 Journal of Cancer Therapy, 2014, 5, 67-73 Published Online January 2014 (http://www.scirp.org/journal/jct) http://dx.doi.org/10.4236/jct.2014.51009 Evidence for Tumour Suppressor Function of DOK7 in Human Breast Cancer James Bracken1, Tamara Ghane m1, Abdul Kasem1, Wen G. Jiang2, Ke fah Mokbel1 1The London Breast Institute, The Princess Grace Hospital, London, UK; 2Metastasis and Angiogenesis Research Group, University Department of Surgery, Cardiff University, Cardiff, UK. Email: kefahmokbel@hotmail.com Received November 25th, 2013; revised December 20th, 2013; accepted December 28th, 2013 Copyright © 2014 James Bracken et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. In accor- dance of the Creative Commons Attribution License all Copyrights © 2014 are reserved for SCIRP and the owner of the intellectual property James Bracken et al. All Copyright © 2014 are guarded by law and by SCIRP as a guardian. ABSTRACT Introduction: Downstream of tyrosine kinase 7 (DOK-7) is a member of the DOK family, which has been associ- ated with the development and progression of various humancancers. Previously, identification of CpG hyper- methylation in DOK-7 promoter was identified in breast cancer. Method: DOK-7 mRNA extr actio n and revers e transcription were performed on fresh frozen breast cancer tissue samples and normal background breast tissue. Transcript levels of expression were analyzed against TNM stage, tumour grade and clinical outcome over a 10-year follow-up period. Results: Levels of DOK-7 expression decreased significantly with increasing TNM stage. Higher DOK-7 expression was correlated with longer disease free and overall survival times. Conclusion: To our knowledge, this is the first study to investigate DOK-7 expression in human breast cancer. We identify a potential DOK-7 tumour suppressor role. DOK-7 as a prognostic biomarker in human breast cancer should be included in future vali dati o n studi e s. KEYWORDS Breast Cancer; DOK-7; CpG Hypermethyl a tion; Tumour Suppressor; Prognostic Marker 1. Introduction Identification of breast cancer biomarkers has shown great promise in not only increasing prognostic ability and molecular understanding at all stages of tumorigene- sis but also aid in the decision of specific clinical inter- ventions, potentially leading to the production of indi- vidualized therapies [1-6]. Clinical impact has begun to be achieved by several biomarkers most notably oestro- gen receptor (ER) and human epidermal growth factor receptor 2 ( HER2/ne u) [7]. The downstream of tyrosine kinase (DOK) family of adaptor proteins consists of 7 members that share a struc- tural topology characterized by an NH2-terminal pleck- strin homology (PH) domain, a central phosphotyrosine- binding (PTB) domain, followed by SH2 target motifs in the carboxyl-terminal [8,9]. Several members of the DOK family are associated with various human cancers; recently DOK-2 has been suggested as a marker for poor prognosis in gastric cancer [10,11]. Whilst two sub- groups exist within the DOK family; DOK 1-3 primarily expressed in haematopoietic tissues [12] and DOK 4-6 predominantly within the nervous system [13,14]; DOK- 7, expressed in skeletal muscle and the heart, plays a distinct role in other members [15]. Mutations in DOK7 are a common cause of congenital myasthenic syndrome (CMS). DOK-7 promotes trans- autophosphorylation and activation of muscle-specific kinase (MuSK) through formation of a dimeric structural unit following PTB domain interaction with the phos- phorylated juxtamembrane region of MuSK. Activation of MuSK results in downstream induction of acetylcho- line receptor (AChR) clustering on the post-synaptic membrane, essential for efficient neuromuscular trans- mission [15-20]. MuSK activation is also reliant on the motor neuron-derived ligand Agrin and suggested phos- phorylation of its intracellular domain by Casein Kinase OPEN ACCESS JCT  Evidence for Tumour Suppressor Function of DOK7 in Human Breast Cancer 2 (CK2) [21,22]. Agrin has previously been linked as a biomarker for colorectal and liver cancer [23] whilst CK2 association with breast cancer is well established through oncogene phosphorylation and over expression correlating with metastatic risk [ 21 ,24]. In a recent methylation profiling study of twins dis- cordant for breast cancer, Heyn et al., identified hyper- methylation of the DOK-7 gene in primary breast cancer tissues, cell lines and whole blood samples [25]. CpG site hypermethylation was observed within the DOK-7 pro- moter region. Recently, the importance of epigenetic mo- difications in cancer development and progression has been well-established [26-28]. In breast cancer, CpG island hypermethylation is associated with down regula- tion of various tumour suppressor genes, controlling all aspects of cellular function [3,29]. One way down regu- lation is thought to be that result is via abrogation of transcription factor binding to methylated promoter re- gions. The Sp1 transcription factor has been demon- strated to activate DOK-7 expression [30]. Sp1 has been found to participate in the expression of several onco- genes and up-regulation of its own expression has been observed in a percentage of breast tumours [31-37]. In view of the association between DOK-7 hyper- methylation and breast cancer, we examined the expres- sion profile of DOK-7 in a cohort of archival normal and breast cancer specimens. Transcript levels were evalu- ated against established pathological and prognostic pa- rameters in addition to clinical outcome. 2. Method 2.1. Patients and Samples Institutional guidelines, including ethical approval and informed consent were followed. Primary breast cancer tissues (n = 112) and adjacent non-cancerous mammary tissue (n = 31) were collected immediately after surgical excision and stored at −80˚C. An independent specialist pathologist examined haematoxylin and eosin stained frozen sections to verify the presence of tumour cells in the collected samples. Where normal non-neoplastic tis- sues were used, no tumour cells were found in the sec- tions. All tissues were randomly numbered and the de- tails were only made known after all analyses were com- pleted. All patients were tre ated according to local algorithms of management following a multidisciplinary discussion. Patients treated with breast-conserving surgery received adjuvant radiotherapy. Those with hormone-sensitive malignancy received tamoxifen. Fit patients with node- positive breast cancer or hormone-insensitive large and/ or high-grade cancer were offered adjuvant chemother- apy. Medical notes and histology reports were used to extract clinico-patholog ical data (Table 1) [38]. 