Paper Menu >>
Journal Menu >>
 Vol.3, No.4, 187-199 (2011) Health doi:10.4236/health.2011.34035 Copyright © 2011 SciRes. Openly accessible at http://www.scirp.org/journal/HEALTH/ Transformations of phosphatidylinositol phosphates in the outer and inner nuclear membrane are linked to synthesis and restitution of cellular membranes Amalia Slomiany, Bronislaw L. Slomiany University of Medicine and Dentistry of New Jersey, New Jersey Dental School, Newark, New Jersey, USA; *Corresponding Author: slomiaam@umdnj.edu Received 10 February 2011; revised 18 March 2011; accept 1 April 2011. ABSTRACT The ultimate goal in phosphoinositides cellular metabolism is to decipher their global func- tional organization and coordinatio n of the com- partmentalized signaling processes. In this re- port we present evidence linking nuclear phos- phoinositides cycle w ith endoplasmic reticulum synthesis and function. The rapid transforma- tion of [3H]inositol-labeled phosphoinositides in the intact nuclei (IN) was captured in chase studies for 0-5 min, followed by examination of phosphatidylinositides in the inner nuclear me- mbrane (INM), the outer nuclear membrane (ONM) and endoplasmic reticulum (ER). We re- vealed that synthesis of phosphatidylinositol phosphates (PIPs) occurs in ONM and the de- phosphorylation takes place in the INM. The rapid transformation of the radiolabeled PIPs in ONM reverberated in their appearance and suc- cessive transformation in INM, and in the 5min chased nuclei was tracked to ONM as the re- emerging radiolabeled phosphatidy linositol (PI). These chase-uncovered changes in ONM and INM PIPs profiles allow us to conclude that the observed conversions in the nuclear membrane continuum are induced by the lateral movement of the membrane and its transit from the cyto- solic to nuclear and back to cytosolic environ- ment. The suggested membrane synthesis-in- duced movement provides the means to trans- port the membrane- and the membrane lipid ligand-associated cytosolic proteins to the in- tranuclear spaces and renewal of INM. Export of the nuclear components interacting with the modified INM, by exiting from nuclear to cyto- solic site, endows ER with a steady influx of the membrane that is conditioned to generate vesi- cles according to the nucleus delivered tem- plates. Keywords: Phosphatidylinositides; PIPs; Transformation ; R estitution; Outer Nuclear Membrane; Inner Nuclear Membrane; Endoplasmic Reticulum; Cellular Membranes 1. INTRODUCTION The array of proteins with affinity to phosphatidy- linositol phosphates (PIPs), including protein kinases, phosphatases, phospholipases, ion channel proteins, scaf- fold proteins, cytoskeletal proteins and the regulators of membrane trafficking [1-3], attest to th ese phospholipids pleiotropism in controlling cellular metabolism and to the fact that the distinct molecules targeted by specific PIPs’ must be compartmentalized in a function-depen- dent manner. Thus, the 18 phosph oinositide interconver- sion reactions identified thus far and mediated by as many as 47 genes encoding 19 phosphatidylinositide kinases and 28 phosphatidylinositide phosphatases [4], must be restricted by cellular space and the synthetic or catabolic action. The cell nucleus phosphoinositides seem to function in the organelle niche that is independent and isolated from the environment of the other cell’s mem- branes signaling events [5,6]. The different spectrum of phosphoinositides in the nuclear membranes than that identified in endoplasmic reticulum (ER) [7-9] or Golgi, provides credibility to their functional separation. But, the findings on uncon- ventional intranuclear localization of PIPs in the nuclear speckles, nuclear matrix, nucleoli and chromatin [2,5,6], are still being challenged, and the current evidence sup- ports the model that nuclear phosphoinositides remain in the nuclear envelope [10-15]. Even the model that in- corporates PIPs into nuclear membranes raises several compelling questions: how is PIPs intranuclear localiza- tion achieved, how their nuclear conversions are linked  A. Slomiany et al. / Health 3 (2011) 187-199 Copyright © 2011 SciRes. Openly accessible at http://www.scirp.org/journal/HEALTH/ 188 to the rest of cellular events, and how is their function expressed and metabolism regulated. The enigma is compounded further by the fact that ER which is a con- tinuum of outer nuclear membrane (ONM), that in turn and beyond nuclear pore, becomes the inner nuclear membrane (INM), is devoid of PIPs [16,17]. Thus, the sudden transition from the nuclear presence of PIPs to their absence in the ER is puzzling. Continuity of the nuclear and ER membrane implies that the processes in nuclear membrane are closely linked to the ER synthetic responses reflected in membrane biogenesis and transport. Yet, whilst the precise and explicit intercalation of pro- tein into biomembrane is only possible when specific mRNA tethered to the INM is translated concomitantly with the synthesis of membrane lipids, the phosphoino- sitides complexity of the nuclear membrane is not being passed to ER membrane or to the newly synthesized bio- membrane used to envelop transport vesicles [7-9,16,17] . The ER membrane and its transport v esicles con sist of phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylinositol (PI) and ceramides (Cer) which mature and acquire phosphatidylinositol phosphates (PIP1s) in Golgi [16,17]. These in congruities in the co m- position of the membrane continuum brought us to spe- culation that the membrane biogenesis in the region of ER transiting into ONM may be defining the outer and inner nuclear biomembrane. In the previous study that demonstrated the phosphatidylinositides aided cytosolic protein transport into the nucleus and the concurrent appearance of the transport engaged phosphatidylinosi- tide in INM [15], we proposed that the event must be the consequent effect of ONM synthesis-induced lateral movement of the membrane that aided the transit of the complex into the intranuclear space. Such a movement of the membrane would provide continuous transport of the membrane- and the membrane lipid-ligand associ- ated cytosolic proteins to the intranuclear spaces, restitu- tion of INM, and the export of the nuclear components interacting with INM [12-14]. Moreover, it would en- dow ER with steady influx of the membrane that would be conditioned to generate the transport vesicles accord- ing to the nucleus-deliv ered templates [18-27]. In order to observe the events that would proceed as a continuous, uninterrupted transition from ONM to INM, to ONM again, and then to ER, we have investigated changes in nuclear membranes phosphoinositides com- position in time. The studies of the phosphatidylinositi- des transformation (conversions) in nuclear