 Journal of Biosciences and Medicines, 2014, 2, 30-35 Published Online June 2014 in SciRes. http://www.scirp.org/journal/jbm http://dx.doi.org/10.4236/jbm.2014.24006 How to cite this paper: Querol-Audí, J., et al. (2014) Catalytically Important Residues in E. coli 1-Deoxy-D-Xylulose 5-Phos- phate Synthase. Journal of Biosciences and Medicines, 2, 30-35. http://dx.doi.org/10.4236/jbm.2014.24006 Catalytically Important Residues in E. coli 1-Deoxy-D-Xylulose 5-Phosphate Synthase Jordi Querol-Audí, Albert Boronat, Josep J. Centelles, Santiago Imperial Department of Biochemistry and Molecular Biology, School of Biology, University of Barcelona, Barcelona, Spain Email: simperial@ub.edu Received March 2014 Abstract 1-deoxy-D-xylulose 5-phosphate synthase (DXS) catalyzes the initial step of the 2-C-m ethyl-D- erythritol 4-phosphate (MEP) pathway consisting in the condensation of (hydroxiethyl)thiamin derived from pyruvate with D-glyceraldehyde 3-phosphate (GAP) to yield 1-deoxy-D-xylulose 5-phosphate (DXP). The role of the conserved residues H49, E370, D427 and H431 of E. coli DXS was examined by site-directed mutagenesis and kinetic analysis of the purified recombinant en- zyme mutants. Mutants at position H49 showed a severe reduction in their specific activities with a decrease of the kcat/KM ratio by two orders of magnitude lower than the wild-type DXS. According to available structural data residue H49 is perfectly positioned to abstract a proton from the do- nor substrate. Mutations in DXS E370 showed that this residue is also essential for catalytic activi- ty. Three-dimensional structure supports its involvement in cofactor deprotonation, the first step in enzymatic thiamin catalysis. Results obtained with H431 mutant enzymes indicate that this re- sidue plays a role contributing to transition state stabilization. Finally, mutants at position D427 also showed a severe specific activity decrease with a reduction of the kcat/KM ratio. A role in bind- ing the substrate and selecting the stereoisomer is proposed for D427. Keywords Active Site, 1-Deoxy- D-Xylulose 5-Phosphate Syn tha se, Isoprenoid Biosynthesi s, Kine tic Param e ter s , MEP P a thway , Methylerythritol Phosphate, Mutagene sis 1. Introduction Isoprenoids, the most diverse group of natural products with thousands of compounds identified [1] derive from two common C5 units: isopentenyl diphosphate (IPP) and its isomer dimethylallyl diphosphate (DMAPP). In most eubacteria IPP and DMAPP biosynthesis proceed exclusively through the methylerythritol 4-phosphate (MEP) pathway [2] [3]. In the first reaction pyruvate and D-glyceraldehyde 3-phosphate (GAP) are condensed to yield deoxyxylulose 5-phosphate (DXP). This involves the decarboxylation of pyruvate and the transfer of the resulting C2 unit to the aldehyde group of GAP (Figure 1) in a reaction dependent on thiamin diphosphate (TDP). This reaction is catalyzed by DXP synthase (DXS) [4]-[6], an enzyme belonging to the superfamily of TDP- dependent enzymes including transketolases and the E1 subunit of pyruvate dehydrogenase. Then, DXP is con-  J. Querol-Audí et al. Figure 1. Reaction catalyzed by deoxyxylulose 5-phosphate synthase (DXS). verted by intramolecular rearrangement and reduction into 2-C-methyl-D -erythritol 4-phosphate (MEP) in