 Advances in Anthropology 2014. Vol.4, No.1, 18-37 Published Online February 2014 in SciRes (http://www.scirp.org/journal/aa) http://dx.doi.org/10.4236/aa.2014.41004 Reconsideration of the “Out of Africa” Concept as Not Having Enough Proof Anatole A. Klyosov The Academy of DNA Genealogy, Newton, USA Email: aklyosov@comcast.net Received November 25th, 2013; revised December 23rd, 2013; accepted January 18th, 2014 Copyright © 2014 Anatole A. Klyosov. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. In accordance of the Creative Commons Attribution License all Copyrights © 2014 are reserved for SCIRP and the owner of the intellectual property Anatole A. Klyosov. All Copyright © 2014 are guarded by law and by SCIRP as a guardian. This is an overview of the “Out of Africa” (OOA) concept and the concept is based upon experimental data. The article shows that said concept is based on data which are—as a rule—interpreted by the OOA proponents in a one-sided manner, that is to “prove” the OOA concept. The Y-chromosomal haplogroup tree in its most ancient part includes a number of nodes-haplogroups, which in the current classification are named A0-T, A1, and A1b. Bearers of those haplogroups are not identified in Africa or elsewhere. However, those three haplogroups are assigned by the proponents of the OOA “by default” to be the “African” haplogroups, which “automatically” makes all non-Africans “African descendants”. In fact, each of the three haplogroups represents a split into African and non-African haplogroups. As a result, the evolution (Y-chromosomal) tree produced at least five waves of migrants to Africa (haplogroups A00, A0, A1a, A1b1, and B), while the rest of the tree consists of four non-African (by origin) haplogroups, that is A0-T, A1, A1b, and BT, along with the downstream subclades of the latter. The tree is confirmed by the analysis employing STR (Short Tandem Repeat) and SNP (Single Nucleotide Polymorphism) mutations of the Y-chromosome. Indeed, according to many data, the tree shows a deep split between African and non-African (by origin) lineages. The last split, from haplogroup A1b (into African A1b1 and non-Afri- can—by origin—B) occurred 160,000 ± 12,000 years before present. Haplogroup B is by origin a non- African haplogroup, which is very distant—time-wise—from haplogroups A, by at leas t 250 - 30 0 thou- sand years, an d could not possibly have descended from haplogroups A 1a or A1b1, or their subclades. A similar pattern is observed with the mtDNA haplogroup tree, which shows a deep split between African L0 and non-African (by origin) haplogroups L1-L 6. T he article sh ow s how recent OOA studies (as well as earlier ones) employ biased interpretations to artificially “prove” the OOA concept. The a r tic le shows that the same data can be—and more justifiably—interpreted as incompatible with the OOA concept, and giving support for a “into Africa” concept. It seems that from times of Neanderthals (seemingly having pale skin and fair hair, based on the identified Neanderthal MCR1 melanocortin receptor), our ancestors, of both Africans and non-Africans current populations, lived outside of Africa, apparently in Eurasia or maybe in Europe. Keywords: Y-Chromosome; mtDNA; Mutations; Haplotypes; Haplogroups; STR; SNP; Out-of-Africa; Into-Africa; SHV-1 (Simplex Herpes Virus) Introduction The origin of humans is one of the most intriguing and ex- citing subjects in science, philosophy, view of life. And, at the same time, it is one of the most complicated ones. The thing is that there was not a single and direct experiment thus far which would firmly and reliably answer where on the planet and when our direct ancestor, fitting the anthropological description of a species Homo sapiens and/or the term “ana- tomically modern human” (AMH), first appeared. Every term in the above sentence is not clearly defined and is in fact “floating”. When fossils are found, how to determine if t hey belong to “the most ancient” Homo sapiens and/or AMH, when tomorrow some more ancient relics might be found? How reli- able are (paleo) archaeological dates, which are commonly not reliable at all, and are always disputed by other specialists (see the last section of this paper)? There are dozens of anthropo- logical features that, one way or another, describe the species Homo sapiens and the AMH; however, the theory is one thing (albeit there is no classification which would have been em- braced by all the experts), b ut to apply all those features con- currently, in reality, is practically impossible. Typically, onl y some skeletal fragments of an ancient hominin are found, and for the most ancient bone remains some archaic features are typically observed (see below). The next step is a matter of interpretation, often a rather arbitrary choice whether it was Homo sapiens or its predecessor or something else . Those in- terpretations are often rather far from being unbiased. Here a judgement and honesty of a scientist step in. Bets are high—every new skeleton or its fragments, that are pronounced OPEN ACCESS 18 A. A. KLYOSOV “the most ancient of all known” and related to Homo sapiens or AMH, becomes the worldwide sensation, with all due conse- quences such as prestigious scientific awards, large financial grants, elections to prestigious academies, etc. Unfortunately, these enticements may foster distortions, incomplete or incor- rect data assessment, and may resu lt in biased conclusions. The new and phenomenal discovery, described in academic and other editions, is then eagerly sensationalized by the popular press. Datings are often exaggerated, archaic features are re- touched, and as a result it becomes comp licated determining where the real data is. Cross-examinations, which are badly needed in many cases, are very rare. This is the most obvious in the area of “Out of Africa”, where for some mysterious reason very m any authors seem to exhibit extrapolated, incomplete, or otherwise overinterpreted data to show that the “Out of Africa” concept has no alternatives, as reflected by standard repeats at the beginning of many papers—“As it is known, anatomically modern humans came out of Africa”. Here are a few out of many examples: “Human origins: out of Africa” (title of the paper; Tattersal, 2009). “Human evolution out of Africa” (from the title of the paper; Stewart & Stringer, 2012). “The origin of patrilineal diversity in Africa” (from the title of the paper; Cruciani et al., 2011). “African origin of modern humans in East Asia” (from the title of the paper; Ke et al., 2001). “…anatomically modern humans arrived in Europe from an African source at least 45,000 years a go” (Moorjani et al., 2011). “Modern humans are generally thought to have originated in eastern Africa” (Henn et al., 2011). “It is now well accepted that anatomically modern humans (AMH) originated in Africa (Hammer et al., 2011). “Africa, the ancestral homeland of all modern humans” (La- chance et al., 2012). “…the expansion of anatomically modern humans out of Africa, at approximately 44,000 years ago” (Underhill et al., 2000). “Modern humans evolved in Africa around 200 thousand years ago” (Campbell and Tishkoff, 2010). “…anatomically modern humans originated in a small, iso- lated population in Africa 150,000-200 thousand years ago” (Patin et al., 2009). “sub-Saharan and northeastern Africa, the most likely source region and corridor to the rest of the world” (Arredi et al., 2004). “an expansion originated in Africa” (Ramachandran et al., 2005). “There is a broad concensus that Africa was the birthplace of Homo sapiens—and what have been referred to as “anatomi- cally moden humans” (AMH) (the first sentence of the paper, Rito et al., 2013). There are d ozens and probably hundreds of others, and there is no need to quote more of them. As this paper shows, they are incorrect, or unjustified, or ungrounded, or j ust uncritically repeat each other. According to the authors, the “Out of Africa” ancient migration might have happened some 50,000 years ago (ya), or 70,000 ya, or 100,000 ya, or maybe 150,000 ya, or 200,000 ya, etc. This paper illustrates what kind of data those authors actually obtained, and how they (data) are b ent to “prove” the “Out of Africa” exodus. Before the 1980s there was almost no discussion on the “African origin of modern man”, the subject was rather mar- ginal. There were two reasons why those discussions could n ot have been considered seriously. First, it was agreed upon that a rather remot e ancestor of the modern man was Homo erectus, which had appeared more than a million years ago, possibly in Africa, however, it was known that it then spread all over Eura- sia. Therefore it was recognized that Homo sapiens could ha ve appeared in any region of Eurasia. Second, it was recognized that Neanderthal, the nearest cousin of AMH, had not lived in Africa. Therefore a common ancestor of the AMH and Nean- derthal, who lived on different accounts between 300 and 600 thousand years ago, could not have lived in Africa as well. Neanderthal had a fair (pale) skin (see below), therefore an African origin of man would require an arrival of a fair-skin direct ancestor of AMH in Africa, say, 300 - 500 thousand years ago, an evolution-driven acquisition of a black skin, oth- erwise he would not have survived in Africa, then his exodus from Africa and an independent back conversion of his black skin to a fair one . To defend such an awkward suggestion a smart hypothesi s has been invented about a rol e of vitamin D in an evolution-driven conversion of black skin to white in the course of the suggested migration of humans out of Africa. Clearly, there could not have been any possible mating of the out-of-Africa black migrants with fair-skin people outside of Africa, since they w ere not supposed to be any at that time. Said hypothesis of vitamin D-driven conversion of blacks to whites has never been tested or experimentally confirmed, and still remains a hypothe sis. The Cann et al. (1987) “Seminal” Paper on “Out of Africa” As a result, un til the middle of 1980s, a talk on the African origin of AMH was not considered really serious. An important event happened in 1987, when Nature published a paper by Rebecca Cann and co-authors (Cann et al., 1987) from Univer- sity of California, Berkeley, entitled “Mitochondrial DNA and human evolution”. The paper was rather weak not only by con- temporary criteria, but also by those, in the 1980s, and o ne can only wonder how that paper got through the Nature reviewers, if there were any. It would be enough just to mention that the Abstract, published prior the main text, said that the mtDNA studied in the paper, “stem from one woman who is postulated (!-AK) to have lived about 200,000 years ago, probably (!-AK) in Africa”. The paper opened the gates, figuratively speaking. An ex- citement of the mass media that the Africans happened to be our ancestors was overwhelming. The ancient African woman, mother of all, was immediately baptized “mitochondrial Eve”, and world leading magazines have provided their glossy covers for that informations and assumed portraits of the “Eve”. Since then a non-stop treatment of t he public opinion continues, if it is not growing. This has become a commonly attested opinion, a consensus, to argue with which is alike arguing over a verdict on the thermodynamically impossible perpetuum mobile. So, what was in the paper by Rebecca Cann et al. in 1987? How it all began? Let us take a look. The experimental part of the paper presents a mapping of mtDNA samples of 145 plac entas (mainly from US hospitals) and two cell lines derived from a B lack American and a South African, as follows: OPEN ACCESS 19 A. A. KLYOSOV -20 Africans (representing the Sub-Sahara region, as the pa- per stated); however, “only two of them were born in sub-Sa- haran Africa, the other 18 are Black Americans, who bear many non-African nuclear genes probably contributed mainly by Caucasian males”, according to the paper, -34 Asians (originated from China, Vietnam, Laos, the Phil- ippines, Indonesia and Tonga), -46 Caucasians (originating from Europe, North Africa, and the Middle East), -21 aboriginal Australians, and -26 aboriginal New Guineans. An average of 370 restriction sites were surveyed, repre- senting about 9% of the 16,569 base-pair human mtDNA ge- nome. The authors estimated the extent of nucleotide sequence divergence for each pair of individuals, which ranged from 0 to 1.3 substitutions per 100 base pairs, with an average sequence divergence of 0.32%. However, the authors wanted to show that these differences were the largest ones in the Africans, t here- fore all five populations were divided into “region-specific clusters”, taking into account pairwise differences in each pop- ulation. It turned out that: -46 European mtDNA split to 36 clusters, -34 Asian mtDNA split to 27 clusters, -21 Australian mtDNA split to 15 clusters, -26 New Guinean split to 7 clusters, -However, 20 African mtDNA have postulated to be in only one cluster, “assuming that Africa is the source, there is only one African cluster” (quotation from the paper). Those who understand what the authors did-probably would not want to read further. Because then the authors began to calculate which population is the oldest one, and this was the main purpose for splitting the populations into clusters. The idea was to calculate age of populations, taking into account the divergence between clusters within each population, and the Africans gave, of course, the highest “age”, since they were postulated to have only one cluster. As a result, the diversity (mean pairwise distance within clusters) was as follows: -Africa 0.36% -Asia 0.21% -Аustralia 0.17% -New Guinea 0.11% -Еurope 0.09% Then the authors translated those “divergencies” into years, namely when the respective territories were populated (average age of clusters). For these, the following “calibrated” chrono- logical indications were taken: a minimum of 30,000 years ago for New Guinea, 40,000 years ago for Australia, and 12,000 ago for the New World; from this, the mean rate of mtDNA divergence between humans occurred between 2% and 4% (that is 2 - 4 mutations per each 100 nucleotides) per million years. From here, the authors calculated the average age of clusters as -Africa 90 - 180 thousand years -Asia 