membranes, prepared from intact nuclei subjected to incubation in cytosol supporting transport, gave the credibility to our contention that polyphosphoinositides assembled in ONM transit into the intranuclear space, and remain in INM. As the constituents of INM, they undergo dephosphory- lation, and then reappear in the ONM and ER as PI. 2. MATERIALS Radiolabeled precursors of phospholipids, glycolipids, glycoproteins and other radiochemicals were purchased from New England Nuclear (Boston, MA). Phospholipid standards were from Avanti (Birmingham, AL), Matreya (Pleasant Gap, PA) or prepared in our laboratory. Creatine phosphokinase, phenylmethylsulfonyl fluoride (PMSF), aprotinin, pepstatin, leupeptin, ATP, CTP, GTP, fatty acyl CoA, glycerol 3-phosphate, RNase, and RNase free sucrose were purchased from Sigma Chemicals (St. Louis, MO). Polyacrylamide gel electrophoresis reagents were from Bio-Rad (Rockville Centre, NY). All other chemicals and reagents were purchased from J.T. Baker Chemical Co. (Phillipsburg, PA), Fisher Scientific (Springfield, NJ), and VWR Scientific (Piscataway, NJ). The anti-rat albumin antibodies were purchased, and the antimucin antibodies were prepared in our laboratory [28]. 2.1. Solution Used in Preparation of Cells, Cell Organelles, Nuclei and Nuclear Membranes Buffered saline, with 10 mM potassium phosphate, pH 6.8 (a), buffered saline containing 0.5 mM MgCl2 and 0.5 mM MgS04, (b), buffered saline (100 ml) con- taining 66 mg collagenase, 80 mg hyaluron idase, and 2 g of albumin (c), MSB, pH 6.9 buffer consisting of 0.1 Pipes, pH 6.9, 2.0 M glycerol, 1 mM Mg acetate, 0.5 mM EGTA and mixture of protease inhibitors consisting of leupeptin, aprotinin and PMSF (d), MSB buffer con- taining 0.2% Tr iton X 100 (e), 50 mM TRI S-HCl, pH 7.4 containing 0.25 M sucrose, 10 mM MgCl2 1 mM DTT, 10 mg/ml leupeptin and 2 mM PMSF (f). 2.2. Isolation and Separation of ER, Golgi, Endosomes, Cellular and Outer and Inner Nuclear Membranes. One volume of the isolated cells was homogenized in 8 volumes of 10 mM potassium phosphate buffer, pH 6.8, 1.3 M sucrose and 1mM MgCl2 to rupture at least 80 % of cells [15]. The unbroken cells were removed by centrifugation at 50 xg for 3 min, and the homogenate centrifuged for 15 min at 1000 xg. The soluble cellular material was saved for isolation of cellular organelles and cytosol while the nuclear pellet was processed fur- ther and the outer and inner nuclear membranes (ONM, INM) were collected [15]. 2.3. Preparation of Intact Nuclei (IN) In the experiments employing IN, and the incubation with radiolabel-free (cold) CC that was followed by  A. Slomiany et al. / Health 3 (2011) 187-199 Copyright © 2011 SciRes. Openly accessible at http://www.scirp.org/journal/HEALTH/ 189189 separation of ONM and INM, the IN samples were sub- jected to additional sucrose gradient purification. The samples of IN were suspended in 40% buffered sucrose by adding 80% sucrose in 150mM NaCl, 25 mM TRIS, pH 7.5 buffer and ov erlaid with 10% - 30% sucro se gra- dient in the same buffer and centrifuged at 29 000 xg for 21 h. The nuclei were recovered from 40% bottom layer by diluting out sucrose with the buffer and centrifugation at 1000 xg for 10min. The nuclear pellet was then sus- pended at a protein concentration of 2 mg/ml in 1% cit- rate and proceeded with separation of ONM and INM. The described sucrose gradient afforded separation of the cell membrane fragments that were trapped with nu- clei, and thus allowed the conclusion that the radio- labeled PIPs were strictly derived from nuclear mem- branes. The isolated on sucrose gradient membranes were used to determine whether the treatment with ice- cold buffered Triton X100 (e) dissolved ONM or INM and/or contained PIPs. 2.4. Preparation of Cells for [3H]Inositol Labeling The cells were prepared from rat gastric mucosa and the liver as described previously [7-9,15]. The single cells that were separated from larger debris with aid of specific cell size nylon mesh were centrifuged at 50 xg for 2 min, washed twice with the enzyme-free medium (c), twice with the Minimum Essential Medium (MEM) and counted in he mocytometer. Thus prepared cells were then incubated in MEM for 3 hours with cold or [3H]inositol and used for preparation of nuclei [15] sub- cellular organelles, cell cytosol [7-9 ,16] and cellular me- mbranes [16,17]. In the experiments dedicated to the determination of lipid synthesis with cell cytosol derived from gastric epithelial cells, hepatocytes, or RNase treated cytosol, the preparations of nuclei, ER, Golgi or other organelles and cell membranes were additionally rinsed with PBS (a) and urea-PBS in order to remove the associated residual cytosolic proteins that otherwise would remain on their membranes. Thus prepared sub- cellular organelles and membranes were used for ex- periments on transport vesicles synthesis [16,17], and the preparation of ONM and the INM [15]. The synthe- sis of phosphatidylinositides, phospholipids and protein was determined using radiolabeled [3H]inositol, [3H]ara- chidonate, [3H]choline, [3H]serine, [3H]palmitate and [32P]ATP [7-9,15-17]. The chase studies on [3H]inositol labeled intact nuclei were performed in medium con- taining cold CC at concentration of 5 mg protein/ml of incubation mixture enrich ed with 50 mM ATP, 250 mM CTP, 50 mM GTP, 5 mM creatine phosphate, 8.0 IU/ml creatine kinase, and where indicated 25 mg/ml RNase, 10 mM UDP-Glc and 10 mM palmitoyl CoA [7-9,15- 17]. The aliquots of intact inositol labeled nuclei were with- drawn at 0, 0.5, 1.0, 3.0 and 5.0 min and diluted into 5 ml of ice cold buffer consisting of 10 mM potassium phosphate buffer pH 6.8, 1.8 M sucrose and 1 mM mag- nesium chloride to rinse off the incubation medium. The IN were recovered by centrifugation for 15 min at 1000 xg and processed further to isolate outer and inner nu- clear membranes. The same preparation of cold cytosol was used in the chase experiments with inositol labeled ER. 2.5. Preparation of Transport-Active Cell Cytosol (CC) The viable cells homogenized for 10 sec at 600 rpm in 3 volumes of buffer containing 0.25 M sucrose; 50 mM TRIS-HCl (pH 7.4); 25 mM magnesium acetate and 10 mM each of aprotinin, leupeptin, chemostatin; and 1 mM phenylmethylsulfonylfluoride, were centrifuged at 5 000 xg for 15 min. The supernatant, diluted with 2 volumes of homogenization buffer, was re-centrifuged at 10 000 xg for 20 min. The resulting supernatant was then sub- jected to centrifugation at 100 000 xg for 1h. Thus ob- tained soluble fraction was adjusted to 15 to 18 mg pro- tein/ml; admixed with an ATP generating system con- sisting of 40 mM ATP, 200 mM creatine phosphate, 2000 units/ml cr eatine phosphokinase, and referred to as transport active cell cytosol or active cytosol (CC). 