a reaction catalyzed by DXP reductoisomerase (DXR) [7]. Since DXP is also the substrate for the synthesis of vi- tamins B1 (thiamin) and B6 (pyridoxol) [8] [9], MEP is the first specific intermediate of the pathway and it is accepted to name it as MEP pathway. Many pathogenic microorganisms, such as M. tuberculosis [10] and the malaria parasite P. falciparum [11], synthesize IPP and DMAPP through the MEP pathway. Given the essential role of isoprenoids in these organisms, and the fact that the MEP pathway is absent in mammals, where these precursors are exclusively synthesized via the mevalonate pathway, all the enzymes participating in the MEP pathway are potential targets for the design of inhibitors that could be used as antibiotic or antimalarial drugs [10] [11]. In this context, DXS is one of the most attractive targets, because of its regulatory role in the biosyn- thesis of isoprenoids in various systems [12] [13], indicating that this enzyme catalyzes a rate-limiting step in the MEP pathway. Identification of catalytically essential amino acid residues is necessary in order to design ef- fective drugs. In the present work we analyzed the role of other conserved residues in DXS activity by site-di- rected mutagenesis. Nine recombinant proteins (wild-type, H49Q, H49A, E370Q, E370A, D427N, D427A, H431Q and H431A) were purified to apparent homogeneity using Ni2+-affinity chromatography and kinetic analysis of the mutant enzymes were performed in order to examine their catalytic role in E. coli DXS. 2. Materials and Methods 2.1. Materials Materials from commercial sources included NADPH, DL-glyceraldehyde 3-phosphate (DL-GAP) and TDP from Sigma. Hi-Trap Chelating columns were from GE Healthcare. The protease inhibitors Pefabloc SC Plus and the protease inhibitor cocktail Complete EDTA-free were obtained from Roche. All other reagents were of analytical grade. Recombinant E. coli DXR was obtained as described in [14]. 2.2. Construction of the E. coli DXS Expression Vector pET-23-DXS The coding region of the E. coli DXS, cloned into the expression vector pT7-7 [15], was amplified by PCR us- ing Pfu DNA polymerase and primers T7 (5’-TAATACGACT CACTATAGG-3’) and pET-23-XhoI (5’-CGCTCG AGT CCT G CC AG C CAGG CCTT G ATTT T GGC-3’). After digestion with Nde I and XhoI, the am- plified DNA fragment was ligated into the same sites of the expression vector pET-23(b). Strain DH5α (Prome- ga) was used as the recipient during this transformation. Positive clones were identified by DNA sequencing. The resulting plasmid, pET-23-DXS, produces a C-terminal histidine-tagged enzyme (DXS-cHi s) . 2.3. Site-Directed Mutagenesis Point mutations H49Q, H49A, E370Q, E370A, D427N, D427A, H431Q and H431A were introduced into the E. coli DXS coding sequence by site-directed mutagenesis using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) as described [15]. Primers used for introducing these point mutations are summarized in Table 1. The underlined letters represent the substituted nucleotides. Presence of the desired mutations and absence of other mutations that could be introduced during PCR were verified by DNA sequencing. 