American Journal of Molecular Biology
Vol.2 No.1(2012), Article ID:16545,7 pages DOI:10.4236/ajmb.2012.21005
Molecular surveillance of dengue virus in Bahia State, Brazil
![]()
1Centro de Pesquisas Gonçalo Moniz, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil
2Santa Cruz State University, Ilhéus, Brazil
3Robert Koch-Institut (ZBS-1), Berlin, Germany
4Laboratório Central do Estado da Bahia (LACEN), Secretaria de Saúde do Estado da Bahia (SESAB), Salvador, Brazil
5Instituto de Salud Carlos III, Madrid, Spain
6Case Western Reserve University, Cleveland, USA
Email: *pmelo11@yahoo.com.br
Received 10 August 2011; revised 4 October 2011; accepted 18 October 2011
Keywords: Dengue; Dengue Hemorrhagic Fever; Molecular Surveillance; DENV Genotypes; E/NS1 Junction
ABSTRACT
Dengue is an important emerging viruses, posing a threat to one-third of the global human population. In 2002, the introduction of DENV-3 in the state of Bahia produced massive epidemic (about 35,000 cases detected) and the first cases of dengue hemorrhagic fever. To understand the nature of the virus circulating at Bahia, E/NS1 sequence was determined for 31 DENV viruses isolated in Bahia during the 2006 and 2007 transmission season, from patients presenting with different degrees of severity. The carboxi-terminal region of the E gene (220 nt) of 31 viruses, isolated from dengue patients with clinical diagnosis of dengue infection were used to determine the genetic variability of dengue 2 (DENV-2) and dengue 3 (DENV- 3). Sequence data were used in phylogenetic comparisons with global samples of DENV-2 and DENV-3. DENV-2 sample grouped in the South East Indian genotype, while DENV-3 samples were grouped within Indian genotype. This study is the first report on Bahia isolates during two transmission seasons. Our data confirms reports from other parts of Brazil and different countries showing the DENV-3 (genotype III) strains circulating in the Americas are closely related, and cluster within the genotype that has been associated with DHF epidemics in different continents.
1. INTRODUCTION
Dengue fever, dengue hemorrhagic fever and dengue shock syndrome are considered to be the most important arthropod-borne viral diseases due to its high prevalence, and significant morbidity and mortality. The global incidences of dengue has now increased enormously and about 50 - 100 million dengue infections are now reported annually from more than 100 tropical and subtropical countries of the world [1]. Infection with any of the dengue viruses can be manifested in a range of symptoms from a relatively mild flu-like syndrome with rash (dengue fever, DF) to a severe and potentially fatal disease characterized by capillary leakage, thrombocytopenia, and sometimes hypovolemic shock (dengue hemorrhagic fever, DHF/dengue shock syndrome, DSS). Immune enhancement mechanisms following a second heterologous dengue virus infection have been associated with a higher risk of developing DHF/DSS [2], however, cases of DHF/DSS in confirmed primary infections have been observed [3]. Different studies have shown that changes in the viral genetic structure may contribute to shifts in the epidemic potential or pathogenicity of dengue virus serotypes [4,5]. These observations point to the importance of viral factors in determining disease severity. In addition, some strains have been associated with increased pathogenic potential [6].
DENV-3 re-appeared in Latin American in 1994 after an absence of 17 years [7]. Before, serotypes DENV-1 and DENV-2 were circulating for more than a decade in Brazil. In 2002, the introduction of DENV-3 in the state of Bahia produced the first cases of DHF. This event transformed Bahia into a hiperendemic area with three serotypes circulating at the same time [8]. DENV-3, after its introduction, was the serotype responsible for the majority of infections in both Brazil and in Bahia; isolates from different parts of Brazil have been analyzed with respect to their genetic variability. Molecular techniques including sequencing of the viral genomic RNA and direct sequencing of reverse transcriptase/polymerase chain reaction (RT/PCR)-amplified genes have provided many information on geographic distribution, origin and evolution among dengue viruses. Many genes have been used as 5′3′UTR regions, E gene and E/NS1 junction [9,10].