2.6. Isolation and Separation of Outer and Inner Nuclear Membranes. The cytosol- and membrane fragments-free intact nu- clei (IN) suspended in buffer (f) at concentration of 2mg protein/ml, were adjusted to 1% (w/v) with sodium cit- rate and incubated on ice with gentle stirring for 30 min and then centrifuged at 500xg for 15 min. The obtained supernatant contained the ONM, whereas the pellet con- tained the INM) [12, 15]. The pellet (INM) was sus- pended in buffer (f) at concentration of 5mg/ml and di- gested with DNase 1 (250 mg/ml for 14h at 4˚C). The digested material was separated on sucrose gradient con- sisting of 0.25/1.6/2.4 M sucrose by centrifugation at 10 000 xg for 2 h. The INM were recovered at 1.6 M su- crose boundary. The ONM was collected from citrate supernatant by centrifugation at 100 000 xg for 20 min. The membrane pellet was suspended in buffer (f) and subjected to the same treatment as INM. On the average, the preparation of INM was 30% larger than ONM. 2.7. Preparation of Cellular Membranes The cell membranes and subcellular organelles (ER, Golgi) were recovered from the radiolabeled cells as described earlier [7-9,15-17]. The organelles sediment  A. Slomiany et al. / Health 3 (2011) 187-199 Copyright © 2011 SciRes. Openly accessible at http://www.scirp.org/journal/HEALTH/ 190 remaining after separation of cell cytosol, was suspended in buffer containing 0.2 M PIPES (pH 6.9), 2 M glycerol, 1 mM EGTA and 1 mM magnesium acetate and applied on the top of discontinuous gradient of 2.0/1.5/1.3/1.0M sucrose and centrifuged at 100 000 xg for 16 h. The cell membranes were recovered from 1.0 M sucrose, Smooth Endoplasmic Reticulum (SER) from 1.3 M sucrose, RER from 1.5 M sucrose and Golgi from the top of the 2.0 M sucrose. Each sucrose-separated fraction was subjected to further purification. The cell membranes were washed with original Pipes buffer and centrifuged at 3 000 rpm for 2 min. To separate apical epithelial membranes, the buffer was adjusted with 0.2% Triton X-100 and the mix- ture incubated at 4˚C for 5 min [16,17]. This treatment resulted in breaking up the phospholipids-rich membranes into smaller segments and that allowed us to separate membranes containing cholesterol, glycosphingolipids and glycoproteins. The latter membranes were recovered by low speed centrifugation at 3 000 rpm for 2 min. 2.8. Generation and Purification of Transport Vesicles ER- and Golgi-derived transport vesicles were gener- ated in the presence of radiolabeled precursors according to procedure described previously [7-9,15-17]. The ER or Golgi membranes incubated with cytosol, ATP-gen- erating system, UTP, CTP GTP, fatty acyl CoA and wa- ter soluble cold or radiolabeled lipids precursors were incubated for 30 min at 37˚C, centrifuged over 0.3M sucrose and treated with stripping buffer at 2˚C for 15 min followed by centrifugation at 10 000 xg for 10min to separate transport vesicles from ER or Golgi mem- branes. The separated from maternal membranes trans- port vesicles were recovered from the supernatant re- sulting from centrifugation of the supernatant mixture at 150 000 xg for 60 min. The crude fraction of the trans- port vesicles was suspended in 55% sucrose, overlaid with 55% - 30% gradient and centrifuged at 150 000 xg for 16 h. The purified transport vesicles were recovered from the gradients as reported earlier [1-9,16,17]. 2.9. Fusion of Transport Vesicles with Membranes One volume of Golgi transport vesicles (1.3-1.5 pro- tein/ml) was suspended in one volume of active cytosol (15 mg protein/ml) and added to one volume of cell membranes (8 mg protein/ml of whole cell membranes or 2 mg protein/ml of apical epithelial membranes). The reaction was allowed to proceed from 0-30 min at 4˚C (control) and at 37˚C in the presence of ATP regenera ing system consisting of 40mM ATP, 200 mM creatine phosphate, 2,000 units /ml of creatine phosphokinase, or in the ATP depleting system containing 5mM glucose and 500 units/ml hexokinase. After incubation, the mem- branes were recovered by centrifugation through three volumes of 0.5 M sucrose at 3000 rpm for 5 min. The free vesicles were recovered from the supernatant and used in fusion experiments with endosomes. The cell membranes sedimented through sucrose were washed with 25 mM Hepes-KOH buffer and treated for 5 min with 0.2% Triton X-100 at 4˚C. The soluble fraction of the membrane was recovered in supernatant, whereas apical the glycosphingolipids- and glycoprotein- con- taining membranes remained in the sediment [16,17,29]. In the experiments estimating en bloc fusion of transport vesicles with the membrane, the associated but not fused vesicles were released from the membrane by subjecting the membrane fraction to treatment with 2 M urea for 30 min at 4˚C and then th e recovered membranes w ere cen- trifuged through 0.5 M sucrose, washed and subjected to lipid analysis. 2.10. Lipid Analysis Preparations of IN, ONM, INM and ER from [3H]ino- sitol labeled cells were subjected to PIPs lipid analysis and chase-induced PIPs transformation analysis. The lipid extracts and high performance thin layer chroma- tography were performed exactly as described in our previous studies [7-9,15-17,29]. The column chroma- tography of PIPs was performed on AG1x8 formate form columns. The lipid extracts were first subjected to deacylation by incubation for 30 min at 53˚C with 0.75 ml of methylamine reagent containing monomethyl- amine/ methanol/water/butanol (5/4/3/1) and after re- moval of methylamine by evaporation to dryness the samples were suspended in water and fatty acids ex- tracted with mixture of n-butanol/light petroleum ether/ ethyl formate (20/4/1). The lower phase was re-extracted with the same solvent and the deacylated lipids were applied to column. The columns were eluted with (a) water, (b) 5 mM disodium tetraborate/60 mM ammo- nium formate,(c) 0.1 M formic acid/0.2 M ammonium formate, (d) 0.1M formic acid /0.4 M ammonium for- mate, (d) 0.1 M formic acid /1.0 M ammonium formate. Each fraction was collected in 12 individual aliquots and from each 0.2 ml was used to determine total radioactiv- ity eluted with individual solvents, whereas the Bertho ld LC radioactivity analyzer was used to determine the en- tire elution profile. 