53 - 105 -Аustralia 43 - 85 -New Guinea 28 - 55 -Еurope 23 - 45 It was done in a rather crude way and using a circular argu- ment, however, the authors obtained a reasonable set of ages (partly due to the circular argument). As it will be shown below, other authors, including the author of this paper, obtained more direct figures for the African lineages to be started at 141,500 ± 15,600 ya (Cruciani et al., 2011), 160,000 ± 12,000 years ago (Klyosov and Rozhanskii, 2012a), plus a few archaic African lineages (haplogroups A0 and A00) at about 180,000 and 210,000 ya, respectively; Asian and European lineages com- bined (haplogroup BT) at about 64,000 ya (ibid.), Australia—at about 45 - 5 0,000 ya, and the most ancient bone residues of AMH in Europe are dated by about 45,000 ya (Benazzi et al., 2011; Higham et al., 2011). C learly, t he authors (Cann et al., 1987) have done their calculations with accuracy plus-minus 100%, however, they got a reasonable pattern. In the same way the authors (Cann et al., 1987) calculated that the common ancestor of all surviving mtDNA types existed 143 - 285 kya, and since the common ancestor of all African mtDNA lived, according to their calculations, 90-180 kya (that is it was younger, though withing the margin of error), then “its founders may have left Africa at about the same time”. That is, the authors (Cann et al., 1987) have calculated t hat non-Africans arose from a more recent common ancestor than the African one, and postulated (sic!) that their ancestor came out of Africa. The authors concluded, as indicated in the Ab- stract, that one woman, a common ancestor of all mtDNA, “is postulated (!) to have lived about 200,000 y ears ago (this is a transformation of the value of 143-285 thousand years ago— AK), probably (!) in Af rica”. This was the beginning of an avalanche of similar papers, similar in types of justifications of their conclusions. Very soon that concept spread widely in the Western media and was cheerfully supported by the mass media, by population genetic- ists, in scien ce in general and among the “masses”. In almost no time a general understanding was that the “Out of Africa” theory was totally, completely, and irreversibly proven by strict scientific means. In reality, there were practically no other ge- netic justifications. Alternative Explanations, and Typical Mistakes of the “Out of Africa” Concept This paper shows that dozens of academic genetic articles which “support” the Out of Africa concept suffer from a simple set of common mistakes. Data in none of them actually support the concept. Most of the papers have good data and at least questionable interpretations. Some h ave erroneous da ta and interpretations. How could it have happened? Apparently, lack of critical thinking. Lack of desire to analyze alternative solu- tions. All which define good science. A principal mistake which constantly haunts population ge- neticists is that they typically take a higher “genetic diversity” as an indication to the ancestry, compared to those populations which ha ve lower “genetic diversity”. For example: -Hellenthal et al. (2008), “the haplotype diversity is highest in Africans,” -Campbell and Tishkoff (2010), “Africa not only has the highest levels of human genetic variation in the world but also contains a considerable amount of linguistic, environmental and cultural diversity.” The list in fact can continue almost infinitely. In reality, highest diversity does not by any means represent a homeland, unless the homeland ha s been totally isolated and has not been affected by populations from outside. New York City has a higher diversity than Boston, but that doesn’t mean that New York City is a homeland for Boston. Diversity quite commonly is a result of mixing, and Africa is a home of highly mixed populations, including quite ancient and rather recent popula- OPEN ACCESS 20 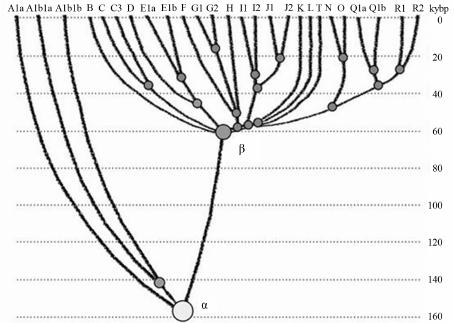 A. A. KLYOSOV tions, migrated to Africa in the last few millennia (see below). “Into Africa” migrations greatly increase the diversity there. Bearers of R1b-V88, which arrived in Africa on its migratory way along the Mediterranean Sea (Cruciani et al., 2010) and their descendants who currently reside in Cameroon and Chad, also contributing to diversity in Africa. So, diversity by itself does not support the idea of a common homeland in this context. Practically the same thing is expressed in a recent paper by Rito et al. (2013): “… the region of highest diversity may differ for different marker systems and, despite having been commonly used as such, is not necessarily informative about the place of speciation.” Some academic papers appear from time to time which de- scribe migrations “into Africa”. For example, a recent paper in Nature in August 2013 (Hayden, 2013) describes migrations into Africa 3000 and 900 - 1800 years ago. Did they add to the “genetic diversity” in Africa? Sure they did. Furthermore, they migrated to the Sub-Saharan region, where Cann et al. (1987) sampled mtDNA and found a “high genetic diversity.” The same is related to a common mistake which is exempli- fied by Atkinson (2011) in a recent article in which he asserts that “human genetic and phenotypic diversity declines with distance from Africa ··· underpinning support for an African origin of modern humans ··· local language diversity ··· points to parallel mechanisms shaping genetic and linguistic diversity and supports an African origin of modern human languages.” This effort to shore up the Out of Africa hypothesis also fails. Since haplogroups A and BT diverged from the same α-haplo- group (Figure 1), this does not mean tha t the younger lineage descended from the older. Our research shows that they parall el to each other such as two te eth in the proverbial fork. They descend not from each other, but from a common ancestor who in this particular case was the α-haplogroup (A1b in the current classification). Haplogroup R1a (20,000 ya) and haplogroup R1b (16,000 ya) (Klyosov & Rozhanskii, 2012b; Klyosov, 2012), arose in Cen- tral Asia (ibid.). Both are younger than haplogroup A (~160,000 ya) but they are not descended from haplogroup A. A boy is not a descendant of hi s olde r brother. They b oth hav e a common ancestor, which is their father. The fact that the In- do-European (IE) languages are younger than the click lan- guages does not prove that IE languages descended from click languages or that Africa is a homeland of the languages of man. Obviously, to consider “genetic diversity” one should know the population genesis, their history, and not formally and “li- nearly” compare their “diversity”. This is a systemic problem with population studies. A lower diversity in non-Africans compared to Africans is explained in Figure 1. About 64,000 ya (64,000 ± 6000 ya) non-Africans went through a severe population bottleneck. Through some cataclysm, yet unidentified, almost all non-Af- ricans have perished, and only a small group of them survived. Y-chromosomal genealogical lineages of practically all present- day non-Africans and some Africans (haplogroup B through T) coalesce to that small group of survivors. Two hypothes es have the highest rank—Toba eruption ~70,000 ya, the most violent volcano eruption on Earth for the last 100,000 years, and cool- ing (probably but not necessarily connected to the eruption) in the Northern Hemisphere. According to climatologists, a cata- strophy rank of the cooling is significantly higher compared to the Toba eruption. Figure 1 shows that the “diversity” of the left branch (the Figure 1. Haplogroup tree of the H. sa piens Y-chromosome derived from haplo- types and subclades. This diagram shows the most recent common ancestors (TMRCAs) of H. sapiens as described in this study. To pre- pare this tree, 7415 haplotypes from 46 subclades of 17 major haplo- groups have been analyzed. The timescale on the vertical axis shows thousands of years from the common ancestors of the haplogroups and subclades. The tree shows the α-haplogroup, which is ancestral to both the African and non-African haplogroups (equivalent to A1b in the curreny classification), its co mmon ancestor lived 1 60,000 ± 12,000 ya), and the β-haplogroup, which is equivalent to haplogroup BT in the current classification (its common ancestor lived 64,000 ± 6000 ya). Haplogroups F through R (which includes T) represent Europeoids (Caucasoids) who arose ~58,000 years before the present. The Mongo- loid and Austronesian haplogroup C split ~36,000 ya; the Middle East- ern haplogroups D and E split ~42,000 ya (Klyosov & Rozhanskii, 2012a). The tree does not show the archaic African lineages A00 and A0 (the last has replaced the lin eag es A1 b o n th e lef t), wh ich are sho wn on the updated tree in Figure 4. Africans today) is higher compared to the right branch (non- African), however, the African branch is not ancestral with respect to non-African branch. Both have a common ancestor, the alpha-haplogroup of about 160,000 ya. As one can see, the figure explains all the data by Cann et al. (1987) without any “Out of Africa” concept. Other proofs of the validity of Figure 1 will be shown. Popularization of the “Out of Africa” Concept Soon after the Cann et al. (1987) publication a figure of 70,000 ya appeared as a date of “Out of Africa” exodus of AMH. That figure became a cliché in almost every article in which the Out of Africa hypothe sis has been mentioned. In fact, the dating was “floating” as well. In reality, none of the dates was a product of serious calculations, if any calculations at all. There was not any methodology of calculations of the “Out of Africa” date, except those with a margin of error plus-minus 100% (at least), and even those were based on crude assump- tions, as it was shown above with the example of the Cann et al. (1987) paper. Here are a number of other examples of the dat- ing: “50 thousand years ago” (Jobling & Tyler-Smith, 2003). “50 thousand years ago” (Thomson et al, 2000). “50 - 60 thousand years ago” (Shi et al., 2010). “50 - 60 thousand years ago” (Mellars, 2011). “50 - 70 thousand years ago” (Hudjasov et al., 2007). “50 - 70 thousand years ago” (Stoneking & Delfin, 2010). “60 thousand years ago” (Li & Durbin, 2011). OPEN ACCESS 21 A. A. KLYOSOV “60 thousand years ago” (Henn et al., 2011). “60 thousand years ago” (Wei et al., 2013). “60 - 70 thousand years ago” (Ottoni et al., 2010). “60 - 80 thousand years ago” (Forster, 2004). “54 ± 8 thousand years ago” (Forster et al., 2001). “60 thousand years ago” (Stewart & Stringer, 2012). “45 - 50 thousand years ago” (Fernandes et al., 2012). “50 - 65 thousand years ago” (Behar et al., 2008). “50 - 60 thousand years ago” (Cann, 2013). “60 thousand years ago” (Chiaroni et al., 2009). “50 - 75 thousand years ago” (Patin et al., 2009). “50 thousand years ago” (Edmonds et al., 2004). “45 thousand years ago” (Moorjani et al., 2011). “50 - 70 thousand years ago” (Xue et al., 2005). “70 - 80 thousand years ago” (Majumder, 2010). “40 thousand years ago” (Campbell & Tishkoff, 2010). “50 thousand years ago” (Poznik et al., 2013). “60 thousand years ago” (Rito et al., 2013). “55 - 70 thousand years ago” (Soares et al., 2009). “between 40 and 70 thousand years ago” (Sahoo et al., 2006). “between 35 a nd 89 thousand year s ago” (Underhill et al., 2000). “between 80 and 50 thousand years ago” (Yotova et al., 2011). “between 50 and 100 thousand years ago” (Hublin, 2011). “between 27 - 53 and 58 - 112 thousand years ago” (Carrigan & Hammer, 2006). “70 - 60 thousand years ago” (Curnoe et al., 2012). “~110 thousand years ago” (Francalacci et al., 2013). “200 thousand years ago” (Hayden, 2013). In fact, there is no date which would have been justified and supported by experimental data. Those datings above do not make any sense. They were just made up. Let us slow down with the critique, and take a look at what do we have, regarding Africans and non-Africans. If present- day non-Africans did not descend from Africans, then who were their ancestors? Where did Africans come from? Whe re did non-Africans come from? Figu re 1 shows that they had a common ancestor (Y-chromosome) around 160,000 years ago. Archaeology and paleoanthropology data of African skeletal residue would not tell us much about African and non-African origins. The y are interesting and educational, however, we do not know if bearers of those bones had offspring, and not just offspring but survived until our times. If not—they are not our ancestors. T he y might belong to terminated lineages. Unless we know haplogroups and haplotypes of those skeletal remains, we know nothing on their contribution to the present d ay human- kind. Besides, we do not know where bearers of those skeletal remains came from. It might well be that their close ancestors have migrated to Africa from elsewhere. Indeed, if “Out of Africa” was possible, equally possible were “into Africa” mi- grations, and we know that they occurred. Many datings of ancient skeletal remains are incorrect, and some examples are given bel ow. Many skeletal residues claimed as Homo sapiens or “Anatomically Modern Humans” have noticeable archaic features, and their assignment to Homo sapiens is questionable or flatly incorrect (more about it in t he last section of this pa- per). Many findings concern not skeletal remains but settle- ments (claimed as those of Homo sapiens), sea or river shells, stone tools. It is often unknown who left those artifacts, and red ochre, found there, also does not mean much, since it is known that Neanderthals used ochre for their purposes. Neanderthals of Eurasia also employed stone tools. The question regarding common ancestors of Africans and non-Africans can be solved preferably by considering their DNA. When data obtained are supported by archaeology and anthropology, it is great, however, there are not much data; nonetheless, let us look at them. Data on DNA in the context of this stu dy can be considered in three ways, which ideally must be compatible. They are: 1) haplotypes and haplogroups of Y-chromosome of m en, 2) mtDNA, and 3) genome dat a . The last set of data is actually a pattern of practically irreversible mutations (SNPs) in the DNA, which can be interpreted with respect of evolution of humans, since it shows dynamics and direction of mutations in the course of evolution. For example, genomes