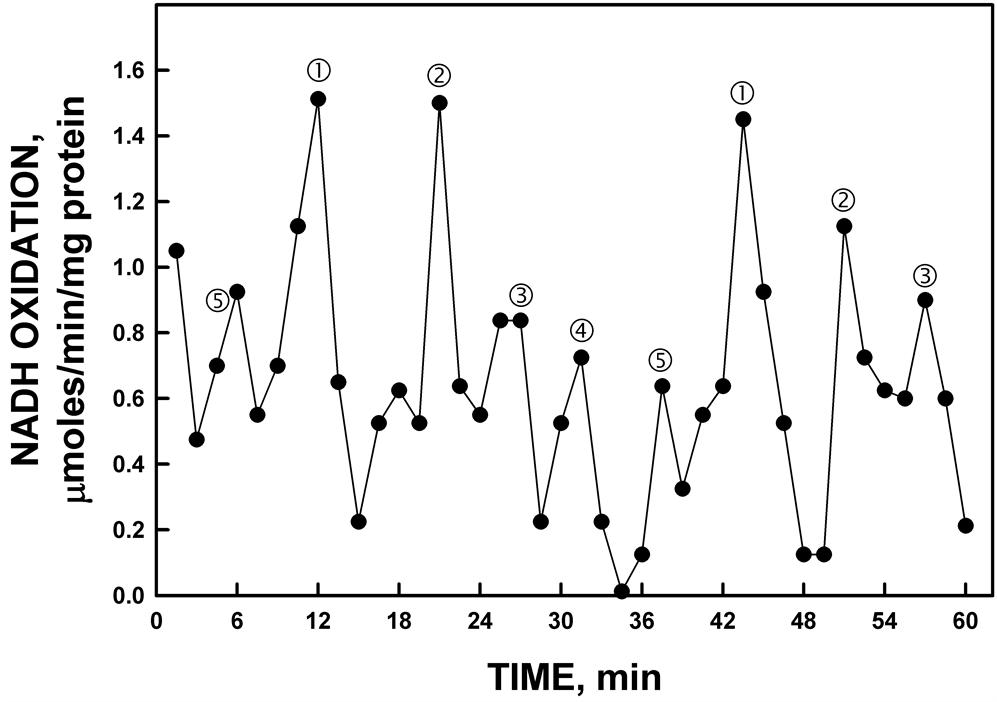
Figure 10. His-tagged YDR005C activity from gel slice assayed in D2O rather than H2O. In the presence of D2O, the period length of the oscillations was lengthened from 25 min to approximately 32 min. The increased period length in response to heavy water is one of the hallmarks of the biological clock (see text).
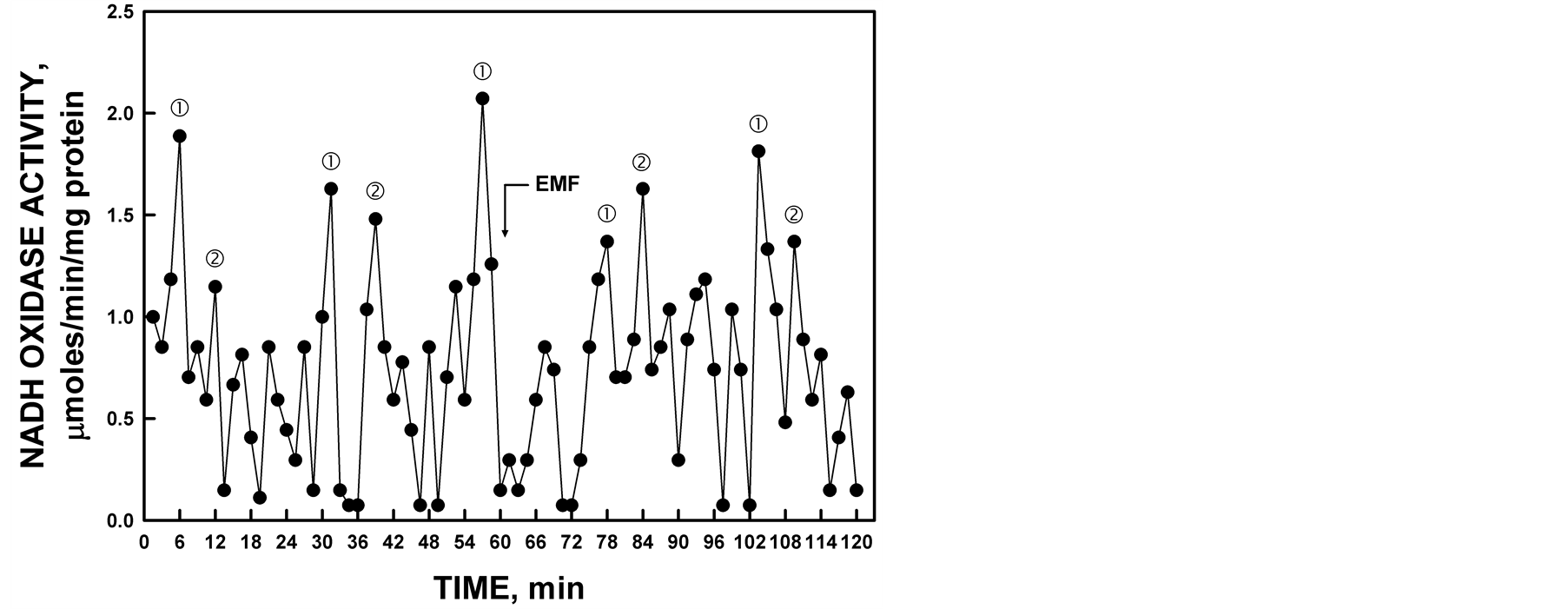
Figure 11. The activity period of NADH oxidation by recombinant YDR005C was phased by the application of low-frequency EMF. All maxima were shifted approximately 2.5 min by the 20 sec EMF exposure to 50 µT at 60 min (arrow).
There is little or no sequence similarity of YDR005C to other known ENOX proteins. However, required functional motifs are conserved. A potential adenine nucleotide binding motif is found in the form of S219- XGXXG and a possible protein disulfide interchange site at C23XXXXXC. Putative copper binding sites are found at Y246PDH and Y335LY. Site-directed mutagenesis studies to confirm these sites have been carried out with the yeast ENOX1 protein [15] and the ENOX1 of human [20] derivation but remain to be completed for YDR005C.
ENOX1 proteins are ubiquitous on the surface of all cells thus far studied [21] . They are protease and heat resistant [22] and responsive to hormones and growth factors [1] [23] . The constitutive ENOX1 serves as an ultradian oscillator of the biological clock [14] and, as such, is important for understanding circadian rhythms.
ECTO-NOX proteins are considered to function as homodimers associated with the external surface of the cell [20] . Each monomer contains two copper atoms. This allows each dimer to transfer four electrons and produce two molecules of water from one oxygen molecule. Both the mammalian ENOX1 [20] and the yeast ENOX1 [16] contain two or more copper binding sites which, when mutated, lead to a loss of ENOX1 activity.
ENOX1 proteins are the terminal oxidases of plasma membrane electron transport and play an important role in oxygen consumption at the cell surface. NADH is an impermeant substrate, so when added to an assay, it is presented to the outside of the cell and only available to enzymes that are on the outside of the cell. Although NADH is used as a substrate in our assays, the natural substrates of the ECTO-NOX proteins are the hydroquinones that reside within the plasma membrane leaflets [12] . When hydroquinones are oxidized by the ECTONOX proteins, they are then available to be reduced by the quinone reductases that reside at the inner surface of the plasma membrane. The quinone reductases use NAD(P)H in the cytoplasm to reduce the hydroquinones, resulting in a net increase in NAD(P)+ in the cytoplasm.