3. RESULTS Our previous study de monstrated that intracellular ve- sicular transport is dependent on the finely tuned synthe- sis of cell specific proteins and lipids that assemble pre-  A. Slomiany et al. / Health 3 (2011) 187-199 Copyright © 2011 SciRes. Openly accessible at http://www.scirp.org/journal/HEALTH/ 191191 cise copies of the biomembrane, which is needed for repair and restitution of the cell membranes and their function [17]. Therefore, the cytosolic protein flux to the nucleus and the signal-prompted nuclear export, are in- timately linked with a specific membrane biogenesis [1-5,15]. In both instances, the transp ort to and from the nucleus are linked to nuclear membrane lipids, particu- larly to PIPs. But, thus far, the site of nuclear PIPs gene- sis and the specific protein affinity to th e variety of PIPs is not determined. As we observed previously, the PIPs which are in- volved in cytosolic protein transport to the nuclear inner space remain in the INM, and are either transformed through dephosphorylation or degraded with aid of phosphatidy l i nosi t ol-specific phosph oli pase C (PIPLC). Hence, in this report, our studies on nuclear mem- brane concentrate on the PIPs transformations in ONM and INM observed during 0-5 min chase performed in the radiolabel-free transport supporting cell cytosol (CC). The data presented in Table 1 focus on the issue whethe r PIPLCs induce changes in PIPs and reveal the amount of labeled inositol released to cold cytosol. The results de- monstrate whether during the chase incubation of la- beled IN the phosphatidylinosito l-specific phospholipase C (PIPLC) releases inositol phosphates (IPs) from ONM. If PIPLC contributes to PIPs metabolism in ONM, the recovered cytosol should contain increasingly larger amount of the labeled inositol. As evident from the pre- sented data, except for the residual counts displaced dur- ing samples processing, the amount of soluble radio- labeled inositol in the cytosol has not increased with time of chase. Thus, the results provide convincing evi- dence that nuclear PIPs of ONM are not metabolized with the aid of cytosolic PIPLC. In Table 2 we present the amount of inositol-labeled lipids extracted from the intact nuclei (IN) follo wing 0-5 min incubation in CC. If nuclear PIPLC releases inositol phosphates (IPs) into nuclear contents, then the total amount of inositol labeled lipids extracted from the in- cubated nuclei would be diminished with time. As dem- onstrated, the amount of labeled PIPs extracted from incubated nuclei has not declined. Table 1. Release of [3H] inositol from IN incubated in an inac- tive (Control) and in transport active CC for 0-5min. Time (min) Control (cpm) % cpm released Sample+ CC (cpm) % cpm released 0 1037 24.6 950 13.7 0.5 1131 19.0 1145 12.0 1.0 1178 23.3 1024 11.5 3.0 1303 24.0 1080 12.0 5.0 806 19.1 555 7.0 Table 2. PIPs extracted from IN subjected to incubation in an inactive (Control) and in transport active CC for 0-5min. Time (min) Control (cpm) % cpm in lipids Sample+ CC (cpm) % cpm in lipids 0 3167 75.3 5995 86.3 0.5 4808 80.9 9545 87.9 1.0 3870 76.7 8920 88.4 3.0 4148 76.1 8965 87.9 5.0 3403 80.9 79.35 92.9 On the contrary, in the aqueous phase, we observed some decrease in the amount of radiolabeled inositol and an increase in an amount of radiolabel extracted in cor- responding samples of nuclear lipids. At this stag e of the investigation we cannot provide other explanation than to suggest that the samples incubated with the transport active cell cytosol retained larger fractions of water phase, and that this inclusion appeared as an increase in nuclear lipids. The spectrum of PIPs present in ONM isolated from [3H]inositol labeled IN is shown in Figure 1 and that in INM in Figure 2. After PIPs deacylation and column chromatography, the glycerylphosphoinositol phosphates (GPIPs) separated into glycerylphosphoinositol (GPI), glycerylphosphoinositol monophosphate (GPIP1), glyc- erylphosphoinositol diphosphate (GPIP2), and glyceryl- phosphoinositol triphosphate (GPIP3). The profiles of the individual GPIPs suggest that each fraction is repre- sented by more than one isomer , and the cis or trans po- sition of the phosphate esters may cause separation of GPIP2 and GPIP3 into two or three subfractions. How- ever, the methods utilized in our study cannot provide precise assignment of the phosphate substitution on the inositol moiety, and therefore in chase study each frac- tion is measured based on the number of phosphate moieties identified (Figures 1 and 2). The results of the nuclear [3H]inositol-labeled PIPs transformations are shown in Figures 3 and 4. The data present qualitative and quantitative changes in PIPs pro- files in ONM and INM isolated from IN subjected to 0-5 min chase in label-free active cytosol. The initial status (0 time) of PIPs labeling in ONM reflects low level (5.7%) of phosphatidylinositol (PI) and the incremental contents of phosphatidylinositol monophosphates (PIP1s), phosphatidylinositol diphosphates (PIP2s) and phospha- tidylinositol triphosphates (PIP3s). The PIP3s represent largest fraction (44.6%) of the labeled PIPs. After 30 seconds, the PIP2s increase to 53.7%, whereas PIP3s decline to 25.2%. While throughout the chase the PIP1s level remains almost constant, the PIP2s decline to the level observed before chase, while PIP3s 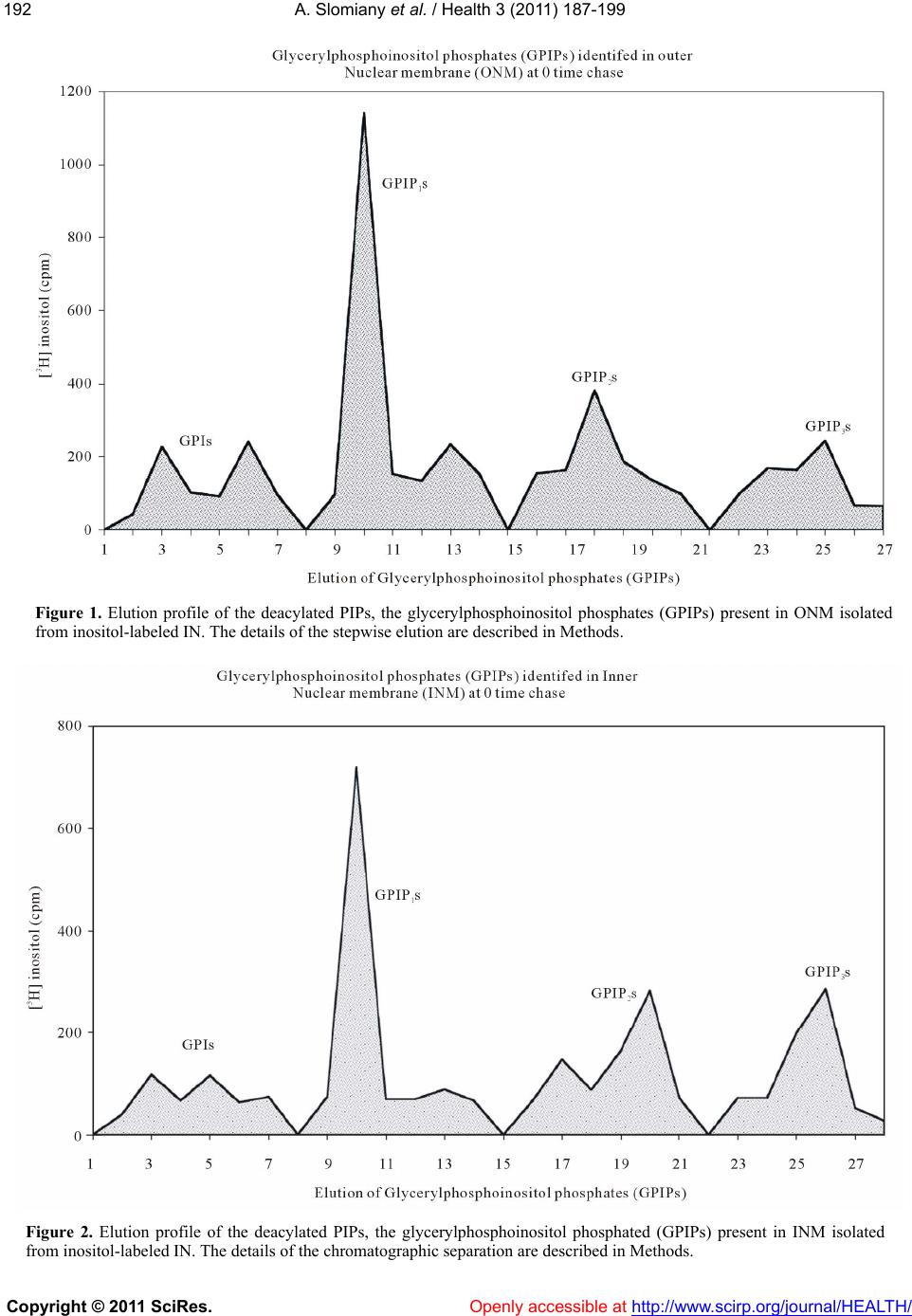 A. Slomiany et al. / Health 3 (2011) 187-199 Copyright © 2011 SciRes. Openly accessible at http://www.scirp.org/journal/HEALTH/ 192 Figure 1. Elution profile of the deacylated PIPs, the glycerylphosphoinositol phosphates (GPIPs) present in ONM isolated from inositol-labeled IN. The details of the stepwise elution are described in Methods. Figure 2. Elution profile of the deacylated PIPs, the glycerylphosphoinositol phosphated (GPIPs) present in INM isolated from inositol-labeled IN. The details of the chromatographic separation are described in Methods.  A. Slomiany et al. / Health 3 (2011) 187-199 Copyright © 2011 SciRes. Openly accessible at http://www.scirp.org/journal/HEALTH/ 193193 Figure 3. Chase studies of the [3H]inositol labeled PIPs of the ONM. The radiolabeled intact nuclei were subjected to incubation in transport active cold CC for 0-5 min. At indicated time the samples of intact nuclei were withdrawn and subjected to isolation of inner and outer nuclear membrane followed by lipid extraction and deacylation. The PIPs con- verted to GPIPs were separated by column chromatography described in Methods. To eliminate quantitative errors, the external standard consisting of 20% each of radiolabeled inositol, PI, PIP1, PIP2 and PIP3 representing 5,000 cpm was subjected to the same procedure and its recovery was used to compensate for errors in procedural recovery and sampling. Figure 4. Chase studies of [3H]inositol labeled PIPs of the INM. The procedural details and quantitation of the PIPs are identical with those described in Figure 3. The INM are derived from the same intact nuclei used for isolation of ONM.  