of Neanderthal and present-day man show many identical mutations, which are also present in the DNA of chimpanzee. It means that those muta- tions were inherited from a common ancestor of chimpanzee and man. Therefore, if we have SNPs which we see in Nean- derthal but not in chimpanzee, it might mean that Neanderthal is our direct ancestor. Those mutations are either not found or they are questionable. The situation with Neanderthal as our ancestor is not clear and conflicting data obtained earlier are currently under reconsideration. Recently announced 1% - 4% as allegedly inherited in modern man from Neanderthal are also under reconsideration. Africans and non-Africans ha ve plenty of common SNP mu- tations from a common ancestor of Homo with chimpanzee. Those SN Ps are not particularly interesting for a question whether non-Africans have descended from Africans, or vise versa. Those common mutations should be filtered out. Another question does present a particular interest: do non-Africans have SNPs which are observed in Africans but not in chimpan- zee? The above is a critical question, an answer to which repre- sents a cornerstone in studying evolution of present-day hu- mankind. An answer thus far is a resounding no. What Do Actual Exp eri men tal Data Tell Us? An Alternative and More Plausible Explanation What do actual experimental da t a tell us on SNP mutations in Africans and non-Africans? Let us begin with haplotypes and haplogroups of man extrapolated to times more than 100,000 years ago; and we have to extrapolate since we do not have experimental data on excavated haplogroups and haplotypes of those times, in Africa and elsewhere. That is why the “Out of Africa” concept and its “supportive” data are always based on conjectures, extrapolations, and as we show in this paper, those conjectures are alway s based on very questionable, or flatly wrong assumptions. Furthermore, “Out of Africa” proponents seem to not ha ve considered eq ually reasonable, alternative explanations, as it is shown below. We do not have experimental data for haplogroups and hap- lotypes at more than 100,000 ya, because it is a very complex technical (and scientific) task; in the course of those times hu- man DNA is degraded almost completely, chemically and par- ticularly under effect of microorganisms. DNA of a Neander- thal (female) of 45,000 ya was isolated and sequenced; however, to isolate and sequence, even partially, D NA of a man of 160,000 ya is a task, orders of magnitude more complex. How do we do those “extrapolations”? One needs to deter- mine haplotypes in a random selection of contemporary men, of OPEN ACCESS 22 A. A. KLYOSOV the Y-chromosome under study. A selection should be taken from among bearers of certain haplogroups or their subclades, that is among men which are united by a certain set of SNPs. They are relatives, according to their common haplogroups or subclades, and a set of their haplotypes allows us to calculate rather reliably when their common ancestor lived. For example, there is a good representative group of males among contemporary Africans, which according to the ISOGG- 2014 (International Society of Genetic Genealogy) classifica- tion form subclade A1b1b2b (third line from the bottom in the tree of haplogroups A00/A0/A, see below). This tree shows the hierarchy of haplogroup A subclades, t hat is, in a way, the evo- lution of haplogroup A. The tree evolves via branching: the first one which split off the trunk was the most ancient haplogroup A00, its sub-branches (subclades) are unknown as yet. The trunk continues with haplogroup A0-T, which split into two subclades, A0 and A1; A1 in its turn split into A1a and A1b; A1b split into A1b1 and BT. Haplogroup BT (beta-haplogroup in Figure 1), as it will be shown below, is very remote from haplogroups of the series “A”, bearers of which now live in Africa. Those a re haplo- groups A00, A0, A1a, and A1b1 with subclades, shown in the haplogroup A tree below (to which haplogroup BT is added in the lower part). Those four haplogroups can indeed be consi- dered as the African ones—by origin or by arrival to Africa more than 100,000 years ago. Other three haplogroups. A0-T, A1, and A1b, have not been found in Africa (or elsewhere), we see them only deep back in time, as ancestral haplogroups, each split into an African lineage (A0, A1a, and A1b1, respectively), and presumably non-African lineages (A1, A1b, and BT). There is no data which allow to call A0-T, A1, A1b, and BT as “African haplogroups”. Let us take a look again, more specifically, how the haplo- group tree branches, how each branch forms a fork, and how one “tooth” of each fork leaves (migrates) to Africa, and anoth- er stays elsewhere, and forms a next fork. In other words, mi- grations to Africa have occurred by waves. The whole pattern can be presented as a non-African “trunk”, which has produced splits, one side of which were leaving to Africa. Those splits can be tentatively numbered: - Split 1: The trunk, which arose from common ancestors with primates (chimpanzee, gorilla, orangutan, macaqua, etc.) splits into Neanderthal (300 - 600 kya) and the rest of the trunk; Neanderthal—as far as we know—was not of an African Homo species, and the rest of the trunk, therefore, was also presumably a non-African Homo species; - Split 2: Around 210 kya the trunk split into the most ancient (thus far) Homo sapiens haplogroup A00 (now African) and the rest of the trunk (presumably non-African); all bearers of A00, identified thus far, belong to an African tribe Mbo in Cameroon, or are Afro-Americans, presumably des- cended from the same tribe (Mendez et al ., 2013); t he last paper did not mention their anthropology; - Split 3: The trunk then reaches haplogroup A0-T (presume- bly no n-African), which around 180 kya split into haplo- groups A0 (now African) and A1 (presumably non-Afri- can); - Split 4: Haplogroup A1 split into А1а (presumably mi- grated to Africa, though A1a bearers now live in Africa, in the Midle East, an in Europe) and A1b (presumably stayed not in Africa); - Split 5: Haplogroup A1b split into A1b1 (presumably mi- grated to Africa) and ВТ (beta-haplogroup in Figure 1), which was not an African haplogroup and it is very remote from them. This series of splits was never under consideration by pro- ponents of the “Out of Africa” concept. It has never been pub- lished and discussed. Now, let us present an important statement. Splits 3, 4, and 5 occurred from haplogroups A0-T, A1, and A1b, respectively. -Split 3 produces A0, bearers of which now live in Africa, and A1, bearers of which are not identified as yet. Non-Afri- cans descended from A1, and have not descended from African A0; A0 mutations are absent in non-Africans. -Split 4 produces A1a, bearers of which now live in Africa, and A1b, bearers of which are not identified as yet. Non-Afri- cans descended from A1b, and have not descended from Afri- can A1a; A1a mutations are absent in non-Africans. -Split 5 produces A1b1, bearers of which now live in Africa (as well as in the Middle East and Europe), and BT, from which all non-African haplogroups descended, including haplogroups the most numerous in Europe-R1a, R1b, I1, I2, N1c1. Most Europeans and Asians descended from BT, and have not des- cended from “African” A1b1. A1b1 mutations are absent in most of non-Africans. So, what is left from the “Out of Africa” concept? In order to “prove” that anatomically modern ancestors of all people on the planet came out of Africa, proponents of the OOA concept claim all the three node haplogroups-A0-T, A1 and A1b “African”. The fact that none of them is found in Africa is of no concern for the proponents. Af ter such a claim the proponents say—look, all non-African haplogroups des- cended from the African A0-T, A1 and A1b. The “Out of Afri- ca” concent is “proven”. This, of course, is not a proof. It is much more likely that those three haplogroups are not African, and their bearers lived elsewhere. Then a connection between an ancestor of a pale skin Neanderthal (see below) and pale skin contemporary non- Africans is easy to explain. Equally easy to explain migrations of bearers of haplogroups A0, A1a, A1b1 to Africa, after they split from the respective non-African haplogroups A0-T, A1, A1b. Equally easy to explain huge time (mutational) distances between African and non-African haplogroups, because they descended from ve ry ancient (160,000 ya) common ancestors, and not from each other; in the latter case the distances between their haplogroups would be around 60 - 70 kya, and in reality they are 250 - 300 kya, as shown below. Non-African lineages principally could not descend from African ones, when the time between them reaches 250 - 300 thousand years. The propo- nents of the OOA concept constantly announce that the OOA exodus was 60 - 70 thousand yea rs ago. They do not know that the actual distance there is 4 - 5 times higher. Therefore when I describe splits/forks above, I write “pre- sumably non-African haplogroup” A0-T, A1, A1b. As it is shown below, haplogroup BT is very remote time- wise from haplogroups A and their subgroups. From the haplo- types analysis it seems that a whole series of haplogroups, namely A00, A0, A1a, and A1b1 ha ve moved to Africa from non-African regions, locations of which remain unknown; most likely they might have been in Europe or Asia. In other words, there have been at least four “into Africa” migrations since some 200,000 ya (haplogroup A00), then about 180,000 ya (haplogroup A0), the n about 160,000 (haplogroup A1a), and OPEN ACCESS 23  A. A. KLYOSOV then about 70,000 ya (haplogroup A1b1). Of course, there have been migrations into Africa at later times, described for exam- ple, by Hayden (2013), which increased a high genetic diversity in Africa. Those haplogroups of the A series and their down- stream subclades, such as A0a, A0b, A0a1, A0a2, A0a1a, A01a1b (see flowchart below), can be considered as “African”, albeit not necessarily by appearance there, but since they live in Africa now. Let us consider subclade A1b1b2b-M13, which in the Hap- logroup A FTDNA Project http://www.familytreedna.com/public/Haplogroup_A/defaul t.aspx?section=yresults is the most numerous. The subclade splits into two main branches of A1b1b2b, Arabic and European. Who was a common ancestor of these two branches and where did he live, we do not know, however, as we have assumed above, he might have lived in Europe or Asia, and his descendants c ould have moved to Africa. Or he could equally have li ved in Africa and his A common ancestor of Homo and chimpanzee (...) • Neanderthal (...) • A00 L1236 • A0-T L1085 • • A0 V148, V149 • • A0a L979 • • • A0a1 V150 • • • • A0a1a V151, V169, Р114 • • • • A0a1b L1289 • • • A0a2 L981, V203 • • • A0b L1035 • • A1 V168 • • • A1a M31, P82, V4 • • • A1b P108, V221 • • • • A1b1 L419 • • • • A1b1a V50 • • • • A1b1a1 M14, L968 • • • • • • A1b1a1a M6, M196 • • • • • • A1b1a1a1 P28 • • • • • • A1b1a1a2 L963 • • • • • • • A1b1a1a2a M114 • • • • • • • A1b1a1a2b P262 • • • • A1b1b M32 • • • • • A1b1b1 M28 • • • • A1b1b2 V160 • • • • • A1b1b2a M51 • • • • • • A1b1b2a1 P291 • • • • • • • A1b1b2a1a P102 • • • • • • • A1b1b2b M13 • • • • • • A1b1b2b1 M118 • BT V31, M91, SRY10831.1 descendants moved to South Arabia and Europe. The general logic of the subclade t ree, described above, makes it unlikely that a co mmon ancestor of A1b1 arose in Africa (see Split 4 and Split 5 above). What we do know is that available haplo- types of the A1b1b2b-M13 subclade represent a rather shallow (time-wise) branch, which apparently has passed a population bottleneck. However, the available haplotypes provide a va lua- ble source of information, since they place their branch into the space of all haplotypes of mankind. Analysis of those A1b1b2b haplotypes, performed with help of very “slow”, most stable 22 markers of Y-chromosome (Klyosov, 2011; Klyosov and Rozhankii, 2012a), shows that a common ancestor of the Arabic branch of the subclade A1b1b2b had the following base haplotype: 12 11 11 9 11 10 10 9 12 12 7 12 8 0 13 11 16 9 14 9 11 11 (A1b1b2b, Arabic) while a common ancestor of the European branch had the fol- lowing: 12 11 11 9 11 10 10 9 12 12 7 10 8 0 13 11 16 10 14 9 11 11 (A1b1b2b, European) Three mutations between them place their common ancestors to 3/0.006 = 500 536 generations apart, that is 13,400 years, and their common ancestor lived about 13,400/2 = 6700 years ago plus an average “age” of the two branches, which finally makes about 7170 years to their common ancestor (principles of calculations are described in [Kly osov, 2011; Kly osov and Roz- hanskii, 2012a]; 0.006 is t he mutation rate constant for the 22 marker haploty pes, in mutations per haplotypes per conditional generation of 25 years), with margins or error pl us -minus 5%. Let us compare the above two base haplotypes with that for the African haplogroup A00 (it was calculated based on 95 marker h aplotypes of men of Mbo tribe, who live in Cameroon [Mendez et al., 2013]): 13 11 12 10 11 16 10 9 14 14 8 8 8 9 12 11 12 8 12 12 11 11 (A00) Here we see a difference in 30 and 29 mutations, respectively, which places their common ancestors (A00 and A1b1b2b) apart by a minimum of 286 - 308 thousand years, therefore, their common ancestor (that is, of haplogroup A00) lived about 210 thousand ya. A number of mutations correlates with time not linearly, but exponentially, due to back mutations, and the cal- culations should include a correction for back mutations (Klyosov, 2009, 2012). If we compare those base haplotypes for A00 and two branches of A1b1b2b with that of haplogroup B (Klyosov and Rozhanskii, 2012a) 11 12 11 11 11 10 11 8 16 16 8 10 8 12 10 11 15 8 12 11 12 11 (B) then we see 29 mutations between A00 and B, and practically the same—29 and 27 mutations, respectively—between B and the A rabic and European branches of A1b1b2b. Thi s again corresponds to 286 - 248 thousand years between common ancestors of haplogroups A and B. This tremendous distance between haplogroups A and B cannot allow haplogroup B to descend from haplogroup A. The two have a common ancestor, who lived about 160,000 years ago (see Figure 1). Thus, haplogroup B is very remote from haplogroup A, by 27-29-30 mutations. However, it is not very remote from Euro- pean (to some extent) haplogroups