The ECTO-NOX proteins also catalyze another reaction, disulfide-thiol interchange, in which they exchange their thiols with disulfide bonds in other proteins and vice versa. This activity is responsible for cell enlargement through breakage and formation of disulfide bonds between membrane proteins essential for growth [21] . In addition to NADH oxidation, both activities catalyze hydroquinone oxidation and protein disulfide-thiol interchange as evidenced by cleavage of the dithio-dipyridine (DTDP) substrate. These activities, NADH or hydroquinone oxidation and protein disulfide-thiol interchange, alternate in a manner unprecedented in the biochemical literature and are unique to the ENOX family of proteins [1][12].
The two yeast ENOX1-like activities are not identical. The activity with a 25 min period is not inhibited by the ENOX1 inhibitor simalikalactone D nor is the 25 min period phased by melatonin. When melatonin is added to the reaction mixture, mammalian ENOX1 begins a pattern of maxima exactly 24 min after melatonin addition. The 24 min yeast ENOX1 period follows this same pattern. In contrast, the 25 min yeast ENOX period does not. It has been shown that hydrogen sulfide and acetaldehyde can synchronize the 40 min period of oxygen consumption in a population of cells in continuous culture [24] so it is possible that the 25 min period may respond to other synchronizing influences. YDR005C (ENOX4) activity does respond to low-frequency EMF by shifting the phase.
It is unclear why the ENOX4 period is slightly longer than the ENOX1 period. The altered period length may be due to differences in the disulfide-thiol interchange motif, as mutations in this site in other ENOX proteins result in altered period lengths [20] [25] .
ECTO-NOX proteins are heat-(80˚C) and protease-resistant [10] [22] [26] -[30] and they form amyloid rods visible with the electron microscope. All ECTO-NOX proteins are prone to aggregation when they are concentrated or in a relatively pure solution.
The statistical validity of the asymmetric non-sinusoidal oscillatory activity pattern has been validated previously by Fast Fourier and decomposition (seasonal forecasting) analyses [19] . Each 24 min period exhibits 5 maxima, 2 of which are separated by 6 min designated as and ‚ and the remainders of which, designated as ƒ, „ and …, are separated from each other and from maximum and ‚ by 4.75 min [6 + 4 × (4.75) = 25 min.
It is interesting that in yeasts, ENOX1 inhibitors do not inhibit growth, as they do in mammalian and plant cells [3] . This attribute is probably due to the difference in the method of enlargement. Plant and mammalian cells enlarge by physical membrane displacement [31] -[33] whereas yeasts enlarge by tip growth or budding [34] [35] . In mammalian cells, the cell enlargement function of ENOX1 is central to its activity and essential for cell viability, so studying the other functions of ENOX1 is not possible. In yeasts, the enlargement function is no longer a primary function, so that other activities of ENOX1 can be studied separately from cell growth.
References
- Morré, D.J. (1998) NADH Oxidase: A Multifunctional Ectoprotein of the Eukaryotic Cell Surface. In: Asard, H., Bérczi, A. and Caubergs, R., Eds., Plasma Membrane Redox Systems and Their Role in Biological Stress and Disease, Kluwer Academic Publishers, Dordrecht, 121-156. http://dx.doi.org/10.1007/978-94-017-2695-5_5
- Morré, D.J. and Morré, D.M. (2003) Cell Surface NADH Oxidases (ECTO-NOX Proteins) with Roles in Cancer, Cellular Time-Keeping, Growth, Aging and Neurodegenerative Disease. Free Radical Research, 37, 795-808. http://dx.doi.org/10.1080/1071576031000083107
- Morré, D.J. and Greico, P.A. (1999) Glaucarubolone and Simalikalactone D, Respectively, Preferentially Inhibit Auxin-Induced and Constitutive Components of Plant Cell Enlargement and the Plasma Membrane NADH Oxidase. International Journal of Plant Science, 160, 291-297. http://dx.doi.org/10.1086/314133
- Morré, D.J., Chueh, P.-J. and Morré, D.M. (1995) Capsaicin Inhibits Preferentially the NADH Oxidase and Growth of Transformed cells in Culture. Proceedings of the National Academy of Science USA, 92, 1831-1835. http://dx.doi.org/10.1073/pnas.92.6.1831
- Morré, D.J., Sun, E., Geilen, C., Wu, L.-Y., de Cabo, R., Krasagakis, K., Orfanos, C.E. and Morré, D.M. (1996) Capsaicin Inhibits Plasma membrane NADH Oxidase and Growth of Human and Mouse Melanoma Lines. European Journal of Cancer, 32A, l995-2003.