A. Slomiany et al. / Health 3 (2011) 187-199 Copyright © 2011 SciRes. Openly accessible at http://www.scirp.org/journal/HEALTH/ 194 within 1 and 3 min chase continue to increase to 38.3% at 1 min, 49.0% at 3 min, and then suddenly at 5 min drop to the lowest level of PIPs (2.7%). That is accom- panied by unexpected increase in PI, which at 3 min represented 1.9% of PIPs and in 5 min chase rises to 53.5%. The results reflecting PI level in 0 to 3 min im- pose that radiolabeled PI is used for synthesis of PIPs and its level is not replenished through dephosphoryla- tion of PIPs and particularly PIP3. The simultaneous and slight decrease in th e level of PI and PIP1s an d the initial increase in PIP2s suggest that PIP1s are further phos- phorylated and their level is not restored through dephosphorylation. In 1 and 3 min chase, we observed even lower but constant level of PIP1s, while phos- phorylative activity concentrated on PIP2s and PIP3s. The PIP2s decrease was in concert with the continued increase in PIP3s. In 5 min chase we observe that the levels of PIP1s and PI P2s seem to stabilize after reaching or diminishing to their initial value found at the zero time, but PI level rises dramatically from 1.9% to 53.5% and PIP3s drops precipitously from 49.0% to 2.7 %. As evident from the values obtained in the chase between 0-3 min PI increase in ONM, in 5 min chase could not be linked to dephosphorylation of PIP3s in ONM, since the process would have to be detected in incremental quantities of PIP2s, PIP1s and then PI. The obtained re- sults suggest that the su dden drop in ONM PIP 3s and th e appearance of PI are reflecting the process that is linked to PIPs transformations in INM. The INM membranes (Figure 4) purified from the same nuclei as ONM contained 6.1%, PI, 37.7% PIP1s, 30.9% PIP2s and 25.3% PIP3s. In the 0.5 to 3 min chase, minimal changes in PI level were detected, although some fluctuations were evident. However, at 5 min the level of PI rose to 15.7%. The PIP1s which represented the largest fraction of PIPs in INM in first 30 seconds rose from 37.7 to 52.1% and that was accompanied by 17.7% decrease in PIP2s, thus suggesting fast PIP2s dephosphorylation to PIP1s, while PIP3s tempered in- crease continued. In the 1 min chase, we observed PIP3s build up to 53.7%. After reaching its highest concentra- tion in 1 min chase, in longer chase (3 min) PIP3s drop precipitously to 4.6% while PIP2s and PIP1s increase to 21,1 % and 57.7%, respectively. Finally, in 5min chase the PIP3s level drop further to 2.7%, PIP2s decrease to 24.1% and PIP1s remain at 57.6%. The sudden decrease in PIP3s, in 5 min chase is accompanied by PI generation, which is not as dramatic as disappearance of PIP3s and constitutes 15.7% of total PIPs. In our interpretatio n the changes in PIPs of INM reflect dephosphorylation reaction as the main process that causes transformation of PIPs with final product of PI. Since the results of our study depict also chase out of phosphorylated radio- labeled PIPs it is reasonable to suggest that the level of PIP3 is continuously being rebuilt by influx of phos- phorylated PIPs from ONM. Again, as in ONM it is dif- ficult to correlate sudden increase of PIP3s in 1min chase while PIP1s decrease, PIP2s level undergoes tem- pered rebuilding and PI is unchanged, except to suggest that PIP3s are introduced from ONM. Such an infusion of PIP3s is then reflected in the increments of PIP2s, PIP1s and finally at 5 min in the accumulation of PI and depletion of PIP3s. Hence, in continuation of the ex- periments that implied that in nuclear membranes PIPs flux was terminated by their complete dephosphorylation and reemergence in form of PI, the studies on PIPs pro- files in ER during 5 min chase were performed (Figure 5). The data presented in Figure 5 revealed that ER membranes contain PI and some traces of PIP1s. Neither PIP2s nor PIP3s were present at the beginning of the chase or throughout the chase. During the chase some decrement of radiolabeled PI was observed, which, in our opinion, could represent the release of ER transport vesicles or might reflect the presence of some Golgi membranes that are highly enriched in PI and PIP1s. If the above suggested model of the nuclear mem- brane movement is operational, one should expect that with chase the amount of inositol radiolabeled lipids would diminish in INM and increase in ONM. Indeed, we have found that the ONM fraction derived from 0.5 min chase contained 5.3 % more radiolabeled PIPs then its matching INM fraction, 3 min chase resulted in 14.3% increase and 5 min chase produced 23.3 % in- crease of radiolabeled PIPs in ONM fraction. Based on the obtained evidence we propose that nuclear mem- branes are subjected to continuous movement, which is afforded by synthesis of PIPs in ONM and their dephosphorylation in INM. The membrane depleted of PIPs, but retaining their PI emerges onto cytosolic site and thus initiates ER. In our interpretation, the described changes in the level of PIPs in ONM during the chase reflect two events, the phosphorylation of PI, and the continuous movement of the membrane containing PIP2s and PIP3s into the nucleus. After 5 min the radiolabeled PIP3s were no longer present in the ONM, but instead the dephos- phorylation products from INM PIPs reentered ONM continuum in the form of radiolabeled PI. Thus, the metabolic processes in nuclear membranes are linked to phosphorylation of PIs in ONM, dephosphorylation events in the INM, and transition from PIPs of nuclear compartment to PI of ER. Collectively, our data suggest that nuclear membrane represents uninterrupted continuum subjected to synthe- sis-induced mov ement, which capacitates the sys tem with  A. Slomiany et al. / Health 3 (2011) 187-199 Copyright © 2011 SciRes. Openly accessible at http://www.scirp.org/journal/HEALTH/ 195195 Figure 5. PIPs) isolated from ER prepared from the same preparations that were used for isolation of IN. The chase studies were performed in the same preparation of the cold transport active CC as used for ONM and INM. The prepa- ration of PIPs, the separation of the deacylated GPIPs and their quantitation was identical with that described in Figure 1 and 3. the ability to transport cytosolic products to nucleus, export of nuclear components to cytosol, and growth of ER memb rane. 4. DISCUSSION Recent literature data indicate that nuclear PIPs par- ticipate in chromatin remodeling, mRNA splicing, mRNA 3’end processing, and the export from nucleus [9-11,22-25]. There is also emerging evidence that some actively transcribed genes localize to nuclear periphery enriched with PIPs in the inner nuclear membrane and the nuclear pore complexes, thereby positioning the products for exit [12,14]. At the same time, the PIPs that aid pre-mRNA processing, including splicing factors, small nuclear ribonucleoproteins (nRNPs) and RNA polymerase II, are not present in the ER that constitutes nuclear membrane continuum [7-9,16,17]. Logically, such an immediate extension of the nuclear organelle should conserve compositional similarity. Since it does not, then