Here we report the analysis of a genomic interval spanning the E/NS1 junction of the dengue genome used for molecular surveillance of this flavivirus and to determine which strains of dengue virus for DENV-2 and DENV-3 were circulating in Bahia state during the 2006- 2007 transmission seasons.
2. MATERIALS AND METHODS
2.1. Sampling
For administrative and epidemiologic purposes, the state is divided into municipalities, which are the basic reporting unit for the State Secretariat of Health (SSH). Dengue is a reportable disease in Brazil. The secretary of health in each municipality is responsible for collecting serum from each suspected case of dengue within five days of onset of symptoms, and sending this on to the Bahia State Central Laboratory (LACEN) for viral isolation and serology. Officially, all cases of dengue in the state should send samples the LACEN-Bahia for viral serotyping. Between 2006 and 2007, 4018 samples were sent to LACEN to be serotyped. 108 samples were positive for isolating in C6/36 Aedes albopictus cells, and 31 positive samples were further analyzed by RT-PCR and sequencing.
2.2. Viral Isolation and Serotyping
Viral isolation was performed in all samples (in 4018 samples) by inoculation of an Aedes albopictus C6/36 cell line with 25 µL of undiluted acute serum. The cell line was then incubated for seven days at 32˚C in Leibovitz’s medium supplemented with 5% fetal calf serum. After fixation to slides in quadruplicate with cold buffered acetone, the cells were incubated with serotypespecific monoclonal antibodies for each of the 4 serotypes followed by fluorescein conjugated anti-mouse antibodies. The slides were then scored as positive or negative by fluorescent microscopy [11].
2.3. RNA Extraction, Amplification and Sequencing
Viral RNA was extracted from serum of infected patients of all positive samples for cell culture using QIAamp Viral RNA Mini kit (Qiagen, CA) following the manufacturer’s instructions. A Mastercycle Eppendorf (model 5331) was used throughout. Five µl of viral RNA solution were added to 45 µl of a medium compatible with both the reverse transcription and PCR amplification steps of the QIAGENOneStep RT-PCR kit. The RTPCR reactions were carried out using an initial reverse transcription step at 41˚C for 45 minutes followed by a denaturation and Hot Star Taq polymerase activation step (94˚C, 15 minutes) and 40 cycles of denaturation (94˚C, 30 seconds), primer annealing (55˚C, 1 minute), and primer extension (72˚C, 30 seconds). A final incubation was carried out at 72˚C for 5 minutes. A second amplification reaction (nested PCR) was seeded with 1 µl of the initial amplification product. The reaction mixture contained 1 buffer B (60 mM Tris-HCl pH 8.5, 2 mM MgCl2, 15 mM (NH4)2SO4, 40 pmol of each sense and antisense primer [9] and 2.5 U of DNA Taq Polymerase (Invitrogen). The samples were subjected to a denaturation step (94˚C, 2 minutes) followed by 40 cycles of denaturation (94˚C, 30 seconds), primer annealing (57˚C, 4 minutes), and primer extension (72˚C, 30 seconds) and a further extension step at 72˚C for 5 minutes.
2.4. Sequence Analysis
Sequencing reactions were performed as recommended in the BigDye Dideoxy Terminator sequencing kit (ABI Prism, Foster City, CA) and the products were analyzed using an automated 3100 DNA sequencer (ABI Prism, Applied Biosystems). Original sequence data were first analyzed by the CHROMAS software (version 1.3, McCarthy 1996; School of Biomolecular and Biomedical Science, Faculty of Science and Technology, Griffith University, Brisbane, Queensland, Australia); forward and reverse sequence data of each sample were aligned using the EDITSEQ program (DNASTAR Inc. Software, Madison, Wisconsin, USA). The consensus sequence was compared and aligned to other samples and DNA database sequences using the software CLUSTAL X, version 1.83 [12]. Program from MEGA package [13] were used to produce phylogenetic trees using neighbor joining as the method to reconstruct the phylogeny and Kimura-2p as nucleotide substitution calculation method. The statistical significance of a particular tree topology was evaluated by bootstrap resampling of the sequences 1000 times. Published sequences used in the comparisons were obtained from the GenBank databases (75 DENV-2 and 73 DENV-3 sequences).