2.4. Overexpression and Purification of Cloned DXS Proteins BL21 (DE3) pLysS cells carrying pET-23-DXS or the corresponding mutants were grown in LB medium sup- plemented with 100 μg/ml ampicillin and 34 μg/ml chloramphenicol at 22˚C to an absorbance at 600 nm of 0.3 - 0.4 and then induced with 0.3 mM isopropyl β-D-thiogalactoside for 18 - 20 h. Bacterial cells were recovered by  J. Querol-Audí et al. Table 1. Primers used in this study for site-directed mutagenesisa. name Sequence H49Q 5’-CCAGCGGGCAGTTCGCCTCC-3’ H49A 5’-TCCAGCGGGGC CT TC GC C TC -3’ E370Q 5’-GC A ATT GCC CAGC A ACACGCG G-3’ E370A 5’-GC A ATT GCC GCGC A ACACGCG-3’ D427N 5’-TGTTGGTGCTAACGGTCAAACCC-3’ D427A 5’-GTTGGTGCTGC CGGTCAAAC CC-3’ H431Q 5’-GGTCAAACCCAGCAGGGTGCTT-3’ H431A 5’-GGTCAAACCGCTCAGGGTGCTT-3’ aThe underlined letters represent the substituted nucleotides. centrifugation and the cell pellet was resuspended in Buffer 1 (40 mM Tris-HCl, pH 8.5, 100 mM NaCl, 10 mM imidazole, 1 mM MgCl2, 5 mM 2-mercaptoethanol) containing 1 mM TDP, lysozyme (1 mg/ml), Pefabloc SC Plus (1 mM) and one tablet of protease inhibitor cocktail Complete EDTA-free. After incubation at 4˚C for 30 min and brief sonication (3 pulses, 30 w, 20 s), lysate was centrifuged at 12,000 ×g for 45 min at 4˚C. Recom- binant DXSs were purified by Ni2+ affinity chromatography (1ml Hi-Trap Chelating column, GE Healthcare) with a linear gradient of 10 to 500 mM imidazole. Fractions containing DXS were pooled, dialyzed by ultrafil- tration (Ultrafree-0.5 and Ultrafree-4 centrifugal filter Biomax-5K, Millipore) against 50 mM glycylglycine buffer (pH 7.8), 1 mM MgCl2, 1 mM TDP, 5 mM 2-mercaptoethanol and stored at −20˚C in 50% glycerol. Puri- fied fractions were analyzed by SDS-PAGE. After dialyzing the samples against (1) glycylglycine buffer 10 mM, pH 7.8, 1 mM DTT, 50 mM EDTA and (2) glycylglycine buffer 10mM, pH 7.8, 1 mM DTT, protein concentra- tion was determined by measuring the absorbance at 280 nm using an Abs 1% = 6.25. 2.5. DXS Activity Measurements and Determination of Kinetic Parameters Specific activity for wild-type and mutant DXS proteins was monitored by a coupled system using purified E. coli DXR [16]. All kinetic measurements were made at 37˚C in triplicate. The standard reaction mixture con- sisted in 50 mM glycylglycine buffer pH 7.8, 1 mM TDP, 1 mM MgCl2, 0.05 mM MnCl2, 1 mM DTT, 0.15 mM NADPH, 1 mM pyruvate, 50 μg DXR and variable amounts (10 μg to 160 μg) of DXS or the corresponding mutants in a final volume of 1 ml. Mixtures were incubated at 37˚C for 10 min and the reaction was initiated by adding 1 mM DL-GAP. Reactions were conducted at 37˚C and initial velocities were monitored at 340 nm due to the oxidation of NADPH in a UNICAM UV-3 spectrophotometer. Specific activities were determined from the slope of a plot of initial velocities versus the enzyme amount. Steady-state kinetic parameters for the wild-type and mutant enzymes were determined at saturating concen- trations of the second substrate, 0.05 mM to 1.6 mM pyruvate and 25 μM to 800 μM DL-GAP, by measuring in- itial rates. KM values for pyruvate and GAP were calculated by fitting the data to the Michaelis-Menten equation using the program GRAFIT. For mutants H49Q, H49A, D427N, D427A and H431Q, kcat/KM values for pyruvate were calculated from initial velocities at unsaturating concentrations of this substrate (30 μM). 