what is the deciding factor that demarcates the nuclear and ER bound aries, while satisfying clear link in the continuous processing of the signal in the ER. Our uncovering of PIPs transformation in the nuclear mem- branes and the nuclear PIPs-associated processes which cease at ER boundary, while the membrane retains PI core, is crucial for compartment’s functional separation with preserved membrane continuum. It demonstrates the assignment of nuclear and ER lipids function, while safeguarding the paramount meaning of the continuity and the fidelity in signaling dialog between the cellular and nuclear membranes resulting in signal-intended re- sponses in ER. While the pre-mRNA processing machinery depends on the presence of PIPs, it is obvious that such function must be terminated with mRNA exit from the nucleus. It has been suggested that this is achieved through the nu- clear PIPLC activity [30]. If indeed this is the case, then during the chase, the IN would release watersoluble ra- diolabeled IPs and the level of radiolabeled PIPs in IN would gradually decline. As we demonstrate in Table 1 and 2, PIPLC activity is not apparent in PIPs transfor- mations of either ONM or INM membrane. With chase time lapse, the labeled cytosol-soluble inositides have not increased, whereas the amount of radiolabeled lipids extracted from the IN increased slightly. In our opinion, the increase in nuclear lipids, could not be factual, but most likely reflects some discrepancies in the separation of the org anic (lipid) an d water-soluble phase. As found,  A. Slomiany et al. / Health 3 (2011) 187-199 Copyright © 2011 SciRes. Openly accessible at http://www.scirp.org/journal/HEALTH/ 196 the turn to unexpected opposite values is an unlikely outcome in the chase experiments, and therefore the re- sults provide ind isputable ev idence th at PIPs metabolism in the nuclei is not attributed to PIPLC activity in the cytosol or nucleus [30,31]. In analyzing the chase data, we considered two se- quences of possible ev ents in PIPs transformations. One, the static scenario where PIPs of ONM and INM are independent of each other and second, the dynamic sys- tem where nucleu s is surrounded by one membrane con- tinuum in which PIPs transformation in ONM and INM are inherently linked. In the static scenario, the contin- ued synthesis of PIPs in two independent membranes would be reflected in the loss of radiolabeled PI in ONM and gain of the labeled PIPs. In contrast, in INM mem- brane, depho sphorylation of PIPs would yield incr easing quantities of PI. Indeed, in our chase study utilizing [3H]inositol labeled nuclei subjected to 0.5-5 min incu- bation in transport active cytosol, we observed the proc- esses that were consistent with PI phosphorylation in th e ONM and dephosphorylation in the INM. In ONM, the initial phase of chase demonstrated PI phosphorylation that was reflected in an increase of PIPs. However, with time of chase, the level of PIPs in ONM declined, while in the matching nuclear samples of INM their level rose incrementally. The decline of PIPs in the ONM could not be attributed to some PIPs-specific cytosolic phos- phatases that could act on ONM at the same time. In a such scenario, all chase results would generate the same as time 0 radiolabeled PIPs profiles. The results of chase studies on the ONM presented in Figure 3, do not reflect neither static sequence of the events, nor phosphatase- specific changes in time-dependent PIPs profiles. Rather, the data express the synthesis of PIPs in the ONM facing cytosolic environment and the active transport of the PIPs to INM facing nuclear inner space. The sudden increase in the amount of PIP3s in the INM and PI in the ONM suggests that the nuclear membrane is a dynamic continuum, which was experimentally separated into independent fractions of the ONM and INM. The chase- generated results capture the membrane motion by reg- istering enrichment in PIP3s in INM and concomitant ejection of PI-containing IMN onto cytosolic site. If nu- clear membranes were fixed in position, the PIP3s build up would remain in ONM and PI in INM. Together, the chase-generated results support the dynamic mode of PIPs’ transitions, that under in situ condition s, prior arti- ficial separation of the nuclear membrane con tinuum, t he PIPs synthesis-induced ONM movement introduces PIPs onto intranuclear site while expels incremental amount PI-containing IMN onto the cytosolic site. Such lateral membrane movement explains reemergence of the INM with dephosphorylated PI onto cytosolic site and transi- tion of the INM into new segment of ER. In previous studies we have found that PIPs of nuclear membrane are involved in transport of cytosolic protein to nucleus and remained in INM while their structural profile suggested that while facing intranuclear envi- ronment they were subjected to dephosphorylation [15]. Thus, the protein transport-associated data and the chase results presented here, imply that transferred PIPs are subjected to dephosphorylation while remaining in the membrane. Moreover, the interpretation that PIPs are confined to nuclear membranes, is also supported by physicochemical properties of phospholipids [5,18-21]. Indeed, it is unquestionable that the amphipathic struc- ture of PIPs render these phosp holipids en ergetically an d thermodynamically unfavorable to move freely within nuclear environment [6,32]. We believe that the findings placing PIPs in the subnuclear sites originate from the literal acceptance of the term ‘membrane stripping’. The detergent aided membrane ‘stripping’ disintegrates phos- pholipid-enriched membrane continuum, but does not displace the lipids interacting with their ligands. There- fore, the findings on the detergent-treated nuclei retain- ing PIPs linked to subnuclear structures reinforce the fact that phosphatidylinositides or other phospholipids that interact with nuclear compon ents remain in the com- plex [33,34]. Also, identification of the so-called phos- phatidylinositides carrier proteins supports the fact that specific complexes of lipid ligands do not easily disso ci- ate, but normally as anchored in membrane they trans- port cytosolic protein to the nucleus and participate in the chromatin remodeling. Apparently, the intranuclear interaction with PIPs remain until the lipids are trans- formed by specific phosphatases and then enter different set of intranuclear reactions [35]. In reality, the data that capture the presence of pools of PIPs associated with an unique nuclear compartments, illustrate which intranu- clear compartments are induced by the signal to interact with a specific and membrane-tethered PIPs ligands. Hence, our data argue against the view that nuclear PIPs function in membrane-free compartments, and suggest that