R1a and R1b, by only 12 and 10 mutations, respectively, from their subclades R1a-Z280 and R1b-M269: 11 12 13 11 11 12 11 9 15 16 8 10 8 12 10 12 12 8 12 11 11 12 (R1b-M269) 12 12 11 11 11 11 11 8 17 17 8 10 8 12 10 12 12 8 12 11 11 12 (R1a-Z280) This again is explained by Figure 1, in which haplogroups B and R sit in the same cluster, and all of them are remote from haplogroups A. The base haplotypes of R1a-Z280 and R1b-M269 are sepa- rated by only 8 mutations, whic h corresponds to t he time to their most recent common ancestor, that is haplogroup R1, of OPEN ACCESS 24 A. A. KLYOSOV about 26,000 ya. A c ommon ancestor of haplogroup B lived about 50,000 ya, and he did not descent from haplogroup A. Haplogrops A and B possess two independent DNA-genealogy lineages, which arose from the alpha-haplogroup 160,000 ya. Debates of the Last Two Years over Figure 1 above When the paper which contained Figure 1 and its interpreta- tion was publi shed in May 2012 in Advances in Anthropology, it was met with total silence in academic media. There was not a single objection or critique in academia, or any positive comment, despite the paper having been downloaded 10,895 times (as of January 18, 2014, according to the AA counter). However, active discussion among population geneticists took place on related Internet forums. Specif ically, four main con- clusions of the paper met negative reception among the popula- tion geneticists: 1) African and non-African DNA lineages split about 160,000 ya, and there is a large corresponding “genetic” (muta- tional) distance between them, 2) non-African DNA lineages did not descend from “African” haplogroups A00, A0, A1a, and A1b1, 3) haplogroups A1, A1b, and BT did not come “Out of Afri- ca”, and 4) there were not a ny “Out of Africa” migrations which would have been derived non-African AMH from African AMH. In reality, they both split from their common AMH about 160,000 ya. There have been many “out of Arica” and “into Africa” reciprocal migrations, as well as many “out of Europe” and “into Europe” migrations, and “out of Asia” and “into Asia” migrations, and none of them can be considered as a deci sive one or the only one in shaping AMH, or Homo sa- piens for that matter. The most active forum critics have announced that Figure 1 and the respective conclusions totally co ntradict the “Out of Africa consensus”, and equally conflict with all published dia- grams, flowcharts, and haplogroup trees, obtained with Y-DNA, mtDNA and genome studies. They hav e also announced that non-African lineages descended from haplogroups A (and their subclades), hence, African. They have announced t hat Figure 1 disagrees with the evolution of mtDNA, in which ancestors of non-African (current) lineages also came out of Africa, which is “logical”, since male and female haplogroups should have been coming from Africa together, concurrently. Each and all of those announcements were mistaken. As it happens often, the opponents did not wa nt or could not under- stand that ther e were not any contradictions or conflicts. The data whic h they quoted were generally right, but the opposition interpretations of the published “Ou t of Africa” studies were incorrect. Let us consider it more attentively. The Latest Book Human Evolutionary Genetics (by Jobling, Hollox, Hurles, Kivisild, and Tyler-Smith, 2014) Presents Right Data but Wrong Conclusions The new book, Human Evolutionary Genetics, by Jobling, Hollox, Hurles, Kivisild, and Tyler-Smith (2014), chapter 9 “Origins of Modern Humans”, section “Mitochondrial DNA phylogeny”, p. 304. Quotation: “This has some striking features: Complete separation of African and non-African lineages”. T he next section—“Y-chromosomal phyloge ny”, p. 305. Quotation: “Although less detailed than the mtDNA phylogeny, its struc- ture shows close parallels: Complete separation of African and non-African lineages”. As we see, there are not any contradictions with Figure 1 above. However, the authors of the book employ old interpreta- tions, by quoting papers of 2000, on both mtDN A and Y- chromosome. They consider Y-chromosomal haplogroup B “African” (see calculations above that haplogroup B could not possibly be “African” by origin, since it is tremendously distant from haplogroups A), and continue that it “contains both Afri- can and non-African lineages”. Is it a valid argument? Haplo- group R1b also contains both African and non-African lineages (Cruciani et al., 2010), and many R1b bearers lived for thou- sands of years in Cameroon and Chad regions. It is not enough to say “African” without defining, did they arise in Africa or migrated there. Figure 1 shows that haplogroup B forms a cluster with non-African haplogroups, and we described above that hapl o- group B is very remote from haplogroups A. So why do the authors of the quoted book call haplogroup B “African”? Only because many haplogroup B bearers live in Africa now. Former Europeans live in America now, however, the y can hardly be defined as Native Americans. If haplogroups contain both Afri- can and non-African lineages, how may that be equated with “Out of Africa”? Why not—equally logically—“into Africa”? TMRCA for the branch conta ining both African and non-Afri- can lineages is described in the book as 52 ± 28 kya (mtDNA) and 31 - 79 kya (Y-DNA), while Figure 1 gives for the origin of BT haplogroup 64 ± 6 kya. There is not any contradiction. The same authors provide with the TMRCA for the entire mtDNA phylogeny of 172 ± 50 kya, for the entire Y-DNA phylogeny of 40 - 140 kya. Cruciani et al. in their recent work (2011) gave the TMRCA for African lineages of 141.5 ± 15.6 kya. Figure 1 above (Klyosov and Rozhanskii, 2012a) shows 160 ± 12 kya for the split between African and non-African lineages. There is not any conflict. With respect to Y-chromosome the authors of the book symmetr ically provide the same interpretations as those in the case of mtDNA. If a branch contains both “African DNA” (since their bearers live there now) and non-African DNA, it is not enough to claim that they migrated “Out of Africa”. The same conclusion, albeit in a more careful form, can be seen at the end of the book’s chapter. The authors write on the OOA— “Interpretations of almost all sources of evidence are hotly debated and it is difficult to identify a consensus view about many topics”. However, after it—“Genetic diversity is higher in Africa than on other continents, which is consistent with a longer period of evolution in Africa…”. Agai n, it is a typical mistake of population geneticists. It should have been add- ed—“… or a mix in Africa of many DNA-lineages from an- cient to recent times”. Mixing gives diversity, not necessarily “a longer period of evolution”. Another quote: “Most phylogenies of individual loci show a root in Africa…”. Do they? Phylogenies never show “a root”. They show a direction of evolution, unrooted. “… this is seen particularly clearly in the well-resolved phylogenies of mtDNA and the Y chromosome”. In reality, it is shown below that the well-resolved phylogenies of both mtDNA and th e Y-chromo- some do not indicate “a root in Africa”. It is a common and unjustified stretch, as Figure 1 above shows. The final conclu- sion of the authors “The current consensus view is therefore … an out-of-Africa model…” Is there really “consensus”? OPEN ACCESS 25 A. A. KLYOSOV An Apologetic Follow-Up of the Cann et al. (1987) Paper It is of interest to follow the development of “ideas” ex- pressed in the Cann, Stoneking and Wilson (1987) paper, which I have discussed above. A follow-up paper came out after four years (Vigilant et al., 1991), with two former authors, Stone- king and Wilson, along with three new co-authors, absent Cann. This new paper (1991) informs, that the Cann et al. (1987) proposal “that all contemporary human mtDNAs trace back… to the ancestral mtDNA present in an African population some 200,000 years ago” was at first “rejected because of confusion over conceptual issues”, and mentions “perceived weaknesses of the Cann et al. study”. Among those weaknesses the authors (Vigilant et al., 1991) count “it used an indirect method of comparing mtDNAs…; used a small sample made up largely of African Americans to represent Native African mtDNAs; used an inferior method … for placing the common DNA ancestor on the tree of human mtDNA types; gave no statistical justification for inferring an African origin of human mtDNA variation; and provided an inadequate calibration of the rate of human mtDNA evolution”. In other words, its authors recognized the weakness of the paper (Cann et al., 1997) tha t formed a ground for the “Out of Africa” concept. However, the concept was already accepted by the “consensus”, and it was too late to turn it back. Therefore, the 1991 paper aimed at throwing the Cann et al. (1987) paper out as a weak one, but justified the concept itself. So what was the “justification” aimed at showing that Afri- can mtDNA is ancestral with respect to the non-Afri can mtDNA in the follow-up paper? This is again a repetition of the common fundamental mistak e by the proponents of the OOA concept, that if one population is more ancient then the other, the firs t must be an ancestral with respect to the second one. My u ncle is old er than me, but he is not my ancestor. In conclusion of the paper (1991), the authors wrote: “our study provides the strongest support yet for the placement of our common ancestor in Africa some 200,000 years ago”. In reality, as the reader will now hopefully appreciate, that was the “strongest support” that the surviving branch of people who now live in Africa is more ancient than that of people who now live not in Africa (see Figure 1). This “proof” has nothing to do with an ancestry. For a verification of an ancestry, one has to compare the SNPs and hapl otypes of the populations. The authors of both the 1987 and 1991 papers did not compare any haplotypes, as well as most population geneticists presently do not. SNP Mutations Show That Non-Africans Are Not Descendants of Africans of Haplogroups A and B. Instead, They Have a Common Ancesto r of About 160,000 Years Ago Let us take a look at a recent paper by Scozzari et al. (2012) that some consider as an exemplary study of African genome and justification of the “O ut of Africa” concept. Indeed, the paper designed a new, upgraded African haplogroup/subclade tree with a transition to the non-African part of the tree, that is to CT haplogroup. The last part of the tree is represented in Figure 1 by the right-hand side containing a set of lineages, related to C to R2 haplogroups. Scozzari et al. call it “the recent ‘Out of Africa’ CT clade”. In conclusion, the authors claim— “Our data provide a highly resolved branching in the Afri- can-specific portion of the Y tree and support the hypothesis of an origin in the north-western quadrant of the African conti- nent for the human MSY diversity”. The Scozzari et al. (2012) tree is shown in Figure 2. Let us pay attention to some important features of the tree in Figure 2. It starts (rather, cont inues the evolution trunk of hu- man Y-chromosome) in the upper left-hand si de of the tree, and immedia tely reaches the first fork at haplogroup A0-T (the name of the haplogroup is not shown on the tree). We see a split of A0-T into haplogroups A1b (it is A0 now according to the new nomenclature) with its subclades, which go “up”, on the top of the t re e, and A1, wh ich goes “down”. In other words, we see a departure of a first African branch (A0) from the “main tree”. None of non-African groups (haplogroup CT, among them) descends from that departed haplogroup A0 (=A1b in the graph above). It should be noted that the Scozzari paper employs an obsolete nomenclature (of 2011), the current nomenclature (2013) uses A0 in place of the old A1b, with its SNPs V148, V149 and others, listed in the upper pa rt of the tree. Their positions are clarified in the haplogroup A tree above in the text (section “What do actual experimental data tell us?). The ne xt fork took place at haplogroup A1, into “African” haplogroups A1a and A1a1 with its downstream SNPs M31, Р82, V4, and others, on the one hand, and the res t of the tree, on t he other. None of non-African groups (haplogroup CT, among them) descend from the second “African” branch (A1a and its subclades). The third fork is represented by appearance of haplogroup A1b in its current nomenclature. It sends asi de “African” hap- logroups А2 and А3 (now they are A1b1a and A1b1b) with downstream subclades; currently both of them are covered by haplogroup A1b1 with its downstream SNPs V249/L419, which further splits apart into subclades A1b1a-V50 (former А2) and A1b1b-M32 (former А3), both are with their subclades. Among the latter is subclade A1b1b2b-M13, haplotypes have considered above, and consists of the Arabic and European DNA lineages. Another branch of said fork of haplogroup A1b is haplogroup BT, shown in the lower part of the tree in Figure 2. It is obvious that haplogroup BT does not descend from the “African” haplogroups having index “A”, since all of them are positioned in the upper part of the tree in Figure 2, which BT is in the lower part. The word “African” we have to place in quo- tation marks, since among their haplogroups and subclades are the A rabic and European haplotypes; Europeans are mainly from England, Ireland, Scotland, Turkey; Arabian branch is composed mainly by haplotypes from Saudi Arabia, as well as by haplot ypes from England, Switzerland, Finland, and other countries. One can notice, of course, that ancestors of bearers of the contemporary European and Asian haploty pes of haplogroup A could have migrated out of Africa; however, it can be equally suggested that they could have migrated into Africa. Therefore, those arguments do not support the “Out of A fr ica” concept, they are rather mute. For some mysterious reason only the “Out of Africa” alternative finds support among proponents of the concept. Figure 2 shows that the non-African haplogroup CT (th e lowest line in Figure 2) does not descend from “African” branches of haplogroup A, nor from haplogroup B with its sub- clades. CT leaves aside all the “African” haplogroups. If we follow the time direction (that is from left to right in Figure 2), the mutation (SNP) flow, leading to haplogroup CT, comes