3. Results and Discussion 3.1. Expression of DXS-cHis Mutants in E. coli The gene encoding E. coli DXS was cloned into the expression vector pET-23. Mutations at positions H49, E370, D427 and H431 were introduced by site-directed mutagenesis. DNA sequencing confirmed that undesired mutations were not introduced during the course of mutagenesis. Over expression of the wild-type and mutant enzymes was carried out in E. coli BL21 (DE3) pLysS. Purification of the recombinant proteins, containing a His-tag at the C-terminus, was performed in a single chromatographic step (Hi-Trap Chelating, GE Healthcare). Wild-type and mutant DXS proteins were obtained in similar amounts (≥10 mg of pure DXS/litre) and purified (>95%) as shown by SDS-PAGE and Coomassie-blue staining (Figure 2(a)). The enzyme purity was compara- ble for the wild-type and the mutant proteins. 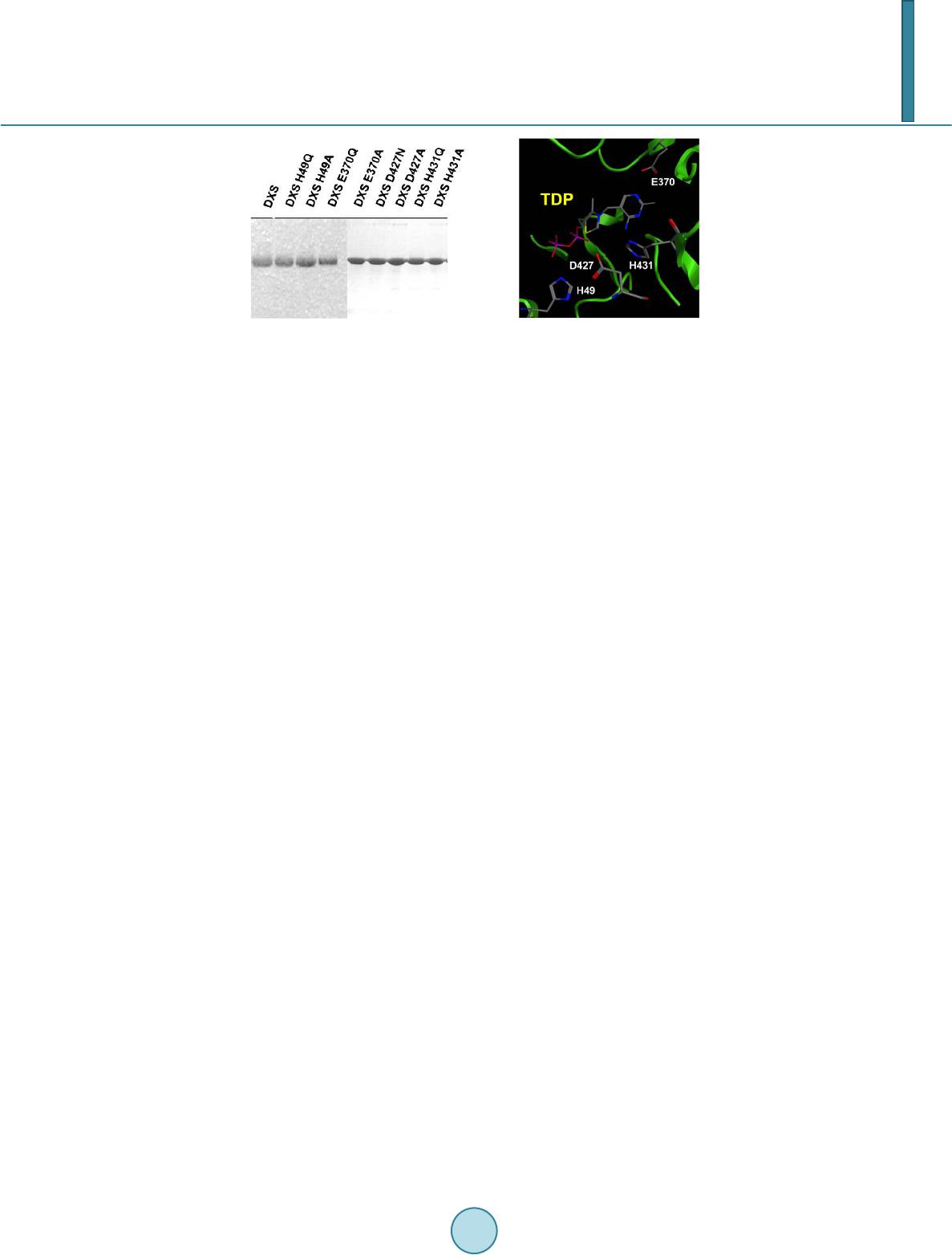 J. Querol-Audí et al. (a) (b) Figure 2. (a) SDS-PAGE analysis of purified E. coli DXS mutants. Purified DXS mutants obtained by using a Hi-Trap Chelating column were analyzed by SDS-PAGE. Proteins were stained with Coomassie brilliant blue R-250. Purified wild type and mutant proteins are indicated in the top of the gel; (b) Pose of the mutated residues and the coenzyme TDP at the catalytic site of E. coli DXS based on structural data reported at [17]. 3.2. Kinetic Studies E370Q and E370A mutations resulted in a completely inactive DXS and the H431A mutant showed only a moderate decrease in its specific activity compared to the wild-type enzyme (0.84 and 1.46 μmol pyruvate/min μg, respectively). The greatest decrease in activity levels were observed with the H49Q, H49A, D427N and D427A mutants which showed only 2%, 6%, 3% and 10% of the activity of the wild-type DXS. H431Q also ex- hibited a reduced catalytic activity (4% of the wild-type) while H431A mutant retained about 57% activity. For mutants at positions H49, D427 and H431Q kcat values were obtained from the plot of initial velocities versus the enzyme amount. kcat/KM values for pyruvate were determined by measuring the initial velocities at subsaturating concentrations of pyruvate (Table 2). We previously reported that mutation of residue H49 by glutamine in E. coli DXS resulted in a mutant en- zyme with undetectable activity [15]. Here we have analyzed the effect of H49Q and H49A point mutations by measuring their kinetic parameters kcat and kcat/KM. Both mutations result in severally impaired catalytic activi- ties. kcat/KM values for mutant H49A was two orders of magnitude lower than the wild-type enzyme. No kinetic parameters could be determined for mutant H49Q due to its reduced specific activity. The H431A mutant re- tained 57% specific activity but increased KM for pyruvate (one order of magnitude compared to the wild-type). No significant change in the KM for GAP was observed for this mutant. When residue H431 was replaced by glutamine, kcat was reduced by two orders of magnitude. Kinetic analysis of the purified D427N and D427A mutants showed reduced kcat values (3.3 min−1 and 10 min−1 respectively, compared to the value of 100.3 min−1 for the wild-type enzyme) (Table 2). Site-directed mutagenesis was used to confirm the essential role of selected residues for E. coli DXS activity. Kinetic parameters of the mutant enzymes H49Q and H49A