3. RESULTS
3.1. Outbreak Molecular Surveillance
Dengue in Brazil is a notified disease, in 2002 (when DENV-3 was introduced) almost 80,000 cases were notified in all Bahia State (data not showed), and being the first time that DHF cases occurred in this region. After four years, between 2006 and 2007, about 25,000 cases were notified. The most important variable influencing this action is the number of cases [8], this agree that during outbreaks the system became more sensitive to detect disease than in inter-epidemic periods. For example, in 2006 Buritirama, a small city in Bahia with 20,495 inhabitants, between September and December notified about 300 cases of dengue, comparing with Salvador with almost 3,000,000 people that notified for all year just 664 cases. Despite that the surveillance is weak, the outbreaks could be detected, and even in municipalities far from the State Central Laboratory (LACEN) the number of cases remains as the more important aspect to a sample be collected and sent to LACEN.
3.2. Isolation and Typing of the Viruses
Between 2006 and 2007 were analyzed 4018 samples by fluorescent microscopy. Only 108 samples had been the serotype determined. RT-PCR was attempted directly in the serum of the patients of the only 31 viruses isolated in C6/36 cells, the positive samples and the RNA isolation was confirmed. The results indicated that 30 samples were positive for DENV-3, and 1 sample was positive for DENV-2 specific RNA.
3.3. Nucleotide Sequence Analysis
We have determined the nucleotide sequences of the E-NS1 gene junction (220 bp) of 31 dengue viruses in the present study. Pair-wise sequence analysis using Needleman-Wunsch global alignment was carried out on the 220 bp sequence for comparison. As expected, comparisons between serotypes showed a low sequence similarity and could be easily grouped (Figure 1). The results were consistent between distance and characterbased methods, with minimal differences in topologies. Samples from different years, for DENV-3, were grouped in the same genotype (INDIA genotype III) with 2006 samples showing more strong clustering (Figure 2). This genotype has been implicated in DHF outbreaks in American and Asian continents. DENV-2 segregated into South East Asia genotype, as Brazilian samples in the last ten years. In both analyzes the bootstraps values were satisfactory, with higher values showed by DENV- 2 sample.
4. DISCUSSION
The regions, in the dengue virus genome, encoding the envelope protein (E) and the nonstructural protein 1 (NS1) seem to be the most appropriate to characterize genotypes within serotypes [14,15]. In this way, for some flaviviruses, as yellow fever viruses, most molecular markers for pathogenicity have been found in the E gene. However for dengue, several studies of cloned or cell culture-passaged wild and attenuated viruses have been performed, but each report described different sites of nucleotide and amino acid changes possibly associated with virulence [16,17].
DENV-3 introduction was responsible, in 2002, for almost 80,000 cases notified in all Bahia State [8], and being the first time that DHF cases occurred in this region. Between 2006 and 2007, about 25,000 cases were notified. Dengue circulation in Bahia (Brazil) from 2006 to 2007 has consisted mainly of a mixture of DENV serotypes. Between two serotypes (DENV-2 and DENV-3), the former has been consistently detected in outbreaks/ epidemics since its introduction 2002. Before this study, dengue virus genotypes in Bahia state were not available or few samples have been analyzed. In this report, we analyzed 31 local DENV isolates collected in a span of two years to further understand the ecology and dynamics of this virus in the region. As known, the potential for causing severe disease has been described for all four serotypes of DENV, and the main factors considered to explain its pathogenicity are host genetic susceptibility [18], antibody dependent enhancement and differences in virulence among strains [19], in this way determinants of virulence have been located in three genomic regions [5] and have been tested in vitro [20], so the genotypes have been more or less related with potential to cause DHF. Different studies concluded that the spread of genotype III, of DENV-3, from the Indian subcontinent to Africa, and then consequently to Americas, was correlated with an increase in severe cases of dengue disease in this region [21,22].