- Pogue, R., Morré, D.M. and Morré, D.J. (2000) CHO Cell Enlargement Oscillates with a Temperature-Compensated Period of 24 Minutes. Biochimica et Biophysica Acta, 1498, 44-51. http://dx.doi.org/10.1016/S0167-4889(00)00076-8
- Morré, D.J., Pogue, R. and Morré, D.M. (2001) Soybean Cell Enlargement Oscillates with a Temperature-Compensated Period Length of Ca. 24 Min. In Vitro Cellular Development and Biology Plant, 37, 19-23. http://dx.doi.org/10.1007/s11627-001-0004-3
- Morré, D.J., Ternes, P. and Morré, D.M. (2002) Cell Enlargement of Plant Tissue Explants Oscillates with a Temperature-Compensated Period Length of Ca. 24 Min. In Vitro Cellular Development and Biology Plant, 38, 18-28. http://dx.doi.org/10.1079/IVP2001249
- Morré, D.J., Brightman, A.O., Hidalgo, A. and Navas, P. (1995) Selective Inhibition of Auxin-Stimulated NADH Oxidase Activity and Elongation Growth of Soybean Hypocotyls by Thiol Reagents. Plant Physiology, 107, 1285-1291.
- Morré, D.M. and Morré, D.J. (2003) Specificity of Coenzyme Q Inhibition of an Aging-Related Cell Surface NADH Oxidase (ECTO-NOX) That Generates Superoxide. BioFactors, 18, 33-43. http://dx.doi.org/10.1002/biof.5520180205
- Morré, D.J., Guo, F. and Morré, D.M. (2003) An Aging-Related Cell Surface NADH Oxidase (arNOX) Generates Superoxide and Is Inhibited by Coenzyme Q. Molecular and Cellular Biochemistry, 254, 101-109.http://dx.doi.org/10.1023/A:1027301405614
- Kishi, T., Morré, D.M. and Morré, D.J. (1999) The Plasma Membrane NADH Oxidase of HeLa Cells Has Hydroquinone Oxidase Activity. Biochimica et Biophysica Acta, Bioenergetics, 1412, 66-77. http://dx.doi.org/10.1016/S0005-2728(99)00049-3
- Morré, D.J., Gomez-Rey, M.L., Schramke, C., Em, O., Lawler, J., Hobeck, J. and Morré, D.M. (1999) Use of Dipyridyl-Dithio Substrates to Measure Directly the Protein Disulfide-Thiol Interchange Activity of the Auxin Stimulated NADH: Protein Disulfide Reductase (NADH Oxidase) of Soybean Plasma Membranes. Molecular and Cellular Biochemistry, 200, 7-13. http://dx.doi.org/10.1023/A:1006916116297
- Morré, D.J., Chueh, P.J., Pletcher, J., Tang, X., Wu, L.Y. and Morré, D.M. (2002) Biochemical Basis for the Biological Clock. Biochemistry, 41, 11941-11945. http://dx.doi.org/10.1021/bi020392h