obtained in this work corroborate that this residue is essential for DXS activity. The structure of the catalytic center of E. coli DXS [17] confirms that residue H49 is perfectly positioned to abstract a proton from the donor substrate. Kinetic analysis of the purified DXS mutant enzymes at the corresponding positions (E370, D427 and H431) were carried out to establish the role of these conserved residues for E. coli DXS catalytic activity. Residue E418, equivalent to E370 in DXS, has been reported to be essential for TK catalytic activity since its implication in cofactor deprotonation, the first step in enzymatic thiamin catalysis [18]. Analysis of mutations in DXS E370 demonstrated that this residue is also essential for catalytic activity. E. coli DXS 3D-structure [17] shows a di- rect hydrogen bond to the pyrimidine of thiamin supporting the catalytic role suggested by Wikner et al. [19]. Site-directed mutagenesis studies of residue H481 in yeast transketolase indicate that this residue contributes to transition state stabilization but is not absolutely required for catalysis [19]. When the E. coli DXS histidine residue at position 431 was replaced by glutamine, a mutant DXS with a severe reduction in enzyme activity (4% of the wild-type) was obtained. In contrast, moderated effects in terms of specific activity and kcat were ob- served when H431 was replaced by alanine. Only KM for pyruvate was increased one order of magnitude (Table 2). Similar results have been obtained for mutants at residue H481 in yeast transketolase [19] or the correspond-  J. Querol-Audí et al. Table 2. Comparison of kinetic parameters of wild-type (DXSwt) and mutant DXS enzymes. Enzyme KMpyr (μM) KMGAP (μM ) kcat (min−1) kcat/KMpyr (min−1∙μM−1) DXSwt 134±8 41±6 100.3 0.7480 H49Q n.d. n.d. n.d. n.d. H49A n.d. n.d. 6.6 0.0091 D427N n.d. n.d. 3.3 0.0 022 D427A n.d. n.d. 10.0 0.0095 H431Q n.d. n.d. 4.4 0.1 600 H431A 1623±81 62±6 57.4 0.0350 E370Q n.d. n.d. n.d. n.d. E370A n.d. n.d. n.d. n.d. ing residue in human transketolase Q428 [20]. Site-directed mutagenesis studies of H481 in yeast transketolase suggested that this residue contributes to transition state stabilization but is not required for catalysis [20]. Our results would indicate that TK H481 and DXS H431 are conserved equivalent residues playing similar roles. The fact that H431Q mutant shows a markedly reduced specific activity could be explained by considering that the increase in side chain size may result in steric clashes with the substrate. Substitution of aspartic acid residue at position 427 by glutamine or alanine yielded a DXS with highly de- creased specific activity with kcat/KM ratio reduced by two orders of magnitude. The hydrogen at the side chain of the corresponding residue, D477, in yeast TK is proposed to be the determinant of the enantioselectivity of the enzyme [21]. In this study we describe the active site architecture of 1-deoxy-D-xylulose 5-phosphate synthase, a target for antimicrobial agents, herbicides, and antimalarial drugs [22] [23]. A role in catalysis has been assigned to con- served amino acid residues and a model of the catalytic site has been generated which can be of great interest to predict drug-enzyme interactions. References [1] Chappell, J. (2002) The Genetics and