The routine laboratory diagnosis of dengue virus infection is primarily achieved by the isolation of virus, detection of IgM/IgG antibodies by serodiagnosis and/or molecular detection by the demonstration of viral RNA by RT-PCR [23,24]. RT-PCR is one of the most important confirmatory tests to confirm dengue infection. But, its positivity is highly dependent of the time in the clinical diagnosis and conditions in the sample maintenance (better when is collected within first five days of onset of the symptoms). The lower rate of the virus isolation may be attributed to the absence of live virus in the sample. This must be due to misclassification of the cases by epidemiological staff and others factors as improper maintenance or storage system. All positive cell culture isolations were confirmed and identified by RT-PCR and nested PCR (31 available samples). Still, we have carried out the molecular epidemiology and genotyping study of the detected DENV-2 and DENV-3 viruses, during 2006 and 2007 circulation seasons. We studied the sequence of these dengue viruses, directly from patient serum sample, as recently advocated by several researchers [5,25]. Various genomic regions of dengue viruses have been selected
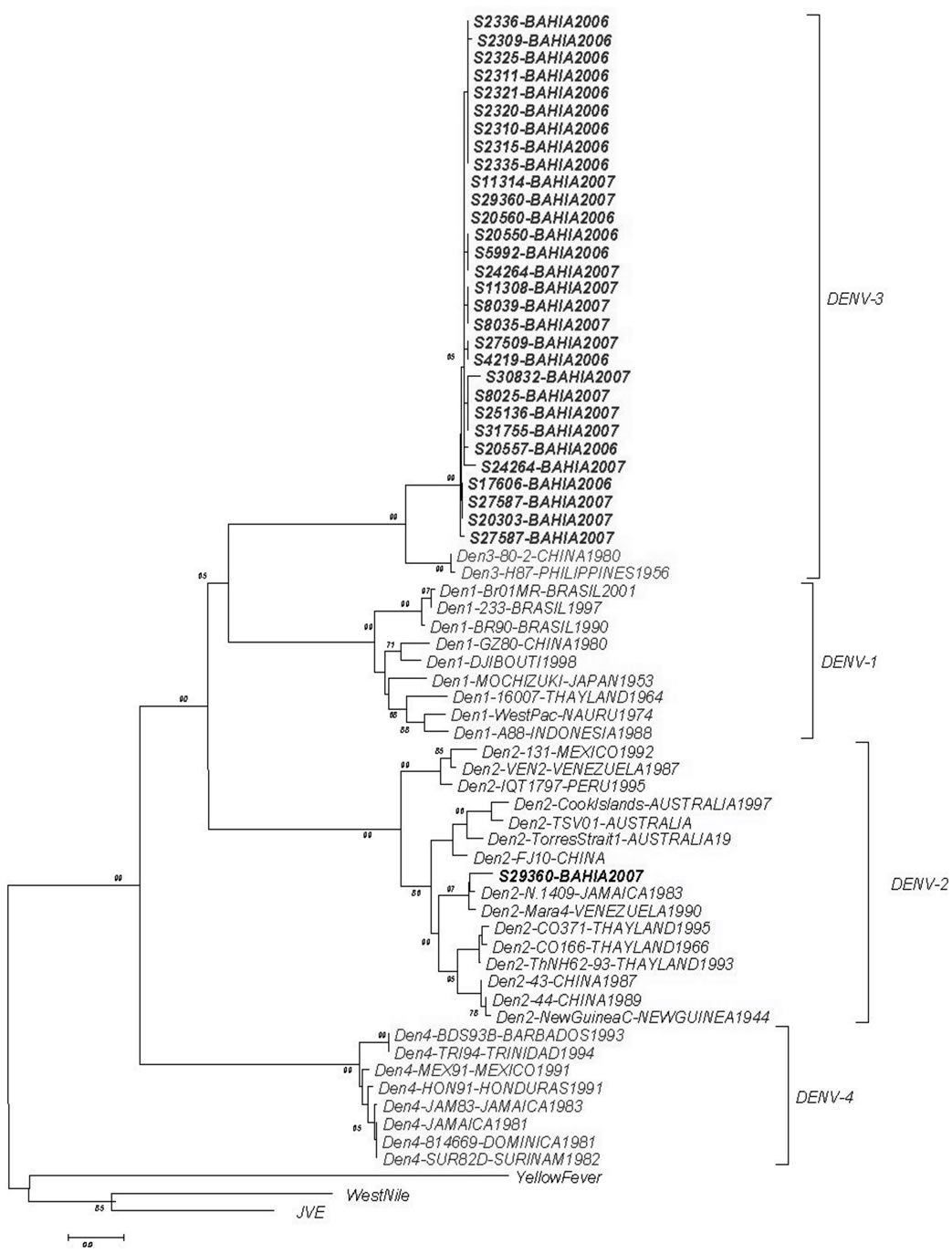
Figure 1. Phylogenetic tree constructed with the E/NS1 fragment which detected the samples in two different dengue serotypes.
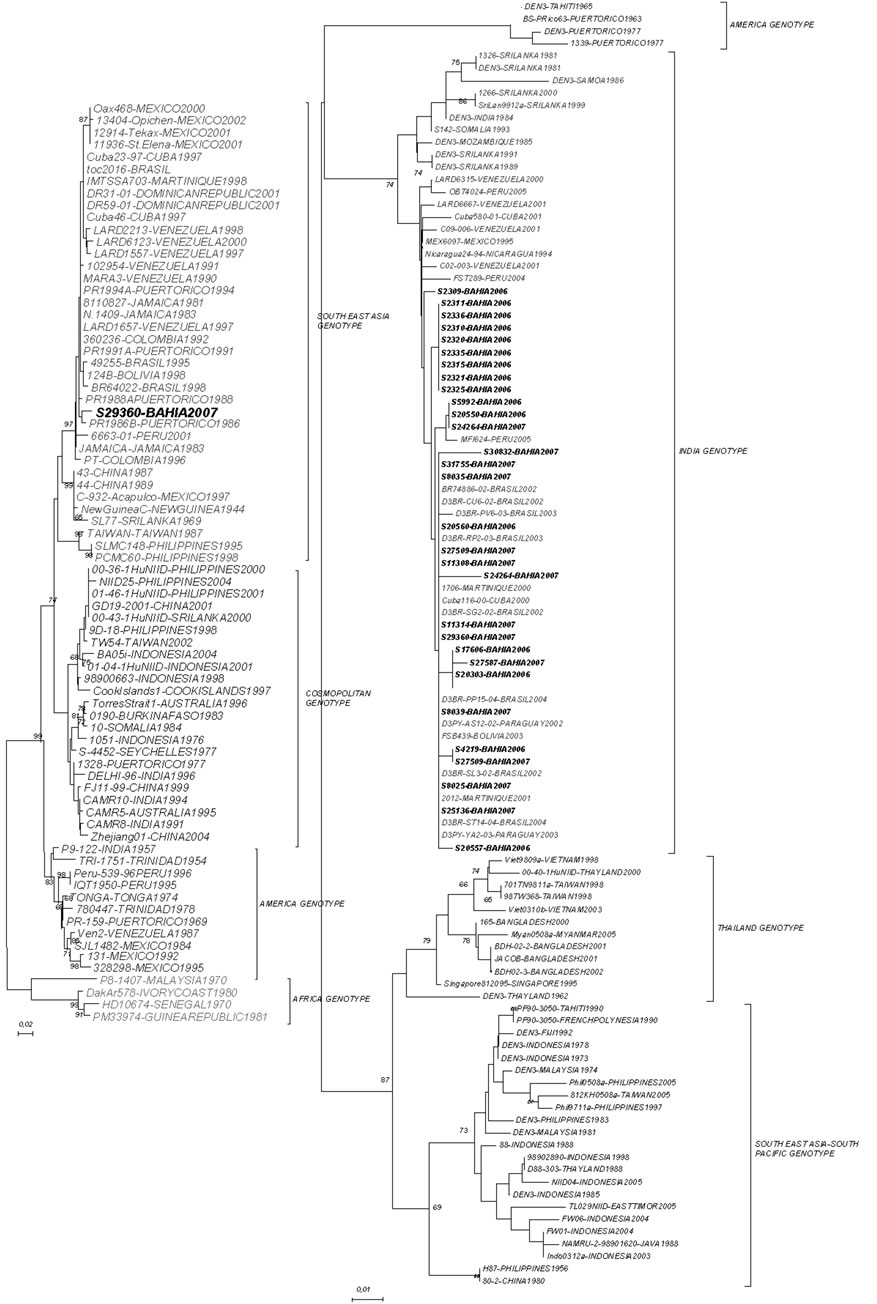