OPEN ACCESS 26 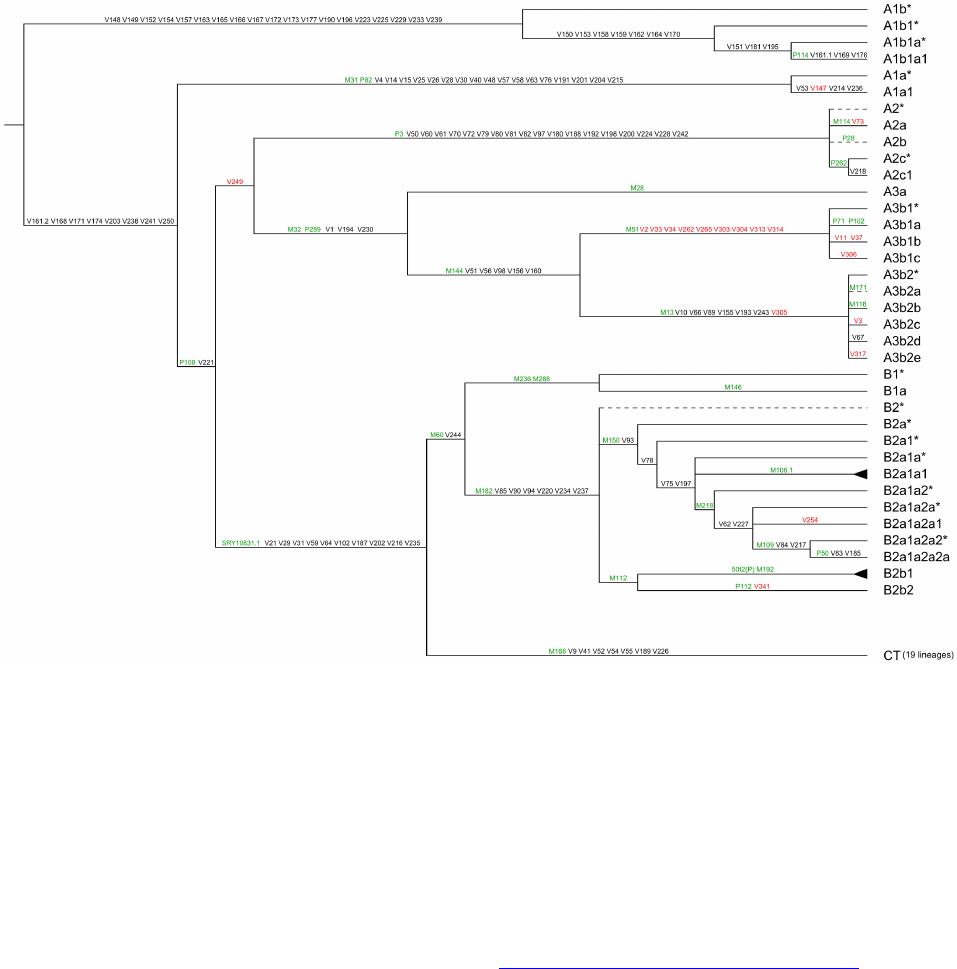 A. A. KLYOSOV Figure 2. The deepest portion of th e hu man Y-chromoso me accord ing to Sco zzari et al.’s (20 12) paper. The internal structure of some haplogroup B sub-clades is not shown (black triangles). One can see that more than a half of lineages are related to haplo group A, which th e autho rs (Scozzari et al., 201 2) call all “African”, apparently by definition. All other subclad es, except one, are related to h aplogroup B, which the authors also call “African”, again by definition. Haplogroup CT in the lower right-hand side contains, according to the authors of the study, 19 DNA lineages, all non-African. From Scozzari et al. (2012) with permission. from the main trunk of the Y-chromosome evolution, that is from the short bar in the upper left part of the tree, then passes through haplogroup A0-T (there are no indications that A0-T is an “African” haplogroup); then through haplogroup A1 (simi- larly, there is no indication that it is an “African” haplogroup); then through haplogroup A1b, which is also non-African; then through non-African haplogroup BT, and appears as haplo- group CT. Thus, all three “African” branches (А0, А1а, А1b1) on this evolution route split off from the “main trunk” (though it is a relative term here) via the respective deviations, forks, etc. We have considered the Scozzari et al. (2012) paper in such detail, because this kind of analysis is practically absent in the literature. The typical manner is to show a tree, such as in Fig- ure 2, and briefly announce that “it supports the ‘Ou t of Africa’ theory”. On the contrary, it does not support the “Out of Africa” concept. There is no contradiction in Figures 1 and 2. They both show the same dynamics of evolutionary development of the haplogroups, namely their repeated split into “African” and “non-African” branches. A difference between the trees in Figures 1 and 2 is only in that the Figure 2 shows subclades of haplogroup A in more detail, and Figure 1 shows haplogroups between B and R2 in more detail. Also, Figure 2 (from Scoz- zari et al.) lacks the chronological scale, that Figure 1 shows. In order to illustrate the similarity of Figures 1 and 2, Figure 3 shows both of them, next to each other, oriented in the same direction. Both trees shown in Figures 1 and 2 do not reveal recently discovered haplogroup A00, which arose around 210,000 ya. It is shown in Figure 4, along with updated nomenclature. The current nomenclature is given in accord with ISOGG-2014. http://www.isogg.org/tree/ISOGG_HapgrpA.html The pattern is now clarified. There is no contradiction or conflict between the trees of Y-chromosome haplogroups shown in Figure 1 (Klyosov and Rozhanskii, 2012a) and in Figure 2 (Scozzari et al., 2012). All data as well as all known others show a deep mutational difference and a deep separation between African and non-Af rican branches (subclades, haplo- groups), leading to a common ancestor of both, when moved back in time. These data do not reveal an “African” origin of AMH, at least during 30,000 - 500,000 ya. Instead, the data show a split between African and non-African lineages about 160,000 years ago. Let us take a look again at the above discussed paper by Scozzari et al. (2012). At which moment did a statement of the African origin of mankind appear there? What was it based upon? That “Out of Africa” statement appears as soon as in the OPEN ACCESS 27 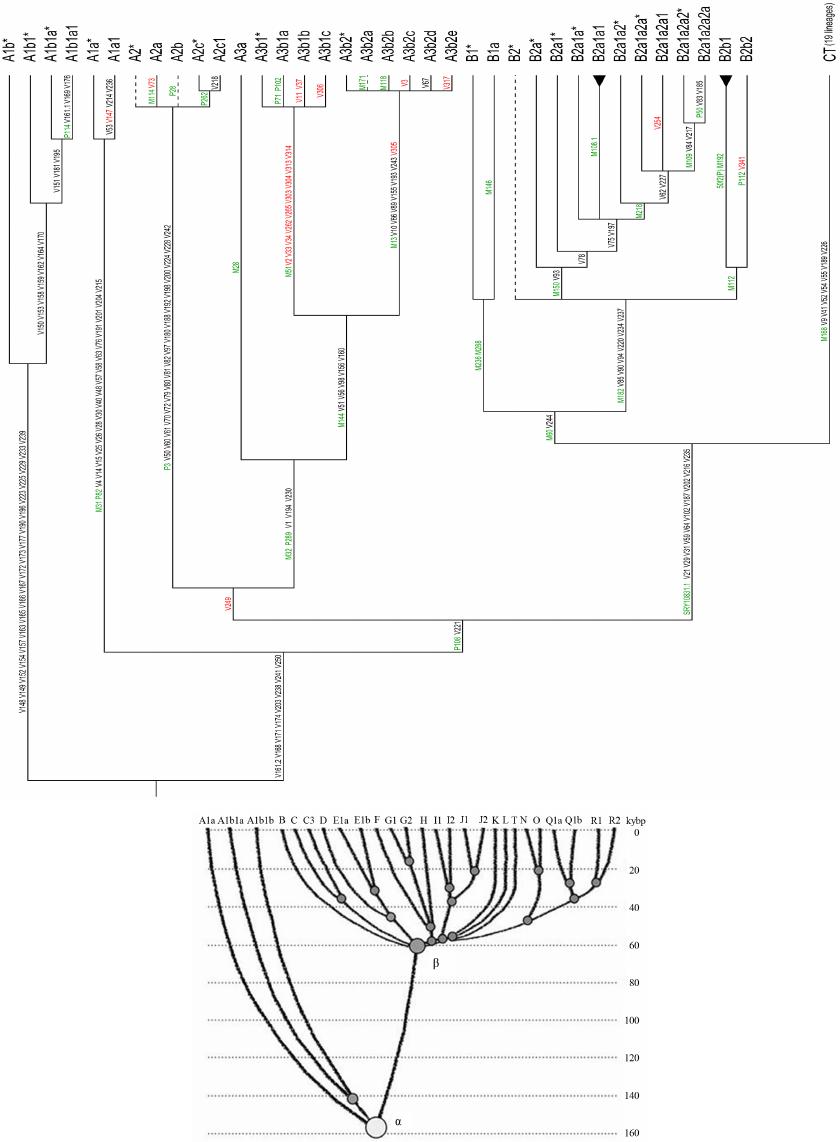 A. A. KLYOSOV Figure 3. A comparison of haplogroup and subclades of the “African” lineages (the left part of both trees, from Figures 1 and 2), and non-African (the tree at the top shows one combined lineage of haplogroup CT, and the tree at the bottom shows the extend ed haplogroup BT). The tree at the bottom was published by Klyosov and Rozhanskii in May 2012, and the tree at the top was pub- lished by Scozzari et al . in November 2012 . The tree at the top s hows that the series of “ African” haplogroup s split three times in a row and depart from the non-African haplogroups, and that non-African haplogroups BT and CT do not descend from the African ones. The tree at the bottom shows the same splits of “African” haplogroups from non-African (haplogroups on the right), and indicates that the split of “African” (by the current location) from non-African occurred about 160, 000 years ago. OPEN ACCESS 28 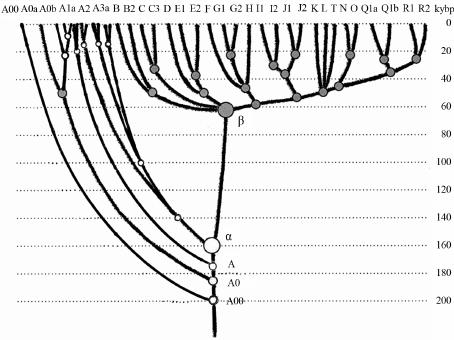 A. A. KLYOSOV Figure 4. Haplogroup tree of the H. sa piens Y-chromosome derived from haplo- types and subclades (see Figure 1) with an addition of the recently discovered haplogroup A00, and with an updated nomenclature of haplogroups and subclades compared with those in Figures 1 and 2. The timescale on the vertical axis shows thousands of years from the common ancestors of the haplogroups and subclades. The tree shows the α-haplogroup, which is apparently equivalent to haplogroup A1b in the current nomenclature, and is ancestral to both the African and non-African haplogroups (its co mmon ancestor lived 160,000 ± 12,000 ya), and the β-haplogroup, which is equivalent to haplogroup BT in the current classification (its common ancestor lived 64,000 ± 6000 ya). second paragraph of the introduction, and informs on “the re- cent ‘out of Africa’ CT clade [15]”. What was the reference [15] which allegedly proved the state ment? It was the paper “The 1000 Genomes Project Consortium (2010) A map of human genome variation from population-scale sequencing. Nature, 467, 1061-1073”, which does not mention “out of Africa” nor Africa at all. It does not mention “CT clade”. The Scorzarri (2012) paper authors (and others they refered to) have identified some African bearers of subclades: A0a1-V150, A0a 1a-P114, A0a1b-V151, A1a-M31, A1a-V53, A1b1a1a2a-M114, A1b1a1a2b-P262, A1b1b1-M28, A1b1b2a-M51, P71, A1b1b2b-M13, A1b1b2b1-M118, which they found in “low frequencies”: one in Algeria, two in Cameroon, one in Ghana, two in Niger, seven in South Africa, three in Ethiopia, two in Kenya. All these subclades are located in the upper part of the tree in Figure 2, and indeed belong to “African” sub- clades (or they migrated to Africa relatively recently), and NONE of BT or CT lineages descend from them. So how can they be claimed as “ancestral” to non-African lineages? None of actual bearers of haplogroups A0-T, A1, A1b have been found in Africa or elsewhere. So how they can be claimed as “African”? Just because they were assigned “A” letter in the index? As we see, all those conjectures by proponents of the “Out of Africa” concept are groundless. They assume that the currently African branches A00, A0, A1a, A1b1 descended from African haplogroups, and those were A0-T, A1, A1b. In reality, this has never been shown. Those haplogroups represented forks from which A0, A1a, A1b1 split and migrated to Africa. The authors (Scozzari et al., 2012) place the origin of haplo- group B to the sub-Saharan region (they write-“essentially a sub-Saharan clade). We have shown above that haplotypes of haplogroup B are tremendously remote from “African” h aplo- types of haplogroup A, and obviously have a non-African ori- gin. CT does not come from haplogroup B, as it is obvious from Figure 2. B and CT haplogroups have a common ancestor, that is haplogroup BT (see Figures 1-3). So, what was the major conclusion the authors (Scozzari et al., 2012) made? As the reader ca n guess, it was that haplo- group CT came from Africa, and not just from Africa, but “from the north-western quadrant of the African continent”. There, as the authors concluded, “an origin for the human MSY diversity” is located. How come? Their subclade tree (Figure 2) shows that CT did not descend from “African sources” that have been identified by the authors in (current) African chro- mosomes. All of them are in the upper segment of the tree. Besides, as Pontius Skoglund (2013) rightly noticed, “Patterns of genetic diversity can never provide strong evidence for t he origin of expansions”. After study ing the genetic relationships of populations of Khoe-San and other African populations such as Central African Pygmies, click-speaking East African popu- lations such as Hadza and Sandawe, the author (ibid.) wrote “this leaves the question of modern human origins in Africa unanswered”. Some consensus. A recent paper by Cruciani et al. (2011) fits the same pattern as that of other OOA proponents beginning with its title—“… The origin of patrilineal diversity in Africa”. On what scientific ground? On the same “ground” as explained above—they showed that African lineages are more diverse compared with those of non-Africans. Their SNP tree is almost the same as those shown in Figures 1-3, and their dating of the split be- tween African and non-African lineages of 141.5 ± 15.6 kya is within the margin of error with 160 ± 12 kya for the tree in Figure 1. The Cruciani et al. (2011) tree shows as follows: about 142 kya there was a split of haplogroup A1b (A0 in the new nomenclature) from the main trunk of the tree, then ap- proximately 108 kya the branch A1a split off, then, about 105 kya, A2 and A3 split off (A1b1a and A1b1b, respectively, in the new nomenclature). Subclades A0, A1a, A1b1a, and A1b1b the authors reasonably assigned to “African”, since they were found in four Africans. There are no objections to that, however, those subclades are not ancestral to non-African subclades. Then, about 75 kya, according to the authors, haplogroup BT arose, and 39 kya haplogroup CT arose. We know that neither BT nor CT descended from “African” haplogroups. However, since the branching points (fork haplogroups) are designated by the authors as “A” (A1a-T, A2-T), the authors consider them— as default—the “African” haplogroups, hence, “Out of Africa”. According to the authors (Cruciani et al., 2011), since “Afr i- can” haplogroups A0 (=A1b in the outdated nomenclature), A1a, A2 and A3 (obsolete nomenclature) arose 105 - 108 kya, and BT arose 75 kya, a (mutation-derived) distance between them should be about 30 - 33 kya. In fact, it is 248 - 286 thou- sand years (see calculations above). It clearly shows that hap- logroup B did not descend from the “African” haplogroups. It descended from their common ancestor about 160,000 ya (see above). The same conclusion follows from Figu re 2 by Scoz- zario et al. (2012), and from Figure 1 (Klyosov and Rozhanskii, 2012a). The above considerations and conclusions were related to Y-chromosome. An overall conclusion is that all available ex- perimental data do not show any “Out of Africa” principal mi- grations of AMH. There are no African SNPs in Y-chromo- somes of non-Africans. Non-African lineages and haplotypes are tremendously distant from the African. As practically all sources admit, there is “complete separation of African and OPEN ACCESS 29 A. A. KLYOSOV non-African lineages” (see above for actual quotes). However, it is interesting that by accepting all of it, population geneticists continue to repeat—“Out of Africa”. On what ground? No an- swer. Cross-Verification of the Haplogroup Tree in Figure 4 Employing SNPs There is one more approach to a verification of the tree in Figure 4, that is by employing SNPs (Single Nucleotide Poly- morphism). SNPs, which are practically irreversible mutations, are accumulating in Y-chromosome (as well in other pa rts of the DNA) in the course of evolution. The longer (time-wise) the distance between certain nodes (points) in the flowchart in Figure 4, the more SNPs are accumulated there. It is a random, statistical process, however, it can be quantified. The longest single evolution line in Figure 4 belongs to haplogroup A00, therefore, we can expect that t he present-day bearers of haplo- group A00 reveal the highest number of SNPs from the pre- sumed time when A00 arose (about 210,000 ya). The runner up should be haplogroup A0. The distance between alpha- and beta-haplogroups (that is between A1b and BT) should be rela- tively short compared to that in A00 and A0, about 100,000 years, and then SNPs kept accumulating upon transition from BT to more recent and contemporary haplogroups. This is what has been confirmed, considering and counting the SNPs. A list of the SNPs is given below; it is not the final one, and new SNP are being discovered from time to time. However, as a certain approximation the list works. Haplogroup A00 is the most ancient, the most archaic, its evolution line is the longest one. Its SNPs were discovered in men of the Mbo tribe in Came roon (Mendez et al., 2013): AF4, AF5, AF7, AF8, AF9, AF10, AF13, L990, L1086, L1087, L1088, L1091, L1092, L1094, L1096, L1097, L1100, L1102, L1103, L1104, L1106, L1107, L1108, L1109, L1110, L1111, L1113, L1114, L1115, L1117, L1119, L1122, L1126, L1131, L1133, L1134, L1138, L1139, L1140, L1141, L1144, L1146, L1147, L1148, L1149, L1151, L1152, L1154, L1156, L1157, L1158, L1159, L1160, L1161, L1163, L1233, L1234, L1236, L1284. Haplogroup A00 contains 59 SNPs. Sinc e A00 arose a bout 210,000 ya, then it can be estimated tha t one SNP-mutation occurs once in 3600 years, on average, in the considered frag- ment of Y-chromosome. Then, as we already know, haplogroup A0-T arose on the main trunk of the haplogroup tree, shown in Figure 4. It split into A0 and A1, the last one continued the trunk. A0 now live mainly in Africa. A0-T does not contain any A00 mutation. That is, A0-T did not descend from the African haplogroup A00. There is no data which would define A0-T as African. Indeed, it is an ancestor of the “African” A0, as well as of the non-African haplogroups, which passed via haplogroups BT, then CT and so on. There are 32 SNPs of haplogroup A0-T: AF3, L1085, L1089, L1090, L1093, L1095, L1098, L1099, L1101, L1105, L1116, L1118, L1120, L1121, L1123, L1124, L1125, L1127, L1128, L1129, L1130, L1132, L1135, L1136, L1137, L1142, L1143, L1145, L1150, L1155, L1235, L1273. Haplogroup А0 has 51 SNPs: L529.2, L896, L982, L984, L990, L991, L993, L995, L996, L997, L998, L999, L1000, L1001, L1006, L1008, L1010, L1011, L1012, L1015, L1016, L1017, L1018, L1055, L1073, L1075, L1076, L1077, L1078, L1080, V148, V149, V152, V154, V157, V163, V164, V165, V166, V167, V172, V173, V176, V177, V190, V196, V223, V225, V229, V233, V239 Haplogroup A0 is 8 SNPs short compared to A00, that is A0 is about 30,000 years younger compared to A00. Indeed, А0 