2.2. Materials RNA extraction kits and reverse transcription kits were obtained from Sigma-Aldrich Ltd (Poole, Dorset, Eng- land, UK). The PCR primers were designed using Bea- con Designer (Palo Alto, CA, USA) and synthesized by Sigma-Aldrich. Custom made hot-start Master Mix for quantitative Polymerase Chain Reaction (PCR) was ob- tained from Abgene (Surrey , England, U K) [38-40]. 2.3. Tissue Processing, RNA Extraction and cDNA Synthesis Frozen sections of tissue were cut at a thickness of 5 - 10 μm and kept for routine histological analysis. Additional 15 - 20 sections were mixed and homogenized using a hand-held homogenizer in ice-cold RNA extraction solu- tion. The concentration of RNA was determined using UV spectrophotometry. Reverse transcription was carried Table 1. Clinical and pathological data. Parameter Category Nu mb e r Node status Positive 54 Negative 73 Tumour grade 1 24 2 43 3 58 Tumour type Ductal 98 Lobular 14 Medullary 2 Tubular 2 Mucinous 4 Other 7 TNM staging 1 70 2 40 3 7 4 4 NPI NPI1 68 NPI2 38 NPI3 16 Clinical outcome Disease-free 90 With local recurrence 5 Alive with metastasis 7 Died of breast cancer 16 Note: missing values reflect discarded/un-interpretable values. OPEN ACCESS JCT  Evidence for Tumour Suppressor Function of DOK7 in Human Breast Cancer out using a reverse transcription kit with an anchored oligo (dT) primer supp lied by Ab gen e, using 1 μg of total RNA in a 96-well plate. The quality of cDNA was veri- fied usin g Cytokeratin 19 (CK19) primers (Table 2) [38]. 2.4. Quantitative Analysis The level of DOK-7 transcripts from the above prepared DNA was determined using real-time quantitative PCR based on the Amplifluor technology, modified from a method reported previously [38,41]. The PCR primers were designed using Beacon Designer software, but to the reverse primer an additional sequence known as a Z sequence (5’-ACTGAACCTGACCGTACA-3’) which is complementary to the universal Z probe (Intergen Inc., Oxford, UK) was added. The product expands one intron. The primers used are detailed in Table 2. The reaction was carried out using Hotstart Q-master mix (Abgene), 10 pmol of specific forward primer, 1 pmol reverse pri- mer which had the Z sequence, 10 pmol of FAM (fluoro- genic reporter dye, carboxyfluorescein) tagged probe (Intergen Inc.), and cDNA from 50 ng of RNA. The re- action was carried out using the IcyclerIQ (Bio-Rad Ltd, Hemel Hempstead, England, UK), which is equipped with an optic unit that allows real-time detection of 96 reactions, under the following conditions: 94˚C for 12 min and 50 cycles of 94˚C for 15 sec, 55˚C for 40 sec, and 72˚C for 20 sec. The levels of the transcript were generated from a standard that was simultaneously am- plified with the samples. The levels of gene expression were then normalized against the housekeeping gene CK19, which was already quantified in these specimens, to correct for varying amounts of epithelial tissue be- tween samples [42]. The primers used for CK19 are de- tailed in Tab l e 2 . With every PCR run, a negative control without a template and a known cDNA reference sample as a positi ve c o nt rol, were i nc luded. 