(a) DENV-2,(b) DENV-3
Figure 2. (a), (b)—Maximum likelihood tree depicting the phylogenetic relationships of dengue samples based on the E/NS1 junction (DENV-2 and DENV-3). Numbers on branches represent bootstrap support for each branch. Genotypes are also indicated.
for molecular phylogenetic analysis [26]. However, we have selected the E/NS1 gene junction as it also harbours epidemiologically important sequence information and sensitive for surveillance application [9].
The results obtained for DENV-3 in this study bring some interesting findings how can samples from two years, 2006 and 2007, display a high identity with isolated from Peru in 2002? And in the same time samples from 2007 showed a high similarity with samples differrent countries in all American Continent. Other important finding was that almost all grouping were with samples from the same area in Bahia state. In some way, the microregion where the virus circulate in any level determine how they become grouped. During 2006 and 2007, the main serotype circulating in Bahia state was DENV-3, in agreement with the displacement of serotypes 1 and 2 of DENV after its introduction in Brazil in 2002 [27]. A study developed in Rondônia and Acre found both genotype I and V [10], while we found just genotype III after two years of surveillance. This confirm the epidemic potential of this genotype by the displacement of serotypes 1 and 2 of DENV when a highly virulent genotype of DENV-3 was introduced in Brazil in 2002 [27]. Since its introduction this genotype have been related with severe disease in Bahia and Brazil [8,28].
In conclusion, the effective worldwide control of dengue virus into Bahia and consequently in all Brazil, requires the definition of the epidemical viral sources, and the precise identification of virus genotypes. A key objective of DF and DHF surveillance programmes is early detection of outbreaks to permit the implementation of control measures by the public health authorities. DHF outbreaks can be anticipated by monitoring the emergence of new genotypes in a region like Bahia State.
5. ACKNOWLEDGEMENTS
This project was funded by FAPESB PPSUS, CNPq and FIOCRUZ.
REFERENCES
- Mackenzie, J.S., Gubler, D.J. and Petersen, L.R. (2004) Emerging flaviviruses: The spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nature Medicine, 10, S98-109. doi:10.1038/nm1144
- Halstead, S.B., Nimmannitya, S. and Cohen, S.N. (1970) Observations related to pathogenesis of dengue hemorrhagic fever. IV. Relation of disease severity to antibody response and virus recovered. Yale Journal of Biology and Medicine, 42, 311-328.
- Barnes, W.J. and Rosen L. (1974) Fatal hemorrhagic disease and shock associated with primary dengue infection on a Pacific island. American Journal of Tropical Medicine and Hygiene, 23, 495-506.