arose 180,000 ya, haplogroup A00 210,000 ya. Haplogroup А1 has only 21 SNP discovered: L985, L986, L989, L1002, L1003, L1004, L1005, L1009, L1013, L1053, L1084, L1112, L1153, P305, V161.2, V168, V171, V174, V238, V241, V250 Unlike the sister haplogroup A0, bearers of A1 apparently did not survive until present time. It might be that they perished in that cataclysm, mentioned above, which caused the bottle- neck of the non-African population (see above). A summary lifetime of haplogroups А0-Т, А1, and A1b (the last one has only two SNPs, Р108 and V221) is equal to 32 + 21 + 2 = 55 SNPs, that is 55 × 3600 = 198 thousand years. Finally, haplogroup ВТ has 30 SNPs: L413, L418, L438, L440, L604, L957, L962, L969, L970, L971, L977, L1060, L1061, L1062, M42, M91, M94, M139, M299, P97, SRY10831.1, V21, V29, V59, V64, V102, V187, V202, V216, V235 This gi ves 30 × 3600 = 108,000 years for t he evolution of the beta-haplogroup since the time of the divergence with the “African” haplogroup A1b1 (160 ± 12 kya, see Figure 4) to the bottleneck of the BT population of 64 ± 6 kya. Those 108 kya is the distance between alpha- and b eta-haplogroups in Figure 4. Those are years which disappeared (between A1b, that is the alpha-haplogroup, and BT, which is the beta-haplogroup), from the evolution of the non-African BT haplogroups. Non-Africans popped up literally from nowhere 64 ± 6 thousand years ago, from a viewpoint of archaeology and anthropology, as fully developed AMH. They did not descend from Africans. It is important that those “lost” millennia in the evolution of the non-African DNA lineages, are cross-reproducible in the analysis of both STRs (haplotypes, based on t hat the flow chart in Figure 4 was composed), and SNPs, as it is shown above. Why we do not see those excavated DNA in Eurasia is still unknown. However, we know of excavated skeletal remains of Homo sapiens in the Middle East, dated between 160 and 60 kya, but their haplogroup was not examined. If it happens hap- logroup BT, with those SNPs listed above, the puzzle will be finally solved. Let us now look at mitochondrial DNA. There, as proponents of “Out of Africa” say, it is more than clear. “Non-African mtDNA all descended from African”. Is it so? Non-African mtDNA Do Not Descend from African mtDN A The recent pap e r by Behar et al. (2012) shows that in the very beginning of the mtDNA evolution tree there is a split of the main “stem” lineage into L0 (on t he right) and L1-L6 (on the left). All other haplogroups in the world descended from L1-L6. Haplogroup L0 embrace a large series of about fifty ancient African haplogroups, which were identified mainly in sub- Saharan Africa, among Khoisan-speaking populations, and also in Ethiopia and Tanzania (East Africa), in Mozambique (South- Eastern Africa), and among Pygmies. All ot her haplogroups are believed to descend from haplogroup L3, with an “age” of OPEN ACCESS 30 A. A. KLYOSOV about 60 - 70 kya, that is approximately the same as that of Y-chromosomal BT haplogroup. Therefore, it is quite plausible that haplogroup L3 did not come out of Africa, but migrated into Africa alo ng with bearers of Y-chromosomal haplogroup BT, or B. However, which conclusion is made by Behar et al. (2012)? Did they consider that altermative, as an equally valid conclusion? Of course not. Their conclusion was—“Out of Africa”. On what ground? Behar et al. (2012) did not even consider a deep separation of the African haplogroup L0 from all others despite their own figure, which shows that the African haplogroup L0 is not an- cestral with respect to all other mtDNA haplogroups (Figure 5 below). In reality, the separation of the African haplogroup L0 at 150 - 170 kya from apparently non-African haplogroups L1-L6 (Figure 5) and the subsequent migration of haplogroup L3 (60 - 70 kya) into Africa fit nicely to the separation (split) of African Y-chromosomal haplogroup of series A (160 kya) and non- African (the apparent age of 64 kya after passing through a population bottleneck, see above) with a subsequent migration of haplogroup B to Africa. More data has accumulated suggesting that there were a mul- titude of ancient migrations into (and apparently from) Africa. A recent paper in Nature (Hayden, 2013) presented data re- garding two migrations from Eurasia to Khoisan tribes, one 3000 ya to East Africa, another 900 - 1800 ya to South Africa. The author did not report which haplogroups were brought to Africa by the migrants. However, there is no doubt that those migrations have increased the “genetic diversity” among Khoi- sans, which is believed to be the highest in Africa. Another important feature of the data is that it points to the possibility of ancient “migrations into Africa”, which was not and could not be in any doubt. Sarah Tishkoff, one of the most active propo- nents of the “Out of Africa” concept, has greeted the “into Africa” new data, and called them “making sense”, since “arc- haeological and linguistic studies support it” (Nature, 29, 2013, p. 514). Herpes Virus Migrations Are Mistakenly Announced as Supporting the “Out of Africa” Concept A history with a recent publication which made a w ide press “Using HSV-1 genome phylogenetics to track past human mi- grations” (Kolb et al., 2013) presents a good ex ample how the aim at “Out of Africa” distorts data interpretations and conclu- sions of a study. The stu dy compared 31 complete and nearly complete “globally derived” (not exactly so) HSV-1 genomic sequences. Sources of the Herpes Simplex Virus were from Keny a (East A f rica), China, South Korea and Japan (East Asia), Seattle, San Francisco and Houston (USA), and Glasgow (UK). The authors have found that the phylogenetic tree forms six clusters (Figure 6). The clusters consist of the following sam- ples: I—10 samples total, of which 7 samples were from Seattle, one from “USA” (no detail provided), one from San Francisco, and one from Glasgow, II—6 samples total, of which one was from China, one from Houston (US), two from South Korea, and two from Japan, III—two samples from Kenya, IV—three samples from Kenya, V—seven samples from Kenya, VI—two samples from Kenya. The authors of the study rounded them up and reported that “the tree revealed a six clade pattern based on the geographical origin of the isolates”. According to the authors, “the Euro- pean/North American viruses fo rmed clade I, East Asian strains formed clade II and the East African viruses comprised clades III, IV, V and VI”. An origin of the “USA” strain was not dis- closed, and it is not clear if they were obtained from Europeans (by descent) or Native Americans. However, since the authors suggested that an outlier strain (Houston) placed into the East Asian cluster “may originally have been from the native Ame- rindian population”, it seems that the other US-based samples were from individuals of European descent. Nevertheless, the authors concluded that those clusters “mirror global human migrations” and support the “Ou t of Africa model” of human migration. The main reason for it, as always, was an argument of diversity, namely that “The East African derived viruses contained the greatest amount of genetic diversity and formed four of the six clades.” Let us consider, as we did above, where that “diversity” is coming from. As we have seen repeatedly above, the diversity is coming from a common ancestor of all those lineages, shown in Figure 6, which did not necessarily live in Africa. Some viruses migrate to Africa with their carriers, others migrate elsewhere. There is no indication that they descended initially from African viruses. Figure 6 shows again a series of cascade splits, and after each split the resulting downstream strains move in different directions. There is no “African origin” here. Let us follow the phylogenetic tree in more detail. A small bar on the very left represents a common ancestor of all the strains of the virus. We do not know where it lived. It could have been anywhere in the world. The first split results in two different strains: the contemporary Kenyan strain splits off, and another strain later underwent a cascade of splits (the lower node in Figure 6). The first “Kenya n” strain (the top two sam- ples in Figure 6) was not ancestral to any of the later strains considered in the study, including the European ones. The n ext split caused again a separation of populations of the virus, and, apparently, their human carriers, and cluster V in Figure 6 was formed, which was also found in Kenya. There were no non-African viruses descended from it; at least, t he y have not been observed in the st udy. The ne xt split produced virus E07, again found in Kenya. There were no non-African viruses descended from it. The authors mistakenly assigned it to cluster IV, however, it does not belong there. The next two splits produced clusters IV and III, again Ke- nya n, according to the authors of the study. European or Asian viruses did not descend from them, as well as, apparently, Eu- ropean and Asian populations which carried those strains. With that split the African (Kenyan) new strains were not observed in the study. There was nothing related to “Out of Africa” there. It was more likely “into Africa”, since the Kenyan strains were not ancestral to the European or Asian strains, as it is obvious from Figure 6. The last split is into European and East-Asian strains of the virus, that is clusters II and I, respectively. There is nothing “American” (with respect to Native Americans) there. It is a European cluster of the virus, carried by migrants from Europe to America. It is surprising that the authors of the study have not seen such a simple explanation of the dynamics of the strains. OPEN ACCESS 31  A. A. KLYOSOV Figure 5. Schematic representation of the human mtDNA phylogeny within hominine, and exemplified with Homo neanderthalinsis (on the left) and Home sapiens (on the right). The ph ylogeny illustrating approximate divergence times of the s pecies. RNRS stands for Reconstructed Nean- derthal Reference Seq uen ce, RSRS – for Reconstru cted Sap ien s Ref eren ce Sequ en ce. Mutate d nu cleo tide p os ition s s eparatin g th e n od es o f the two basal human haplogroups L0 and L1’2’3’4’5’6 and their derived states as compared to the RSRS are shown. Please notice a huge split between L0 and L1-L6 on the right. From Behar et al. (2012) with permission. OPEN ACCESS 32 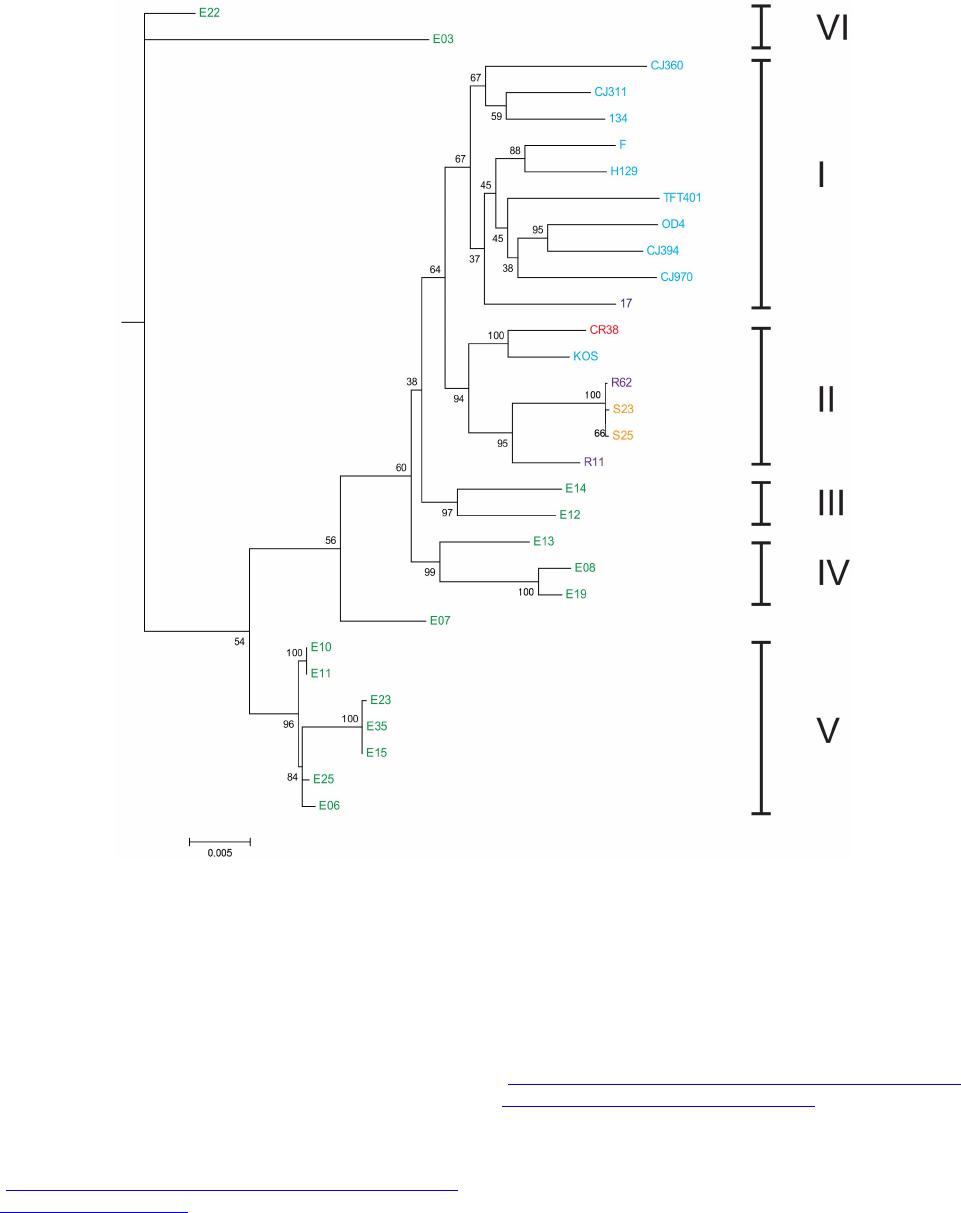 A. A. KLYOSOV Figure 6. Phylogenetic tree featuring HSV-1 strains which depict the formation of six clades based on geographic origin. Clade I includes European/North American strains (Glasgow [#17], Seattle, San Francisco, “USA”—all US are marked in blue color), Clade II comprises East Asian strains (China [red], South Korea [purple], Japan [yellow], and Houston, US [blue]), and III, IV, V and VI are East African (Kenya [green]). From Kolb et al. (2013) with permission. Apparently, they were too mu ch focusing at bendi ng the data to fit “Out of Africa”. The senior author of the study, Kurtis Brandt, a professor of medical microbiology and ophthalmology, sta ted in his inter- view: “What we found follows exactly what the … molecular geneticists who have analyzed the human genome have told us, about wh ere humans originated and how they spread across the planet.” (http://www.sci-news.com/genetics/science-herpes-virus-out-of -africa-theo ry-01482.html). This was exactly the main problem of the study. In another interview Kurtis Brandt says that “the results were quite “stunning.” He goes on to describe their findings: “We found that all of the African isolates cluster together, all the viruses from the Far East, Korea, Japan, China clustered to- gether, all the viruses in Europe and America, with one excep- tion, clustered together.” (http://guardianlv.com/2013/10/herpes-simplex-virus-confirms- out-of-africa-migration-of-ancient-humans/). Generally, it is true, despite some stretches. However, it does not support the “Out of Africa” concept at all, as it was explained above. The paper (Kolb et al., 2013) provides one more illustration how experimental data a re bent in order to show the desir able “Out of Africa” conclusion by all means. This concerns calcu- lations of t he “divergence time” for the strains. The authors wanted to obtain 50,000 ya from the initial divergence, which would fit the desired “Out of Africa” time, and they obtained it. Here is how th ey came to those 50,000 ya. This is rather educa- OPEN ACCESS 33 A. A. KLYOSOV tional and entertaining. The authors (Kolb et al., 2013) have reported that the re are three significantly varying figures for the mutation rate con- stants for Simplex and other herpes viruses, equal to 18.2 × 10−9, 30 × 10−9 and 3 × 10−9 substitutions/site/year. To employ all of them would hav e resulted in divergence times that dif- fered by 30 times. The authors did not know which constant of the three would have been a matter of choice. They decided to take such a constant which would give them a desired and “consentual” 50,000 ya. As a first step, the authors considered a suggested divergence time between European and Asian popu- lations as 23 - 45 thousand yea rs ago, as four literature sources have claimed. The authors took an average figure from all the four values, making it 34,000 ± 10,500 years ago. Those are obviously lower figures compared to historical ones, since Eu- ropean and Asian haplogroups separated not later than 55 - 60 ky (see Figure 1); however, all those calculations in the paper were so crude—and datings, and the mutation rate constants— that it could not have been taken seriously. Since the authors took “reference” divergence times lower than t he y should have been, the mutation rate constants must be much higher tha n they should have been. Indeed, the adjusted mutation rate constant was much higher than the all the four cited from the literature, namely, 134 × 10−9 substitutions/site/ year, with an upper and lower limits of 214 × 10−9 and 74.8 × 10−9, respectively. This can be compared to the literature’s 3×, 18.2× and 30 × 10−9 substitutions/site/year. This apparently was not a matter of concern to the authors. The ultimate goal was 50,000 years. Now, using this grossly inflated figure for the mutation rate constant, the authors obtained th at the initial divergence time for the virus occurred 