- Tang, X., Parisi, D., Spicer, B., Morré, D.M. and Morré, D.J. (2013) Molecular Cloning and Characterization of Human Age-Related NADH Oxidase (arNOX) Proteins as Members of the TM-9 Super Family of Transmembrane Proteins. Advances in Biological Chemistry, 3, 187-197. http://dx.doi.org/10.4236/abc.2013.32024
- Dick, S.S., Ryuzoji, A., Morré, D.M. and Morré, D.J. (2013) Identification of the Constitutive Ultradian Oscillator of the Circadian Clock (ENOX1) in Saccharomyces cerevisiae. Advances in Biological Chemistry, 3, 320-328.http://dx.doi.org/10.4236/abc.2013.33036
- Dick, S.S., Ryuzoji, A., Morré, D.M. and Morré, D.J. (2013) Ultradian Oscillators of the Circadian Clock Saccharomyces cerevisiae. Advances in Biological Chemistry, 3, 59-69. http://dx.doi.org/10.4236/abc.2013.31008
- Smith, P.K., Krohn, R.I., Hermanson, G.T., Mailia, A.K., Gartner, F.H., Provenzano, M.D., Fujimoto, E.K., Goeke, N.M., Olson, F.J. and Klenk, D.C. (1985) Measurement of Protein Using Bicinchoninic Acid. Analytical Biochemistry, 150, 76-85. http://dx.doi.org/10.1016/0003-2697(85)90442-7
- Foster, K., Anward, N., Pogue, R., Morré, D.M., Keenan, T.W. and Morré, D.J. (2003) Decomposition Analyses Applied to a Complex Ultradian Biorhythm: The Oscillating NADH Oxidase Activity of Plasma Membranes Having a Potential Time-Keeping (Clock) Function. Nonlinearity in Biology, Toxicology and Medicine, 1, 51-70.
- Jiang, Z., Gorenstein, N.M., Morré, D.M. and Morré, D.J. (2008) Molecular Cloning and Characterization of a Candidate Human Growth-Related and Time-Keeping Constitutive Cell Surface Hydroquinone (NADH) Oxidase. Biochemistry, 47, 14028-14038. http://dx.doi.org/10.1021/bi801073p
- Morré, D.J. and Morré, D.M. (2012) ECTO-NOX Proteins. Springer, New York, 507 p.
- Kim, C. and Morré, D.J. (2004) Prion Proteins and ECTO-NOX Proteins Exhibit Similar Oscillating Redox Activities. Biochemical and Biophysical Research Communications, 315, 1140-1146. http://dx.doi.org/10.1016/j.bbrc.2004.02.007
- Bruno, M., Brightman, A.O., Lawrence, J., Werderitsh, D., Morré, D.M. and Morré, D.J. (1992) Stimulation of NADH Oxidase Activity from Rat Liver Plasma Membranes by Growth Factors and Hormones in Decreased or Absent with Hepatoma Plasma Membrane. Biochemistry Journal, 284, 625-628.