2.5. Statistical Analysis The Mann-Whitney U-test and two-sample t-test were used for statistical analysis of absolute and normalised gene copy number. The transcript levels within the breast cancer specimens were compared to normal background Table 2. DOK-7 and CK19 Primers. DOK-7 Forward gagtaggtggctggtgct Z Reverse actgaacctgaccgtacacagatgtcctctagcgtca CK19 Forward caggtccgaggttactgac Reverse actgaacctgaccgtacacactttctgccagtgtgtcttc tissues and analyzed against conventional pathological parameters and clinical outcome over a 10 year follow- up period. The statistical analysis was carried out using Minitab version 14.1 (Minitab Ltd. Coventry, England, U.K.) using a custom written macro (Stat 2005. mtw). For purposes of the Kaplan-Meier survival analysis, the samples were divided arbitrarily into two groups, “high transcript level” or “low transcript level”, for the DOK-7 gene. The cut-off was guided by the Nottingham Prog- nostic Index (NPI) value, with which the value of the moderate prognostic group was used as the dividing line at the start of the test. Disease Free Survival (DFS) and Overall Survival (OS) analyses were performed using SPSS version 12.0.1 (SPSS Inc. Chicago, IL, USA). For multivariate analysis using the Cox regression model, PASW Statistics 18 Software (Chicago, IL, USA) was used. 3. Results DOK-7 expression profile was determined via quantita- tive PCR in both absolute terms and normalized against CK19. DOK-7 was found to be expressed in both nor- mal/benign breast tissue and breast cancer specimens. Overall, no difference was found between DOK-7 ex- pression in breast cancer specimens and its expression in normal background tissue. The expression of DOK-7 mRNA was demonstrated to significantly decrease with increasing TNM class; TNM- 1 vs. TNM-4 [mean copy number 21,895 vs. 1239, 95% CI (3787, 37,526), p = 0.02] and TNM-2 vs. TNM-4 [mean copy number 7982 vs. 1239, 95% CI (−58, 13,544), p = 0.05]. DOK-7 expression in TNM-4 speci- mens was also significantly lower than normal breast samples [mean copy number 1239 vs. 39,810, 95% CI (−75,665, −1478), p = 0.04] (Table 3). A noticeable trend in decreasing DOK-7 expression with increasing Nottingham Prognostic Index (NPI) was observed; however, this did not reach statistical signifi- cance (NPI-3 compared to NPI-1 and NPI-2, p = 0.08 and p = 0.16, respectively). Transcript levels were sig- nificantly lower in Grade-1 tumour specimens than Grade-2 [mean copy number 752 vs. 32,796, 95% CI (−58,069, −6019), p = 0.02] although no overall trend existed amongst tumour grades (Table 3). After a median follow up of 10 years, DOK-7 mRNA expression levels were significantly higher in women that remained disease free compared to those who developed local recurrence [mean copy number 21,675 vs. 2310, 95% CI (3134, 35 ,596), p = 0.02] or those that died from breast cancer [mean copy number 216,75 vs. 1835, 95% CI (3790, 35,890), p = 0.02]. Furthermore, expression levels of women who developed lo cal recurrence or died from breast cancer were significantly lower than normal breast samples [mean copy number 2310 vs. 39,810, 95% OPEN ACCESS JCT 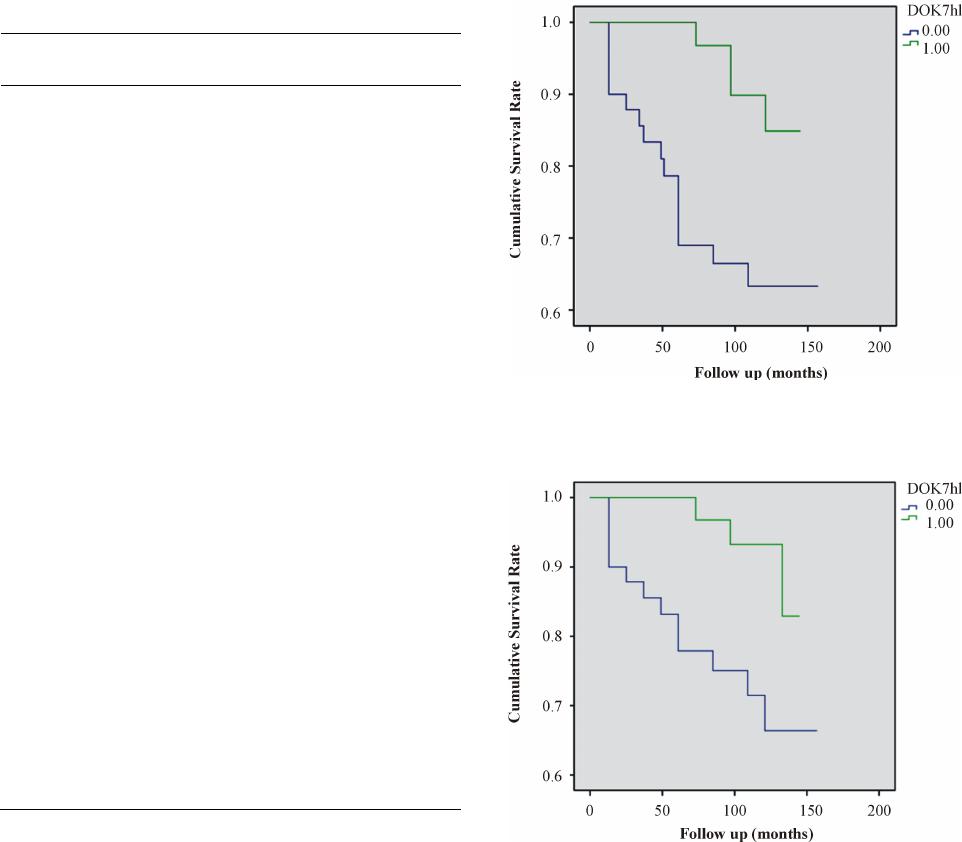 Evidence for Tumour Suppressor Function of DOK7 in Human Breast Cancer Table 3. DOK-7 mean mRNA expression level. Patient and tumour Characteristics DOK-7 mean (SD) p NPI NPI 1 vs. 2 23538 (81191) vs. 20632 (67371) 0.85 NPI 1 vs. 3 23538 (81191) vs. 4267 (8882) 0.08 NPI 2 vs. 3 20632 (67371) vs. 4267 (8882) 0.16 Tumour Grade Grade 1 vs. 2 752 (1584) vs. 32796 (80234) 0.02 Grade 1 vs. 3 752 (1584) vs. 14742 (72914) 0.17 Grade 2 vs. 3 32796 (80234) vs. 14742 (72914) 0.27 TNM TNM 1 vs. 2 21895 (65670) vs. 7982 (20072) 0.13 TNM 1 vs. 3 21895 (65670) vs. 79286 (199804) 0.48 TNM 1 vs. 4 21895 (65670) vs. 1239 (1253) 0.02 TNM 2 vs. 3 7982 (20072) vs. 79286 (199804) 0.38 TNM 2 vs. 4 7982 (20072) vs. 1239 (1253) 0.05 TNM 3 vs. 4 79286 (199804) vs. 1239 (1253) 0.34 Survival DF vs. LR 21675 (70979) vs. 2310 (5029) 0.02 DF vs. DR 21675 (70979) vs. 80140 (173108) 0.49 DF vs. D 21675 (70979) vs. 1835 (5438) 0.02 CI (−74,775, −226), p = 0.05] and [mean copy number 1835 vs. 39,810, 95% CI (−75,165, −785), p = 0.05], respectively (Table 3). There is a trend for specimens with lower levels of DOK-7 expression to associate with shorter disease-free (DFS) and overall survival (OS) times. Survival curves (DFS and OS) for women with tumours expressing “high levels” of DOK-7 differed significantly from those clas- sified as having “low levels”. The survival curves show higher levels of DOK-7 were of significant benefit in predicting higher DFS (p = 0.006) and better OS (p = 0.009) (Figures 1 and 2). 4. Discussion Here we present the mRNA expression profile of DOK-7 in breast cancer specimens and demonstrate decreased expression levels with increasing pathological and prog- nostic statuses. We have observed a significant decrease in DOK-7 Figure 1. Kaplan Meier Di se ase Fr ee Sur vi va l (DSF) Cur ve s for DOK-7. Survival times are expressed as mean number of months with 95% confidence interval. DFS (p = 0.006). Figure 2. Kaplan Meier Overall Survival (OS) Curves for DOK-7. Survival times are expressed as mean number of months with 95% confidence interval. OS (p = 0.009). expression level with increasing TNM stage, raising the potential for a novel tumour suppressor function outside its essential role in neuromuscular synaptogenesis. In addition, DOK-7 expression in TNM-4 stage tumours was significantly lower than that of normal breast tissue. Prior to this study, Heyn et al. detailed hyper-methyla- tion of a CpG site within the DOK-7 promoter in twins discordant for breast cancer [25]. CpG promoter hyper- methylation is associated with down regulation of gene expression, concurrent with our results. The scale of epi- genetic modifications associated with tumour develop- ment and progression is beginning to be appreciated with particular efforts placed in the identification of methyla- tion signatures that could serve as prognostic/predictive markers in breast cancer [7,27]. OPEN ACCESS JCT 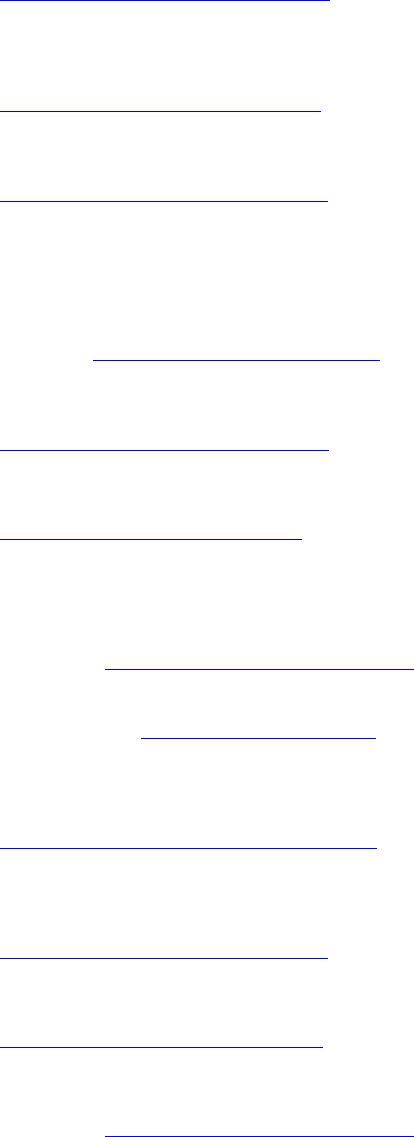 Evidence for Tumour Suppressor Function of DOK7 in Human Breast Cancer Other members of the DOK family of adaptor proteins have been identified to possess tumour suppressor roles; none more so than DOK-1, which is down regulated in several human cancers as a result of hyper-methylation of its promoter region [43]. Whilst other members of the DOK family modulate proliferative signalling pathways, DOK-7 is currently seen to have a distinct expression pattern and role in MuSK activation to promote AChR clustering [16], making it difficult to posit the nature of any potential DOK-7 tumour suppressor function. How- ever, several proteins employed in the Agrin/MuSK pathway that harbour