50,300 ± 16,700 ya, and claimed that it “supports the “out of Africa model” of human migrations”. A similarly calculated date for the Eurasian strains divergence time was 32,800 ± 10,900 ya, and time for divergence of a sin- gle Chinese virus sample and a single Texas sample of an un- known origin results in a divergence time of 15,760 ± 5300 ya. The authors concluded that the last date “fits with the estimated time period in which the North American continen t was popu- lated from Asia”. No comments. Based on all of it the authors claimed that “the data supports the “out of Africa model” of human migrations”. As we have explained above, this has nothing to do with the “Out of Africa” concept. The mutation rate constant for the Simplex Herpes Virus, “calculated” by the authors, exceeded three other litera- ture values by 4.5 to 45 times. There we re no explanations from the authors for such a discrepancy. There were no other verifi- cations of t he obtained mutation rate constant. The authors obtained, using that mutation rate constant, that HSV-1 and HSV-2 diverged 2.184 ± 0.753 million years ago (please notice the accuracy of the bot h figures in the estimate!), but this also says practically nothing; it could have come up as 20 million years, and it would have been a good number too. There could have been said that it was macaque as a carrier of the virus. In other words, any data goes in such a work. If the literature mu- tation rate constants (or some of them) are valid, then the initial divergence time of the HSV-1 was not 50,300 ya, but between 220 thousand and 2.2 million years ago, and also could have been interpreted in terms of evolution of man. Indeed, every- thing goes. But then there would not have been “Out of Africa” “support’. A conclusion: the experimental data by Kolb et al. (2013) are valuable, but manipulations, “interpretations”, and conclusions, particularly regarding “Out of Africa” have no value at all. Anthropological Data and Datings A peculiar situation was formed in the last few years, when many anthropologists doubted experimental anthropology data on evolution of man as very controversial, but pointed at gene- ticists, that the latter “have real data” and, as they claim, it con- firmed the “Out of Africa” concept. Many geneticists, on the other hand, doubt experimental genetic data on evolution of man as very controversial, unreliable and conflicting with each other, but point at anthropologists, that they “have real data”, and believe that anthropologists confirmed the “Out of Africa” concept. In reality none of them confirmed the dubious (and, actually, false) concept, however, those reciprocal bows kee p the concept afloat. The cornerstone mutual belief is that AMH allegedly arose in Africa and it happened about 200,000 ya. By definition, AMH was Homo sapiens who did not have noticeable archaic skeletal features. Now, let us list several actual experimental observa- tions in that regard: -All African excavated skeletal remains dated before 36,000 ya posess noticeable archaic features, -Ancient skeletal remains very often are so fragmentary, that to reconstruct an appearance of ancient people even in a mi- nimal degree is typically close to impossible, -An anthropological pattern of skeletal remains is often sim- ilar in Africa and elsewhere, so it is close to impossible to firmly conclude who went where—out of Africa or into Africa, -Very often ancient settlements produce no skeletal remains at all, and conclusions (or rather suggestions) about “anatomi- cally modern humans” are often made based on view and dating of the settlements and some st one tools, however, they could be left by archaic humans, and not AMHs or even Homo sapiens, -Datings of ancient bones are often so questionable that few specialists accept them literally or uncritically, or even serious- ly. Unfortunately, radiocarbon datings practically do not work on times more than 40,000 years , and a recent record in that regard was set at 60,000 years. The reason is well unders- tood—a half-life of the radioactive isotope 14С equals to 5730 years, that is 40,000 years is equivalent to seven half-life times, 60,000 years takes more than ten half-life times. The method is based on measurements in biological samples of a rati o of the stable isotope 12C (and a little of 13С, concentration of which is about 1% compared to that of 12C) and radioactive 14С, an ini- tial content of which equals about 1 × 10−10 per cent in the tested biological sample, and decreases with time according to its half-life time. After 60 thousand years the content of 12C drops from the initial 1 × 10−10 per cent by 210 times, that is by 1024 times. Modern detectors are unable to measure such low concentrations of 12C, at least those detectors whic h are in pos- session of archaeologists. This corresponds to 1 click per hour per gram of the tested carbon. A natural radioactivity back- ground is much higher. This opens a wide probability for errors, and no t only for “innocent errors”. Many anthropologists remember an embar- rassing story with dishonoring and firing of a German anthro- pologist, Professor Reiner Protsch for systematic, as it was revealed, falsifications of dates for numerous ancient and not so ancient skeletal relics. After cross-examination, a dating by OPEN ACCESS 34  A. A. KLYOSOV Protsch of 36,000 ya turned out to be 7500 ya, another dating of 21,300 ya occurred in fact 3300 ya, and a skeleton dated by Protsch by 29,400 ya happened to be that of a man who died in 1750, 255 years before (Gardian, http://www.theguardian.com/science/2005/feb/19/science.scien cenews). The same in kind story was presented by the interna- tional journal Archaeology http://archive.archaeology.org/0505/newsbriefs/insider.html. Similar stories happen in archaeology and anthropology, and even if not through obvious (and verified and proven) falsifica- tions, which, of course, occur in very rare cases, still a likelih- ood for a mistake is rather high. It should be noted here that the most ancient biological artifacts are dated not by radiocarbon, but employing other methods, such as the argon method, in which a ratio of 40Ar/39Ar is measured. Overall, t h ere is not any anthropological of archaeological proof of appearance of anatomica lly modern human in Africa, or the “Out of Africa” migration. There is not any proof that stone tool types found in Europe and Eurasia were brought from sub-Sahara regions. All announcements on discoveries of skeletal remains of AMH older than 50 kya, and more so, older than 150 kya, in the sub-Saharan area, are distorted or incorrect in the first place. All of them expose rather noticeable archaic features. A good overview of this issue was recently published in Advances in Anthropology by Australian anthropologist Ro- bert Bednarik. A number of studies tell of absence of such findings from south of the Sahara (e.g., Grine et al., 2007; Grine et al., 2010). All known olde s t fossils, such as Omo Ki- bish 1 (195 kya, Ethiopia, fragments of skull with the face, very incomplete), Omo-2 (having a number of primitive features), Herto (the Middle Awash region, 160 - 154 kya, very robust and thus distinguishable from AMH), dated between 200 - 100 kya, possess archaic features (Rightmire, 2009) and cannot be considered AMH. Many excavated remains do not have facial bones. Even the Hofmeyr skull from South Africa, dated to 36 kya (though has no recorded archaeological associations, and dated by application of optically stimulated luminescence and U-series methods to sediments filling of the braincase) is fea- tured a robust face, close to crania associated with Upper Pa- leolithic industries in Eurasia (cited by Rightmire, 2009). Over- all, the Hofmeyr skull posesses noticeable archaic features (Grine et al., 2007; Rightmire, 2009; Tattersall, 2009). An American anthropologist Rightmire (2009) noted that “neither the Herto hominins, nor others from La te Pleistocene sites such as Klasies River in southern Africa and Skhu_l/Qafzeh in Israel, can be matched in living populations. Skulls are quite robust, and it is only after ~35,000 years ago that people with more gracile, fully modern morphology make their appearance”. Righmire continues that the process or events underlying AMH emergence “are poorly understood”. This conclusion was re- peated by Hammer et al. (2011)—“…hominins with a combina- tion of archaic and modern features persist in the fossil record across sub-Saharan Africa and the Middle East until after ~35 kya”. In other words, multiple references by proponents of the “Out of Africa” concept that many fossils of AMH were found in Africa with dates between 200 and 160 kya , are at least ques- tionable, if not untrue. Manipulations and distortions continue. Michael Hammer, a recent proponent of the “Out of Africa” concept, however, expressed a doubt as late as in 2013, when the situation with “Out of Africa” became increasingly con- fused. His recent paper in Scientific American (May 2013) Hammer concludes as follows: “Many loose ends remain. Yet one thing is clear: the roots of modern humans trace back to not just a single ancestral population in Africa but to popula- tions throughout the Old World”. This does seem quite reasonable. Studying history of the an- cient world, we see plenty of mobile migrations, including very long migrations, long in terms of distance and time. When pro- ponents of the “Out of Africa” hypothesis insist that it took only 10 thousand years for the “Out of Africa” humans to reach Australia, then it is hard to imagine that for the preceding 150 thousand years humans sat still in Africa and did not spread all over the world, out of Africa and into Africa, multiple times. How come that such a shaky concept of the one-sided “Out of Africa” so aggressively and forcefully promoted was accepted (but not quite) by present-day educated population in modern countries? It should cause alarm—regarding both that aggres- sive and overwhelming delivery of the false concept, falsely claiming “consensus among scientists” (there was none), and such a sheepish acceptance of that false concept by a general population. The most alarming is the fact that the concept did not have any proof, it was based on lies, fantasies, conjectures, wishful thinking. How could science, largely funded by the general population, have turned by a handful of manipulators into an abusive tool? This is a real thing to be concerned about. Conclusion Ancestors of the most present-day non-Africans did not come from Africa in the last 30,000 - 600,000 years at least. In other words, those who migrated from Africa, or were forcefully taken out as slaves, are not ancestors of the contemporary Eu- ropeans, Asians, Native Americans, Australians, Polynesians. This follows from the whole multitude of data in anthropology, genetics, archaeology, DNA genealogy. Study of the DNA of excavated bones of Neanderthals has shown in them MCR1 melanocortin rece ptor, in the same variant as that in modern humans, which makes pale skin and red ha ir, observed in mod- ern humans (Lalueza-Fox et al., 2007), though, according to the study authors, humans did not inherit MCR1 from Neanderthals. There was not any dat a that Neanderthals were Black Africans. Indeed, no Neanderthals were found in Africa. Since modern humans and Neanderthals (our cousins) have had a common ancestor, that common ancestor likely had fair skin and light- color hai r, and did not live in Africa. That was between 300 and 600 ky a. There is still a puzzle how our fair skin ancestors, which split about 160,000 ya and one branch of them migrated to Africa (see Figure 1), adjusted to African climate (which, though, was different from today’s) and acquired a black skin color. An answer lies, apparently, in regulation of melanin bio- synthesis. However, it is a different sto ry. REFERENCES Arredi, B., Poloni, E. S., Paracchini, S., Zerjal, T., Fathallah, D. M., Makrelouf, M., Pascali, V. L., Novelletto, A., & Tyler-Smith, C. (2004). A predominantly neolithic origin for Y-chromosomal DNA variation in North Africa. American Jour nal of Human Genetics, 75, 338-345. http://dx.doi.org/10.1086/423147 Atkinson, Q. D. (2011). Phonemic diversit y supports a serial found er effect model of language expansion from Africa. Science, 332, 346-349. http://dx.doi.org/10.1126/science.1199295 Behar, D. M., van Ov en, M., Rosset, S., Metspa lu, M., Loogval i, E. L., Silva, N. M., K ivisild, T., Torro ni, A., & Villems , R. (2012). A “co - pernican” reassessment of the human mitochondrial DNA tree from OPEN ACCESS 35  A. A. KLYOSOV its root. American Journal of Human Genetics, 90, 675-684. http://dx.doi.org/10.1016/j.ajhg.2012.03.002 Behar, D. M., Villems, R., Soodyall, H., Blue-Smith, J., Pereira, L., Metspalu, E., Scozzari, R. et al. (2008 ). The dawn of human matrili- neal diversity. American Journal of Human Genetics, 82, 1130-1140. http://dx.doi.org/10.1016/j.ajhg.2008.04.002 Benazzi, S., Douka, K., Fo rnai , C., B au er, C. C ., Ku ll mer, O ., Sv ob oda , J. et al. (2011). Early dispersal of modern humans in Europe and im- plications for Neanderthal behaviour. Nature, 479, 525-528. http://dx.doi.org/10.1038/nature10617 Campbell, M. C., & Tishkoff, S. A. (2010). The evolution of human genetic and phenotypic variation in Africa. Current Biology, 20, R166-R173. http://dx.doi.org/10.1016/j.cub.2009.11.050 Cann, R. (2013). Y weigh in again on moden humans. Science, 341, 465-467. http://dx.doi.org/10.1126/science.1242899 Cann, R. L., Stoneking, M., & Wilson, A. C. (1987). Mitochondrial DNA and human evolution. Nature, 325, 31-36. http://dx.doi.org/10.1038/325031a0 Carrigan, D., & Hammer, M. F. (2006). Reconstructing human origins in the genomic era. Nature Reviews Genetics, 7, 669-680. http://dx.doi.org/10.1038/nrg1941 Chiaroni, J., Underhill, P. A., & Cavalli-Sforza, L. L. (2009). Y-chro- mosome diversity, human expansion, drift, and cultural evolution. Proceedings of the National Academy of Sciences of the United Sta tes of America, 106, 20174-20179. http://dx.doi.org/10.1073/pnas.0910803106 Cruciani, F., Trombetta, B., Massaia , A., Destro-Bisol, G., Sellitto, D., & Scozzari, R. (2011 ). A revised root for the h uman Y chromosomal phylogeneti c tree: The orog in of patrilineal diversity in Af rica. Ameri- can Journal of Human Genetics, 88, 