the nuclear transformations positioned in the prox- imity of PIPs-containing inner leaflet of the membrane, just as in cell cytosol, allow the specific interaction be- tween lipid ligand and the nuclear complex. Our conten- tion that PIPs are confined to nuclear membranes and that outer leaflet phosphoinositides of the ONM trans- port cytosolic protein to nucleus [15], has been elabo- rated further. With chase experiments performed on IN and ER in the presence of active cytosol we have de- tected that the nuclear membrane movement associated with PIPs assembly, dephosphorylation is inherently connected with initiation of the membrane that becomes part of ER. At this time, we cannot pinpoint the specific  A. Slomiany et al. / Health 3 (2011) 187-199 Copyright © 2011 SciRes. Openly accessible at http://www.scirp.org/journal/HEALTH/ 197197 interactions that take place while PIPs are transformed, except to imply that such an interactions are terminated once the PI is generated, and the reentrance of the INM to the cytosolic environment is evident as the radio- labeled PI is becoming part of ONM fraction. Therefore, it seems more than plausible, that the membrane reen- tering cytosolic environment is the one transiting into ER compartment, and which is conditioned to synthesize lipids, translate specific mRNA, and synthesize specific transport vesicles and their cargo [36]. The support for this conclusion emanates from several facts; firstly the ER from the same cells that were used to isolate inosi- tol-labeled nuclei contained PI only. Secondly, as we have shown previously, the ER transport vesicles con- tained PI in their membrane, which upon delivery to Golgi was subjected to specific phosphorylatio n to PIP1s known to direct cargo to apical, basolateral or en- dosomal membranes [16,17]. Thirdly, the vesicular cargo and its destination was dictated by the mRNA of the cytosol used in the ER transport vesicles synthesis. Finally, the ER transport vesicles were not assembled in the RNA depleted cytosol [16,17]. As demonstrated ear- lier, the ER membrane contains PI only [7-9,16,17]. If the nuclear and ER phosphoinositides were degraded through PIPLC activity, the water soluble polyphospho- inositides would be released from the membrane and be reflected in the loss of inositol labeled phosphatidy- linositides in the nuclear and ER membranes. But, on the other hand, if the processing engages dynamic act of membrane PIPs transformation through phosphoryla- tion/dephosphorylation reactions, the nuclear membrane would retain their modified lipids. Such a turn of events satisfies termination of the pre-mRNA processing at INM, PI lipid retention in the intact nuclei, and the no- ticeable increase of labeled PI following 5 min chase in ONM. In situ, this is an ideal transit from nucleus mem- brane to ER, where the compartment-specific transfor- mation of the membrane completed nuclear signal which in turn is translated into coordinated response in th e ER. Compiled results of our study on the biomembrane bio- genesis in cellular network and the nuclear membranes, lead us to advance the hypothesis on the specificity of the cellular membranes production and repair. All these processes are intimately linked to PIs transformation and movement across the nuclear space, the reappearance of their core PI as a part of the ER conditioned to perform the task of synthesis and transport of the vesicles, thus restoring cellular membranes’ structure and function. The presence of PI in the exiting inner nuclear space INM, recovered in the membranes preparation as out- growing ONM, provides superb transition from the phosphoinositides metabolic transformations in the nu- cleus to ER. As we demonstrated earlier, and again in this study (Figure 5), ER is engaged in the synthesis of specific cellular membranes whose delivery to multiplic- ity of cellular sites is first dictated in ER by the protein intercalated into the membrane and then the process is completed in Golgi, an organelle responsible for mem- brane maturation, protein modification and PI-specific phosphorylation [7-9,16,17]. Considering that the above enumerated criteria of the dynamic progression of mem- brane synthesis and signal transfer have been satisfied, it is apparent that the nuclear envelope constitute uninter- rupted membrane demarcated by pore complex. The challenge remains to determine when the pore complex allows movement of the membrane from outer to inner nuclear space [37], and to identify and coordinate phos- phatidylinositides-regulated nuclear events with the translational and synthetic events executed in the ER. Thus, identification of the PI-sensitive components in the nucleus and passage of the massage to the ER is cen- tral to understanding how the cell nucleus dictates post- transcriptional cellular responses. REFERENCES [1] Di Paolo, G. and De Camilli, P. (2006) Phosphoinositi- des in cell regulation and membrane dynamics. Nature, 443, 651-657. doi:10.1038/nature05185 [2] Vicinanza, M., D’Angelo, G., Di Campli, A. and De Matteis, M. A. (2008) Function and disfunction of the PI system in membrane trafficking. EMBO J., 27, 2457- 2470. doi:10.1038/emboj.2008.169 [3] Sasaki, T., Sasaki, J., Sakai, T., Takasuga, S., and Suzuki, A. (2007) The physiology of phosphoinositides. Bio- logical & Pharmaceutical B u ll etin, 30, 1599-1604. doi:10.1248/bpb.30.1599 [4] Sasaki, T., Takasuga, S., Sasaki, J., Kofuji, S., Eguchi, S., Yamazaki, M. and Suzuki, A. (2009) Mammalian phos- phoinositide kinases and phosphatases. Progress in Lipid Research, 48, 307-343. doi:10.1016/j.plipres.2009.06.001 [5] Barlow, C.A., Laishram, R.S., and Anderson, R.A. (2009) Nuclear Phosphoinositides: a signaling enigma wrapped in a compartmental conundrum. Trends in Cell Biology, 20, 25-35. doi:10.1016/j.tcb.2009.09.009 [6] Boronenkov I,V. Loijens, J.C., Umeda, M., and Ander- son, R.A. (1998) Phosphoinositide signaling pathways in nuclei are associated with nuclear specles containing pre-mRNA processing factors. Molecular Biology of the Cell, 9, 3547-3560. [7] Slomiany, A., Grzelinska, E., Grabska, M., Yamaki, K., Tamura, S. and Slomiany, B.L. (1992) Intracellular proc- esses associated with glycoprotein transport and proc- essing. Archives of Biochemistry and Biophysics, 298, 167-175. doi:10.1016/0003-9861(92)90108-9 [8] Slomiany, A., Grabska M., Piotrowski, E., Morita, M. and Slomiany, B.L. (1994) Intracellular processes asso- ciated with vesicular transport from endoplasmic reticu- lum to Golgi and exocytosis; Ethanol-induced changes in membrane biogenesis. Archives of Biochemistry and  A. Slomiany et al. / Health 3 (2011) 187-199 Copyright © 2011 SciRes. Openly accessible at http://www.scirp.org/journal/HEALTH/ 198 Biophysics, 310, 247-255. doi:10.1006/abbi.1994.1164 [9] Slomiany, A., Sano, S., Grabska, M., Yamaki, K. and Slomiany, B.L. (2004) Gastric mucosal cell homeostatic physiome. Critical role of ER-initiated membrane restitu- tion in the fidelity of cell function renewal. Journal of Physiology and Pharmacology, 55, 837-860. 