Molecular Genetics of Terpene and Sterol Origami. Current Opinion in Plant Bi- ology, 5, 151-157. http://dx.doi.org/10.1016/S1369-5266(02)00241-8 [2] Zhao, L.S., Chang, W.C., Xiao, Y.L., Liu, H.W. and Liu, P.H. (2013) Methylerythritol Phosphate Pathway of Isopre- noid Biosynthesis. Annual Review of Biochemistry, 82, 497-53 0. http://dx.doi.org/10.1146/annurev-biochem-052010-100934 [3] Rohmer, M., Knani, M., Simonin, P., Sutter, B. and Sham, H. (1993) Isoprenoid Biosynthesis in B acteria: A Novel Pathway for Early Steps Leading to Isopentenyl Diphosphate. The Biochemical Journal, 295, 517-524. [4] Eubanks, L.M. and Poulter C.D. (2003) Rhodobacter capsulatus 1-Deoxy-D-Xylulose 5-Phosphate Synthase: St eady- State Kinetics and Substrate Binding. Biochemistry, 42, 1140-1149 . http://dx.doi.org/10.1021/bi0205303 [5] Sprenger, G.A., Schörken, U., Wiegert, T., Grolle, S., De Graaf, A., Taylor, S., Begley, T., Bringer-Meyer, S. and Sahm, H. (1997) Identification of a Novel Thiamin-Dependent Synthase in Escherichia coli Required for the For ma - tion of the 1-Deoxy-D-Xylulose 5 -Phosphate Precursor to Isoprenoids, Thiamin, and Pyridoxol. Proceedings of the National Academy of Sciences of the USA, 94, 12857-12862. http://dx.doi.org/10.1073/pnas.94.24.12857 [6] Lo is, L.M., Campos, N., Putra, S.R., Danielsen, K., Rohmer, M. and Boronat, A. (1998) Cloning and Characterization of a Gene from Escherichia coli Encoding a Transketolase-Like Enzyme That Catalyzes the Synthesis of D-1-Deo xy- xylulose 5-Phosphate, a Common Precursor for Isoprenoid, Thi amin, and Pyridoxol Biosynthesis. Proceedings of the National Academy of Sciences of the USA, 95, 2105-2110. http://dx.doi.org/10.1073/pnas.95.5.2105 [7] Proteau, P.J. (2004) 1-Deoxy-D-Xylulose 5-Phosphate Reductoisomerase: An Overview. Bioorganic Chemistry, 32, 483-493. http://dx.doi.org/10.1016/j.bioorg.2004.08.004 [8] Himmeldirk, K., Kennedy, I.A., Hill, R.E., Sayer, B.G. and Spenser, I.D. (1996) Biosynthesis of Vitamins B1 and B6 in Escherichia coli: Concurrent Incorporation of 1-D eo xy-D-Xylulose into Thiamin (B1) and Pyridoxol (B6). Ch e m i - cal Communications, 10, 1187-1188. http://dx.doi.org/10.1039/cc9960001187  J. Querol-Audí et al. [9] Mukherjee, T., Hanes, J., Tews, I., Ealick, S.E. and Begley, T.P. (2011) Pyridoxal Pho sph ate: Biosynthesis and Cata- bolism. Biochimica et Biophysica Acta, 1814, 1585-15 96. http://dx.doi.org/10.1016/j.bbapap.2011.06.018 [10] Obio l-Pardo, C., Rubio-Martinez, J. and Imperial, S. (2011) The Methylerythritol Phosphate (MEP) Pathway for Iso - prenoid Biosynthesis as a Target for the Development of New Drugs against Tuberculosis. Current Medicinal Chemi- stry, 18, 1325-1338. http://dx.doi.org/10.2174/092986711795029582 [11] Wies n er, J. and Jomaa, H. (2007) Isoprenoid Biosynthesis of the Apicoplast as Drug Target. Current Drug Targets, 8, 3-13. http://dx.doi.org/10.2174/138945007779315551 [12] Lois, L.M., Rodríguez-Concepción, M., Gallego, F., Campos, N. and Boronat, A. (2000) Carotenoid Biosynthesis dur- ing Tomato Fruit Development: Regulatory Role of 1-Deoxy-D-Xylulose 5-Phosphate Synthase. The Plant Journal, 22, 503-513. http://dx.doi.org/10.1046/j.1365-313x.2000.00764.x [13] Banerjee, A., Wu, Y., Banerjee, R., Li, Y., Yan, H. and Sharkey, T.D. (2013) Feedback Inhibition