814-818. http://dx.doi.org/10.1016/j.ajhg.2011.05.002 Cruciani, F., Trombetta, B., Sellitto, D., Massaia, A., Destro-Bisol, G., Watson, E., Co lomb, E. B., Dugoujon, J. M., Moral, P., & Scozzari, R. (2010). Human Y chromosome haplogroup R-V88: A paternal ge- netic record of early mid Holocene trans-Saharan connections and the spread of Chadic languages. European Journal of Human Genetics, 18, 800-807. http://dx.doi.org/10.1038/ejhg.2009.231 Curnoe, D., Xuepin g, J., Herries, A. I. R., Kan ning, B., Tacon , P. S. C ., Zhende, B., Fink, D. et al. (2012). Human remains from the Pleisto- cene-holocene transition of South west China suggest a co mplex evo- lutionary history for East Asians. PLOS ONE, 7, Article ID: e31918. http://dx.doi.org/10.1371/journal.pone.0031918 Edmonds, C. A., Lillie, A. S., & Cavalli-Sforza, L. L. (2004). Muta- tions arising in the wave front of an expanding population. Proceed- ings of the National Academy of Sciences of the United States of America, 101, 975-979. http://dx.doi.org/10.1073/pnas.0308064100 Fernandes, V., Alshamali, F., Alves, M., Costa, M. D., Pereira, J. B., Silva, N. M., Cherni, L., Harich , N., Cerny, V., Soares, P., Richards , M. B., & Pereira, L. (2012). The Arabian cradle: Mitochondrial re- licts of the first step s along the Southern route out of Af rica. Ameri- can Journal of Human Genetics, 90, 347-355. http://dx.doi.org/10.1016/j.ajhg.2011.12.010 Forster, P. (2004). Ice Ages and the mitochondrial DNA chronology of human dispersals: A review. Philosophical Transactions of Royal Society B, 359, 255-264. http://dx.doi.org/10.1098/rstb.2003.1394 Forster, P., Torroni, A., Renfrew, C., & Rohl, A. (2001). Phylogenetic star contraction applied to Asian and Papuan mtDNA evolution. Mo- lecular Biology and Evolution, 18, 1864-1881. http://dx.doi.org/10.1093/oxfordjournals.molbev.a003728 Francalacci, P., Morelli, L., Angius, A., Berutti, R., Rein ier, F., Atzeni, R., Pilu, R. et al. ( 2013). Low-pass DNA sequencing of 1200 Sardi- nians reconstructs Euro pean Y-chromosome phylogeny. Science, 341, 565-569. http://dx.doi.org/10.1126/science.1237947 Grine, F. E., Bailey, R. M., Harvati, K., Nathan, R. P., Morris, A. G., Henderson, G. M., Ribot, I., & Pike, A. W. G. (2007). Late Pleisto- cene human skull from Hofmeyr, South Africa, and modern human origins. Science, 315, 226-229. http://dx.doi.org/10.1126/science.1136294 Grine, F. E., Gunz, P., Betti-Nash, L., Neubauer, S., & Morris, A. G. (2010). Reconstruction o f the late Pleistocene human skull from Hof - meyr, South Africa. Journal of Human Evolution, 59, 1-15. http://dx.doi.org/10.1016/j.jhevol.2010.02.007 Hammer, M. (2013). Human hybrids. Scientific American, 308, 66-71. http://dx.doi.org/10.1038/scientificamerican0513-66 Hammer, M. F., Woe rner, A. E., Mend ez, F. L., Watk ins , J. C., & Wall , J. D. (2011). Genetic evidence for archaic admixture in Africa. Pro- ceedings of the Nation al Academy of Sciences of the United States o f America, 108, 15123-15128. http://dx.doi.org/10.1073/pnas.1109300108 Hammer, M. F ., W o ern er, A. E., Mendez, F. L. , Watk in s , J . C., & Wall, J. D. (2011). Genetic evidence for archaic admixture in Africa. Pro- ceedings of the Nation al Academy of Sciences of the United States o f America. Hayden, E. C. (2013). African genes tracked back. Nature, 500, 514. http://dx.doi.org/10.1038/500514a Hellenthal, G., Auton, A., & Falush, D. (2008). Inferring human colo- nization history using a copying model. PLOS Genetics, 4, Artic le I D: e1000078. http://dx.doi.org/10.1371/journal.pgen.1000078 Henn, B. M., Gignoux, C . R., Jobin, M., Granka, J . M., Macphers on, J. M., Kidd, J. M., Rodríguez-Bo tigué, L., Ramachand ran, S., Hon, L., Brisbin, A., L in, A. A., Underhil l, P. A., Co mas, D., Kidd, K. K. , Nor- man, P. J., Parham, P., Bustamante, C. D., Mountain, J. L., & Feldman, M. W. (2011). Hunter-gatherer genomic diversity suggests a sou thern African origin for modern humans. Proceedings of the National Academy of Sciences of the United States of America, 108, 5154- 5162. http://dx.doi.org/10.1073/pnas.1017511108 Higham, T., Compton, T., Stringer, C., Jacobi, R., Shapiro, B., Trinkaus, E., Chandler, B., Grönin g, F., Collins, C., Hillson, S., O’Higgin s, P., FitzGerald, C., & Fagan, M. (2011). The earliest evide nce for anato- mically modern humans in North-Western Europ e. Nature, 479, 521- 524. http://dx.doi.org/10.1038/nature10484 Hublin, J. J. (2011). Africa n origin. Nature, 476, 395. http://dx.doi.org/10.1038/476395a Hudjashov, G., Kivisild , T., Underhil l, P. A ., En dico tt, P., Sanchez, J . J., Lin, A. A., Shen, P., Oefner, P., Renfrew, C., Ville ms, R., & Forster, P. (2007). Revealing the prehistoric settlement of Australia by Y- chromosome and mtDNA analysis. Proceedings of the National Aca- demy of Sciences of the United States of America, 104, 8726-8730. http://dx.doi.org/10.1073/pnas.0702928104 Jobling, M. A., & Tyler-Smith, C. (2003). The human Y-hromosome: An evolutionary marker comes of age. Nature Reviews Genetics, 4, 598-612. http://dx.doi.org/10.1038/nrg1124 Ke, Y., Su, B., Song, X., Lu, D ., Chen, L., Li, H., Qi, C. et al. (2001). African origin of modern humans in East Asia: A tale of 12,000 Y chromosomes. Science, 292, 1151-1153. http://dx.doi.org/10.1126/science.1060011 King, T. E., Parkin, E. J., Swinfield, G., Crucian i, F., S co zzari, R. , Ros a, A., Lim, S. K., Xue, Y. L., Tyler-S mit h, C., & Jo bli ng , M. A. (2 00 7). Africans in Yorkshire? The d eep est-rootin g clade o f the Y ph ylogeny within an English genealogy. European Journal of Human Genetics, 15, 288-293. http://dx.doi.org/10.1038/sj.ejhg.5201771 Klyosov, A. A. (2009). DNA genealogy, mutation rates, and some historical evidences written in Y-chromosome: I. Basic principles and the method. Journal of Genetic Genealogy, 5, 186-216. Klyosov, A. A. (2011). The slowest 22 marker haplotype panel (out of the 67 marker panel) and their mutation rate constants employed for calculations timespans to the most ancient common ancestors. Pro- ceedings of the Russian Academy of DNA Genealogy, 4, 1240-1257. Klyosov, A. A. (2012). Ancient h istory of the Arbins, bearers of haplo- group R1b, from Central Asia to Europe, 16,000 to 1500 years before present. Advances in Anthropology, 2, 87-105. http://dx.doi.org/10.4236/aa.2012.22010 Klyosov, A. A., & Rozhanskii, I. L. (2012a). Re-examining the “Out of Africa” theory and the o rigin of Europeoids (Caucasoids) in light of DNA genealogy. Advances in Anthropology, 2, 80-86. http://dx.doi.org/10.4236/aa.2012.22009 Klyosov, A. A., & Rozhanskii, I. L. (2012b). Haplogroup R1a as the Proto Indo-Europeans and the legendary Aryans as witnessed by the DNA of their current descendants. Advances in Anthropology, 2, 1- 13. http://dx.doi.org/10.4236/aa.2012.21001 Kolb, A. W., Ane, C., & Brandt, C. R. (2013). Using HSV-1 genome phylogenetics to track p ast human migrations. PLoS ONE, 8, Article OPEN ACCESS 36  A. A. KLYOSOV ID: e76267. http://dx.doi.org/10.1371/journal.pone.0076267 Lachance, J., Vernot, B., Elbers, C. E., Ferwerda, B., Froment, A., Bodo, J. M., Lema, G., Fu, W.Q., Nyambo, T. B., Rebbeck, T. R., Zhang, K., Akey, J. M., & Tishkoff, S. A. (2012). Evolutionary his- tory and adaptation fro m high-coverage whole-genome sequences of diverse African hunter-gatherers. Cell, 150, 457-469. http://dx.doi.org/10.1016/j.cell.2012.07.009 Lalueza-Fox, C., Rö mpler, H., Caramelli, D., Stäubert , C., Cat alano, G., Hughes, D., Rohland, N., Pilli, E., Longo, L., Condemi, S., de la Ra- silla, M., Fortea, J., Rosas, A., Stoneking, M., Schöneberg, T., Ber- tranpetit, J., & Hofreiter, M. (2007). A Melanocortin 1 receptor allel e suggests varying pigmentation among Neanderthals. Science, 318, 1453-1455. http://dx.doi.org/10.1126/science.1147417 Li, H., & Durbin, R. (2011). Inference of human population history from individual whole genome sequences. Nature, 475, 493-496. http://dx.doi.org/10.1038/nature10231 Majumder, P. P. (2010). The human genetic history of South Asia. Current Biology , 20, R184-R187. http://dx.doi.org/10.1016/j.cub.2009.11.053 Mellars, P. (2011). The earliest modern humans in Europe. Nature, 479, 483-485. http://dx.doi.org/10.1038/479483a Mendez, F. L., Krahn, T., Schrack, B., Krahn, A. M., Veeramah, K. R., Woerner, A. E., Fomine, F. L. M., Bradman, N., Thomas, M. G., Kara- fet, T. M., & Hammer, M. F. (2013). An African American paternal lineage adds an extremely ancient root to the human y chromosome phylogenetic tree. American Journal of Human Genetics, 92, 454- 459. http://dx.doi.org/10.1016/j.ajhg.2013.02.002 Moorjani, P., Patterso n , N., Hirsch h orn, J. N., Keinan, A., Hao, L., Atz - mon, G., Burns, E., Ostrer, H ., Price, A. L. , & Reich, D. (2 011). The history of African gene flow into Southern Europeans, Levantines, and Jews. PLOS Genetics, 7, Article ID: e1001373. http://dx.doi.org/10.1371/journal.pgen.1001373 Ottoni, C., Primativ o, G., K ash ani, B. H., Achilli, A ., M artin ez-Labarga, C., Biondi, G. , Torroni, A., & Ric kards, O. (2010). Mitochondrial ha- plogroup H1 in No rth Africa: An early holocene arrival f rom Iberia. Plos ONE, 5, Article ID: e13378. http://dx.doi.org/10.1371/journal.pone.0013378 Patin, E., Laval, G., Barreiro, L. B., Salas, A ., Semino, O., Santachiara- Benerecetti, S., Kidd, K. K. et al. (2009). Inferring the demographic history of African farmers and Pygmy hunter-gath erers u sin g a multi- locus recequencing data set. PLOS Genetics, 5, 1-13. http://dx.doi.org/10.1371/journal.pgen.1000448 Poznik, G. D., Henn , B. M., Yee, M. C., Sliwerska, E., Euskirchen, G. M., Lin, A. A., Snyder, M., Quintan a-Murci, L., Kid d, J. M., Und er- hill, P. A., & Bustamante, C. D. (2013). Sequencing Y chromosomes resolves discrepancy in time to common ancestor of males versus females. Science, 341, 562-565. http://dx.doi.org/10.1126/science.1237619 Ramachandran, S., Deshpande, O., Roseman, C. C. , Rosenberg, N. A., Feldman, M. W., & Cavalli-Sforza, L. L. (2005). Support from the relationship of genetic and geo graphic distance in hu man populations for a serial founder effect originating in Africa. Proceedings of the National Academy of Sciences of the United States of America, 102, 15942-15947. http://dx.doi.org/10.1073/pnas.0507611102 Rightmire, G. P. (2009). Middle and later Pleistocene hominins in Africa and Southwest Asia. Proceedings of the National Academy of Sciences of the United States of America, 106, 16046-16050. http://dx.doi.org/10.1073/pnas.0903930106 Rito, T., Richards, M. B., Fernandes, V., Alshamali, F., Cerny, V., Pereira, L., & Soares, P. (2013). The first modern human dispersals across Africa. PLoS ONE, 8, Article ID: e80031. Sahoo, S., Singh, A., Himabindu, G., Banerjee, J., Sitalaximi, T., Gaik- wad, S., Trivedi, R., End icott, P., Kivisild , T., Metspalu, M., Vill ems, R., & Kashyap, V. K. (2006). A prehistory of Indian Y-chromosomes: Evaluating demic diffusion scenarios. Proceedings of the National Academy of Sciences of the United States of America, 103, 843-848. http://dx.doi.org/10.1073/pnas.0507714103 Scozzari, R., Massaia, A., D’Atanasio, E., Myres, N.M., Perego, U.A., Trombetta, B., & Cruciani, F. (2012). Molecular dissection of the basal clades in the human Y chromosome phylogenetic tree. PLoS ONE, 7, Article ID: e49170. http://dx.doi.org/10.1371/journal.pone.0049170 Shi, W., Ayub, Q., Vermeulen, M., Shao, R. G., Zuniga, S., van der Gaag, K., de Knijff, P., Kayser, M ., Xu e, Y., & Tyler-Smith, C. (2010). A worldwide survey of human male demographic history based on Y-SNP and Y-STR data from the HGDP-CEPH populations. Mole- cular Biology and Evolution, 27, 385-393. http://dx.doi.org/10.1093/molbev/msp243 Skoglund, P. (2013). Reconstructing the human past using ancient and modern genomes. Uppsala Dis ssertation from the Faculty of Science and Technology, Uppsala Universitet, Acta Univ ersitatis Up saliensis, Uppsala, 52 p. Soares, P., Ermini, L., Tho mpson, N., Nor mina, M., Rito, T., Roh l, A., Salas, A., Opp en h eimer, S., Macau l a y, V ., & Richards, M. B. (2009). Correcting for purifying selection: An improved human mitochon- drial molecular clock. American Journal of Human Genetics, 84, 740-759. Stewart, J. R., & String er, C. B. (20 12 ). Hu man ev o lutio n o u t of Af rica: The role of refugia and climate change. Science, 335, 1317-1321. http://dx.doi.org/10.1126/science.1215627 Stoneking, M., & Delfin, F. (2010 ). The human genetic history of East Asia: Weaving a complex tapestry. Current Biology, 20, R188-R193. http://dx.doi.org/10.1016/j.cub.2009.11.052 Tattersall, I. (2009). Human origins: Out of Africa. Proceedings of the National Academy of Sciences of the United States of America, 106, 16018-16021. http://dx.doi.org/10.1073/pnas.0903207106 Thomson, R., Pritchard, J. K., Shen, P., Oefner, P. J., & Feldman, M. (2000). Recent common ancestry of human Y chromosomes: Evidence from DNA sequence data. Proceedings of the National Academy of Sciences of the United States of America, 97, 7360-7365. http://dx.doi.org/10.1073/pnas.97.13.7360 Underhill, P. A., Shen, P., Lin, A. A ., Jin, L., Passarino, G., Yang, W. H., Kauffman, E. et al. (2000). Y-chromosome sequence variation and the history of human populat ions. Nature Genetics, 26, 358-361. http://dx.doi.org/10.1038/81685 Vigilant, L., Stonek ing, M., Harpen ding, H., Hawkes, K., & Wilson, A. C. (1991). African populations and the evolution of human mito- chondrial DNA. Science, 253, 1503-1507. http://dx.doi.org/10.1126/science.1840702 Wei, W., Ayub, Q., Xue, Y., & Tyler-Smith, C. (2013). A co mparison of Y-chromosomal lineage dating using either resequencing or Y-SNP plus Y-STR genotyping. Forensic Science Internatio nal: G enetics, 7, 568-572. http://dx.doi.org/10.1016/j.fsigen.2013.03.014 Xue, Y., Zerjal , T., B ao, W., Zhu , S., Lim, S. K., S hu, Q ., Xu, J. J., Du , R. F., Fu, S. B., Li, P., Yang, H. M., & T yler-Smith, C. (2005). Re- cent spread of a Y-chromosomal lineage in Northern China and Mon- golia. American Journal of Human Genetics, 77, 1112-1116. http://dx.doi.org/10.1086/498583 Yotova, V., Lefebvre, J. F., Moreau, C., Gbeha, E., Ho vhannesyan, K., Bourgeois, S., Bedarida, S. et al. (2011). An X-linked haplotype of Neandertal origin is presen t among all non-African populations. Mo- lecular Biol ogy and Evolution, 28, 1957-1962. http://dx.doi.org/10.1093/molbev/msr024 OPEN ACCESS 37
|