15 [10] Gonzales, M.L. and Anderson, R.A. (2006) Nuclear phosphoinositides kinases and inositol phospholipids. Journal of Cell Biochemistry, 97, 252-260. doi:10.1002/jcb.20655 [11] Irvine, R.F. (2003) Nuclear lipid signaling. Nature Re- views Molecular Cell Biology, 4, 349-361. doi:10.1038/nrm1100 [12] Ruault, M., Dubarry, M. and Taddei, A. (2008) Re-posi- tioning genes to nuclear envelope in mammalian cells; impact on transcription. Trends in Genetics, 24, 574-581. doi:10.1016/j.tig.2008.08.008 [13] Akhtar, A. and Gasser, S.M. (2007), The nuclear enve- lope and transcriptional control. Nature Reviews Genetics, 8, 507-517. doi:10.1038/nrg2122 [14] Moore, M.J. and Proudfoot, N.J. (2009) Pre-mRNA processing reaches back to transcription and ahead of translation, Cell, 136, 688-700. doi:10.1016/j.cell.2009.02.001 [15] Slomiany, A., Grabska, M. and Slomiany, B.L. (2006) Homeostatic restitution of cell membranes. Nuclear membrane lipid biogenesis and transport of protein from cytosol to intranuclear spaces. International Journal of Biological Sciences, 2, 216-226. [16] Slomiany, A., Nowak, P., Piotrowski, E. and Slomiany, B.L. (1998) Effect of ethanol on intracellular vesicular transport from Golgi to apical cell membrane: Role of phosphatidylinositol 3-kinase and phospholipase A2 in Golgi vesicles association and fusion with the apical membrane, Alcoholism: Clinical and Experimental Re- search, 22, 167-175. doi:10.1111/j.1530-0277.1998.tb03634.x [17] Slomiany, A. and Slomiany, B.L. (2010) Cell membranes composition is defined in ER and their restitution pro- ceeds by en bloc fusion of ER generated transport vesi- cles. Health, 2, 1444-1454. doi:10.4236/health.2010.212214 [18] Bunce, M.W., Bergendahl, K. and Anderson, R.A. (2006) Nuclear PI(4,5)P2 : A new place for an old signal. Bio- chemica et Biophysica Acta, 1761, 560-569. [19] Garnier-Lhomme, M., Byrne, R.D., Hobday, T.M.C., Gschmeissner, S., Woscholski, R.,Poccia, D.L., Dufourc, E.J., and Larijani, B. (2009) Nuclear envelope remnants: fluid membranes enriched in sterols and phosphatidy- linositides. Plo S ONE 4, e4255. 16 [20] Tanaka, K., Horiguchi, K., Yoshida,T., Fujisawa, H., Takeuchi, K., Umeda,, M., Kato, S., Ihara, S., Nagata, S. and Fukui, Y. (1999) Evidence that 3,4,5- triphosphate –binding protein can function in nucleus. Journal. Bio- logical Chemistry, 274, 3919-3922. doi:10.1074/jbc.274.7.3919 [21] Ye, K. and Ahn, J.Y. (2008) Nuclear phosphoinositides signaling. Frontiers in Biosciences, 13, 540-548. doi:10.2741/2699 [22] Osborne, S.L., Thomas, C.L., Gschmeissner, S. and Schiavo, G. (2001) Nuclear PtdIns(4,5)P2 assembles in a mitotically regulated particle involved in pre-mRNA splicing. Journal of Cell Science, 114, 2501-2511. [23] Gozani, O.,Karuman, P., Jones, D.R., Lugovskoy, A.A., Baird, C.L, Zhu,H., Field, S.J.,Lessnick, S.L., Villasenor, J.,Mehrotra, B., Chen, J., Rao, V.R., Brugge, J.S., Fer- guson, C,G., Payrastre. B., Myszka, D.G., Cantley, L.C., Wagner, G., Divecha, N. Prestwich, G.D. and Yuan, J. (2003) The PHD finger of the chromatin associated pro- tein ING2 functions as a nuclear phosphoinositides re- ceptor. Cell, 114, 99-111. doi:10.1016/S0092-8674(03)00480-X [24] Macbeth, M.R., Schubert, H. L., VanDemark, A.P., Lin- gam, A.t., Hill, C.P. and Bass, B.L. (2005) Inositol- hexakisphosphate is bound in ADAR2 core and required for RNA editing. Science, 309, 1534-1539. doi:10.1126/science.1113150 [25] Yu, H., Fukami, K., Watanabe, Y., Ozaki, C. and Take- nawa, T. (1998) Phosphatidylinositol 4,5-bisphosphate reverses the inhibition of RNA transcription caused by histone H1. European Journal of Biochemistry, 251, 281-287. doi:10.1046/j.1432-1327.1998.2510281.x [26] Zhao, K., Wang, W., Rando, O.J., Xue, Y., Swiderek, K., Kuo, A. and Crabtree, G.R. (1998) Rapid and phos- phatidylinositol-dependent binding of the SWI/SNFlike BAF complex to chromatin after T lymphocyte receptor signaling. Cell, 95, 625-636. doi:10.1016/S0092-8674(00)81633-5 [27] Mellman, D.L., Gonzales, M.L., Song, C., Barlow, C.A., Wang, P., Kendziorski, C., and Anderson, R.A. (2008) A PtdIns4,5 P2-regulated nuclear poly(A) polymerase con- trols expression of select mRNAs. Nature, 451, 1013-1017. [28] Slomiany, A., Tamura, S., Grzelinska, E.,Piotrowski, J., and Slomiany, B.L. (1992) Characterization of the ‘link’ component of submandibular mucus glycoprotein, Inter- national Journal of Biochemistry, 24, 1003-1015. doi:10.1016/0020-711X(92)90111-D [29] Slomiany, A. and Slomiany B.L. (2003) Lipidomic proc- esses in homeostatic and LPS- modified cell renewal cy- cle. Role of phosphatidylinositol 3-kinase pathway in biomembrane synthesis and restitution of apical epithe- lial membrane . Journal of Physiology and Pharmacology, 54, 533-551. [30] Visnjic, D. and Banfic, H. (2007) Nuclear phospholipid signaling : Phosphatidylinositol-specific phospholipase C and phosphoinositide 3-kinase. Pflugers Archives, 455, 19-30. doi:10.1007/s00424-007-0288-1 [31] Sekine, T. and Raben, D.M. (2004) Nuclear production and metabolism of diacylglycerol. European Journal of Histochemistry, 48, 77-82. [32] Alcazar-Roman, A.R. and Wente, S.R. (2008) Inositol polyphosphates: a new frontier for regulating gene ex- pression. Chromosoma, 117, 1-13. doi:10.1007/s00412-007-0126-4 [33] Brown, D.A. (2006) Lipid rafts, detergent-resistant membranes and raft targeting signals. Physiology, 21, 430-439. doi:10.1152/physiol.00032.2006 [34] Snoek, G.T. (2004) Phosphatidylinositol transfer protein: emerging roles in cell proliferation, cell death and sur- vival. IUBMB Life, 56, 467-475. doi:10.1080/15216540400012152 [35] Yokogawa, T., Nagata, S., Nishio, Y., Tsutsumi, T., Ihara, S., Shirai, R., Morita, K., Umeda, M., Shirai, Y.,  A. Slomiany et al. / Health 3 (2011) 187-199 Copyright © 2011 SciRes. Openly accessible at http://www.scirp.org/journal/HEALTH/ 199199 Saitoh, N. and Fukui, Y. (2000) Evidence that 3’- phos- phorylated polyphosphoinositides are generated at the nuclear surface: use immunostaining technique with spe- cific monoclonal antibodies specific for PI3,4P2. FEBS Letters, 473, 222-226. doi:10.1016/S0014-5793(00)01535-0 [36] Fagone, P. and Jackowski, S. (2009) Membrane phos- pholipid synthesis and endoplasmic reticulum function. Journal of Lipid Research, 50, S311-S316. doi:10.1194/jlr.R800049-JLR200 [37] Theerthagiri, G., Eisenhardt, N., Schwarz, H. and An- toninin, W. (2010) The nucleoporin Nup188 controls passage of membrane proteins across the nuclear pore complex. Journal of Cell Biology, 189, 1129-1142. doi:10.1083/jcb.200912045 |