of Deoxy-D-Xylu- lose-5-Phosphate Synthase Regulates the Methylerythritol 4 -Phosphate Pathway. The Journal of Biological Chemistry, 288, 16926-16936. http://dx.doi.org/10.1074/jbc.M113.464636 [14] Hoeffler J.F., Tritsch, D., Grossdemange-Billiard, C. and Rohmer, M. (2002). Isoprenoid Biosynthesis via the Met hy- lerythritol Phosphate Pathway: Mechanistic Investigations on the 1-Deoxy-D-Xylulose 5-Phosphate Reductoisomerase. European Journal of Biochemistry, 269, 4446-4457. http://dx.doi.org/10.1046/j.1432-1033.2002.03150.x [15] Querol, J., Rodriguez-Concepcion, M., Boronat, A. and Imperial, S. (2001) Essential Role of Residue H49 for Activity of Escheri chia coli 1-D eoxy-D-Xylulose 5 -Phosphate Synthase, the Enzyme Catalyzing the First Step of the 2-C-Me- thyl-D-Erythritol 4-Phosphate Pathway for Isoprenoid Synthesis. Biochemical and Biophysical Research Communica- tions, 289, 155-160. http://dx.doi.org/10.1006/bbrc.2001.5957 [16] Altincicek, B., Hintz, M., Sanderbrand, S., Wiesner, J., Beck, E. and Jomaa, H. (2000) Tools for Discovery of Inhibi- tors of the 1-Deoxy-D-Xylulose 5 -Phosphate (DXP) Synthase and DXP Reducto isomerase: An Approach with En- zymes from the Pathogenic Bacterium Pseudomonas aeruginosa. FEMS Microbiology Letters, 190, 329-333. [17] Xiang, S., Usunow, G., Lange, G., Busch, M. and Tong, L. (2007) Crystal Structure of 1-Deoxy-D-Xylulose 5-P ho s- phate Synthase, a Crucial Enzyme for Isoprenoids Biosynthesis. The Journal of Biological Chemistry, 282, 2676-2682. http://dx.doi.org/10.1074/jbc.M610235200 [18] Wikner, C., Meshalkina, L., Nilsson, U., Nikkola, M., Lindqvist, Y., Sundstrom, M. and Schneider, G. (1994) Analysis of an Invariant Cofactor-Protein Interaction in Thiamin Diphosphate-Dependent Enzymes by Site-Directed Mutagene- sis. Glutamic Acid 418 in Transketolase Is Essential for Catal ysis . The Journal of Biological Chemistry, 269, 32144- 32150. [19] Wikner, C., Nilsson, U., Meshalkina, L., Udekwu, C., Lindqvist, Y., Schneider, G. and Schneider, G. (1997) Identifica- tion of Catalytically Important Residues in Yeast Transketolase. Bio chemistry, 36, 15643-15649. http://dx.doi.org/10.1021/bi971606b [20] Singleton, C.K., Wang, J.J., Shan, L. and Martin, P.R. (1996) Conserved Residues Are Functionally Distinct within Transketolases of Different Species. Bi ochemistry, 35 , 15865-15869. http://dx.doi.org/10.1021/bi9616920 [21] Nilsson, U., Hecquet, L., Gefflaut, T., Guerard, C. and Schneider, G. (1998) Asp477 Is a Determinant of the Enanti- oselectivity in Yeast Transketolase. FEBS Letters, 424, 49-52. http://dx.doi.org/10.1016/S0014-5793(98 )001 36 -7 [22] Matsue, Y., Mizuno, H., Tomita, T., Asami, T., Makoto Nishiyama, M. and Kuzuyama, T. (2010) The Herbicide Keto- clomazone Inhibits 1-Deo x y-D-Xylulose 5-Phosphate Synthase in the 2-C-Methyl-D-Erythritol 4-Phosphate Pathway and Shows Antibacterial Activity against Haemophilus Influenza. The Journal of Antibiotics, 63, 583-588. http://dx.doi.org/10.1038/ja.2010.100 [23] Handa, S., Ramamoorthy, D., Spradling, T.J., Guida, W.C., Adams, J.H., Bendinskas, K.G. and Merkler, D.J. (2013) Production of Recombinant 1 -Deoxy-D-Xylulose-5-Phosphate Synthase from Plasmodium vivax in Escherichia coli. FEBS Open BIO, 3, 124-129. http://dx.doi.org/10.1016/j.fob.2013.01.007
|