- Murray, D.B., Klevecz, R.R. and Lloyd, D. (2003) Generation and Maintenance of Synchrony in Saccharomyces cerevisiae Continuous Culture. Experimental Cell Research, 287, 10-15. http://dx.doi.org/10.1016/S0014-4827(03)00068-5
- Chueh, P.J., Kim, C., Cho, N., Morré, D.M. and Morré, D.J. (2002) Molecular Cloning and Characterization of a Tumor-Associated, Growth-Related and Time-Keeping Hydroquinone (NADH) Oxidase (tNOX) of the HeLa Cell Surface. Biochemistry, 41, 3732-3741. http://dx.doi.org/10.1021/bi012041t
- Sedlak, D., Morré, D.M. and Morré, D.J. (2001) A Drug-Unresponsive and Protease-Resistant CNOX Protein from Human Sera. Archives of Biochemistry and Biophysics, 386, 106-116. http://dx.doi.org/10.1006/abbi.2000.2180
- Chueh, P.J., Morré, D.J., Wilkinson, F.E., Gibson, J. and Morré, D.M. (1997) A 33.5 kDa Heatand Protease Resistant NADH Oxidase Inhibited by Capsaicin from Sera of Cancer Patients. Archives of Biochemistry and Biophysics, 342, 38-47. http://dx.doi.org/10.1006/abbi.1997.9992
- del Castillo-Olivares, A., Yantiri, F., Chueh, P.-J., Wang, S., Sweeting, M., Sedlak, D., Morré, D.M., Burgess, J. and Morré, D.J. (1998) A Drug-Responsive and Protease-Resistant Peripheral NADH Oxidase Complex from the Surface of HeLa S Cells. Archives of Biochemistry and Biophysics, 358, 125-140. http://dx.doi.org/10.1006/abbi.1998.0823
- Kelker, M., Kim, C., Chueh, P.J., Guimont, R., Morré, D.M. and Morré, D.J. (2001) Cancer Isoform of a Tumor-Associated Cell Surface NADH Oxidase (tNOX) Has Properties of a Prion. Biochemistry, 41, 11941-11945.
- Markert, C., Morré, D.M. and Morré, D.J. (2004) Human Amyloid Peptides Aß1-40 and Aß1-42 Exhibit NADH Oxidase Activity with Copper-Induced Oscillations and a Period Length of 24 Min. BioFactors, 20, 221-235.http://dx.doi.org/10.1002/biof.5520200405
- Morré, D.J., Kim, C. and Hicks-Berger, C. (2006) ATP-Dependent and Drug Inhibited Vesicle Enlargement Reconstituted Using Synthetic Lipids and Recombinant Proteins. BioFactors, 28, 105-117. http://dx.doi.org/10.1002/biof.5520280205
- Hicks-Berger, C. and Morré, D.J. (2006) Inside-Out but Not Right Side-Out Plasma Membrane Vesicles from Soybean Enlarge When Treated with ATP+ 2,4-D as Determined by Electron Microscopy and Light Scattering: Evidence for Involvement of a Plasma Membrane AAA-ATPase. BioFactors, 28, 91-104. http://dx.doi.org/10.1002/biof.5520280204
- Auderset, G. and Morré, D.J. (2006) ATPand Growth Substance-Dependent Cell-Free Enlargement of Plasma Membrane Vesicles from Soybean. BioFactors, 28, 83-90. http://dx.doi.org/10.1002/biof.5520280203
- Adams, A.E. and Pringle, J.R. (1984) Relationship of Actin and Tubulin Distribution to Bud Growth in Wild-Type and Morphogenetic-Mutant Saccharomyces cerevisiae. Journal of Cellular Biology, 98, 934-945.http://dx.doi.org/10.1083/jcb.98.3.934
- Cabib, E., Bowers, B., Sburlati, A. and Silverman, S.J. (1988) Fungal Cell Wall Synthesis: The Construction of a Biological Structure. Microbiology Sciences, 5, 370-375.
Abbreviations
BCA: Bicinchoninic acid;
DTDP: Dithiodipyridine;
EMF: Electromagnetic field;
ENOX: ECTO-NOX;
ENOX1: Constitutive ENOX (CNOX);
ENOX2: Tumor-associated ENOX (tNOX);
ENOX3: Age-related NADH oxidase;
ENOX4: Yeast-specific ENOX (YNOX) protein with a 25 min period insensitive to semilikilactone D inhibition and melatonin;
GSH: Reduced glutathione;
NI-NTA: Nickel nitrilotriacetic acid;
PMSF: Phenylmethylsulfonyl fluoride;
SDS: Sodium dodecyl sulfate SDS-PAGE: Sodium dodecyl sulfate polyacrylamide gel electrophoresis;
YEPD: Yeast extract peptone dextrose
NOTES

*Corresponding author.