- Gubler, D.J., Suharyono, W., Lubis, I., Eram, S. and Gunarso, S. (1981) Epidemic dengue 3 in central Java, associated with low viremia in man. American Journal of Tropical Medicine and Hygiene, 30, 1094-1099.
- Leitmeyer, K.C., Vaughn, D.W., Watts, D.M., Salas, R., de Chacon, I. V. and Rico-Hesse R. (1999) Dengue virus structural differences that correlate with pathogenesis. Journal of Virology, 73, 4738-4747.
- Dash, P.K., Parida, M.M., Saxena, P., Abhyankar, A., Singh, C.P., Tewari, K.N., Jana, A.M., Sekhar, K. and Rao, P.V. (2006) Reemergence of dengue virus type-3 (subtype-III) in India: Implications for increased incidence of DHF & DSS. Virology Journal, 3, 55. doi:10.1186/1743-422X-3-55
- CDC (1995) Dengue type 3 infection. Nicaragua and Panama, October-November 1994. Weekly Epidemiological Record, 70, 41-43.
- Melo, P.R., Reis, E.A., Ciuffo, I.A., Goes, M., Blanton, R.E. and Reis, M.G. (2007) The dynamics of dengue virus serotype 3 introduction and dispersion in the state of Bahia, Brazil. Memórias do Instituto Oswaldo Cruz, 102, 905-912. doi:10.1590/S0074-02762007000800003
- Domingo, C., Palacios, G., Niedrig, M., Cabrerizo, M., Jabado, O., Reyes, N., Lipkin, W.I. and Tenorio, A. (2004) A New Tool for the Diagnosis and Molecular Surveillance of Dengue Infections in Clinical Samples. Dengue Bulletins, 28, 87-95.
- Nogueira, M.B., Stella, V., Bordignon, J., Batista, W.C., Borba, L., Silva, L.H., Hoffmann, F.G., Probst, C.M. and Santos, C.N. (2008) Evidence for the co-circulation of dengue virus type 3 genotypes III and V in the Northern region of Brazil during the 2002-2004 epidemics. Memó- rias do Instituto Oswaldo Cruz, 103, 483-488. doi:10.1590/S0074-02762008000500013
- Gubler, D.J., Kuno, G., Sather, G.E., Velez, M. and Oliver, A. (1984) Mosquito cell cultures and specific monoclonal antibodies in surveillance for dengue viruses. American Journal of Tropical Medicine and Hygiene, 33, 158-165.
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F. and Higgins, D.G. (1997) The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 25, 4876-4882. doi:10.1093/nar/25.24.4876
- Kumar, S., Tamura, K., Jakobsen, I.B. and Nei, M. (2001) MEGA2: Molecular evolutionary genetics analysis software. Bioinformatics, 17, 1244-1245. doi:10.1093/bioinformatics/17.12.1244
- Rico-Hesse, R. (1990) Molecular evolution and distribution of dengue viruses type 1 and 2 in nature. Virology, 174, 479-493. doi:10.1016/0042-6822(90)90102-W
- Wang, E., Ni, H., Xu, R., Barrett, A.D., Watowich, S.J., Gubler, D.J. and Weaver, S.C. (2000) Evolutionary relationships of endemic/epidemic and sylvatic dengue viruses. Journal of Virology, 74, 3227-3234. doi:10.1128/JVI.74.7.3227-3234.2000
- Jennings, A.D., Whitby, J.E., Minor, P.D. and Barrett, A.D.T. (1993) Comparison of the nucleotide and deduced amino acid sequences of the wild-type French viscerotropic strain of yellow fever virus and the live vaccine strain, French neurotropic vaccine derived from it. Virology, 192, 692-695. doi:10.1006/viro.1993.1090
- Mangada, M.N. and Igarashi, A. (1997) Sequences of terminal non-coding regions from four dengue-2 viruses isolated from patients exhibiting different disease severities. Virus Genes, 14, 5-12. doi:10.1023/A:1007914520454
- Blanton, R.E., Silva, L.K., Morato, V.G., Parrado, A.R., Dias, J.P., Melo, P.R., Reis, E.A., Goddard, K.A., Nunes, M.R., Rodrigues, S.G., Vasconcelos, P.F., Castro, J.M., Reis, M.G., Barreto, M.L. and Teixeira, M.G. (2008) Genetic ancestry and income are associated with dengue hemorrhagic fever in a highly admixed population. European Journal of Human Genetics, 16, 762-765. doi:10.1038/ejhg.2008.4
- Holmes, E.C. and Burch, S.S. (2000) The causes and consequences of genetic variation in dengue virus. Trends in Microbiology, 8, 74-77. doi:10.1016/S0966-842X(99)01669-8
- Cologna, R. and Rico-Hesse, R. (2003) American genotype structures decrease dengue virus output from human monocytes and dendritic cells. Journal of Virology, 77, 3929-3938. doi:10.1128/JVI.77.7.3929-3938.2003
- Guzman, M.G., Vazquez, S., Martinez, E., Alvarez, M., Rodriguez, R., Kouri, G., de los Reyes, J. and Acevedo, F. (1996) Dengue in Nicaragua, 1994: Reintroduction of serotype 3 in the Americas. Boletín de la Oficina Sanitaria Panamericana, 121, 102-110.
- Messer, W.B., Gubler, D.J., Harris, E., Sivananthan, K. and de Silva, A.M. (2003) Emergence and global spread of a dengue serotype 3, subtype III virus. Emerging Infectious Diseases, 9, 800-809.
- Guzman, M.G. and Kouri, G. (2004) Dengue diagnosis, advances and challenges. International Journal of Infectious Diseases, 8, 69-80. doi:10.1016/j.ijid.2003.03.003
- Wu, S.J., Hanson, B., Paxton, H., Nisalak, A., Vaughn, D.W., Rossi, C., Henchal, E.A., Porter, K.R., Watts, D.M. and Hayes, C.G. (1997) Evaluation of a dipstick enzyme-linked immunosorbent assay for detection of antibodies to dengue virus. Clinical and Diagnostic Laboratory Immunology, 4, 452-457.
- Dash, P.K., Parida, M.M., Saxena, P., Kumar, M., Rai, A., Pasha, S.T. and Jana, A.M. (2004) Emergence and continued circulation of dengue-2 (genotype IV) virus strains in northern India. Journal of Medical Virology, 74, 314- 322. doi:10.1002/jmv.20166
- Uzcategui, N.Y., Comach, G., Camacho, D., Salcedo, M., Cabello de Quintana, M., Jimenez, M., Sierra, G., Cuello de Uzcategui, R., James, W.S., Turner, S., Holmes, E.C. and Gould, E.A. (2003) Molecular epidemiology of dengue virus type 3 in Venezuela. Journal of General Virology, 84, 1569-1575. doi:10.1099/vir.0.18807-0
- Nogueira, R.M., Schatzmayr, H.G., de Filippis, A.M., dos Santos, F.B., da Cunha, R.V., Coelho, J.O., de Souza, L.J., Guimaraes, F.R., de Araujo, E.S., De Simone, T.S., Baran, M., Teixeira Jr., G. and Miagostovich, M.P. (2005) Dengue virus type 3, Brazil, 2002. Emerging Infectious Diseases, 11, 1376-1381.
- Miagostovich, M.P., dos Santos, F.B., Fumian, T.M., Guimaraes, F.R., da Costa, E.V., Tavares, F.N., Coelho, J.O. and Nogueira, R.M. (2006) Complete genetic characterization of a Brazilian dengue virus type 3 strain isolated from a fatal outcome. Memórias do Instituto Oswaldo Cruz, 101, 307-313. doi:10.1590/S0074-02762006000300015
NOTES
*Corresponding author.