additional roles, such as Ck2, have been associated with human breast cancer [21,22,44]. Moreover, one mechanism by which methylation can down-regulate expression, is by blocking transcription factors from accessing target-binding sites within the promoter region. Amongst its broad spectrum, Sp1 tran- scriptionally activates DOK-7 and is up-regulated in a percentage of breast cancers [30,45]. Furthermore, our results demonstrated significantly lower DOK-7 expression levels in women who devel- oped local recurrence or died from breast cancer follow- ing a median 10-year follow up period compared to both normal breast tissue and women that remained disease free over the same period of time. Disease free survival (DFS) and overall survival (OS) curves revealed that a higher DOK-7 expression level was a significant predic- tor of superior DFS and OS, supporting the suitability of DOK-7 as a prognostic biomarker for breast cancer. Bio- marker prediction of recurrence after curative resection is useful for determining intensity of clinical surveillance and adjuvant therapies. Limitations of the present study included the use of background parenchyma from breast cancer patients to provide “normal tissue” for comparison. Ideally, such material should be derived from patients without breast cancer in order to avoid any “field change” that may ex- ist within cancer bearing tissues. Although the follow-up period was substantial, sample size was relatively small and it is possible that a larger cohort may have influ- enced several results that approached, but failed to reach, statistical significance. Furthermore, the protein expres- sion and epigenetic modifications were not analysed in the present study and should be included in future inves- tigations. 5. Conclusion To our knowledge, this is the first study to investigate DOK-7 expression in human breast cancer and identify a potential tumour suppressor role. We also present data to support the value of DOK-7 as a prognostic biomarker in breast cancer. Restoring or mimicking the function of DOK-7 could provide a novel therapeutic modality against cancer. REFERENCES [1] S. Mehta, et al., “Predictive and Prognostic Molecular Markers for Cancer Medicine,” Therapeutic Advances in Medical Oncology, Vol. 2, No. 2, 2010, pp. 125-148. http://dx.doi.org/10.1177/1758834009360519 [2] A. W. Opstal-van Winden, et al., “Searching for Early Breast Cancer Biomarkers by Serum Protein Profiling of Pre-Diagnostic Serum; a Nested Case-Control Study,” BMC Cancer, Vol. 11, 2011, p. 381. http://dx.doi.org/10.1186/1471-2407-11-381 [3] S. Y. Park, et al., “Promoter CpG Island Hypermethyla- tion during Breast Cancer Progression,” Virchows Arch, Vol. 458, No. 1, 2011, pp. 73-84. http://dx.doi.org/10.1007/s00428-010-1013-6 [4] P. Taneja, et al., “Classical and Novel Prognostic Markers for Breast Cancer and their Clinical Significance,” Clini- cal Medicine Insights: Oncology, Vol. 4, 2010, pp. 15-34. [5] M. T. Weigel and M. Dowsett, “Current and Emerging Biomarkers in Breast Cancer: Prognosis and Prediction,” Endocrine-Related Cancer, Vol. 17, No. 4, 2010, pp. R245-R262. http://dx.doi.org/10.1677/ERC-10-0136 [6] F. Zhang, et al., “Recursive SVM Biomarker Selection for Early Detection of Breast Cancer in Peripheral Blood,” BMC Medical Genomics, Vol. 6, Suppl. 1, 2013, p. S4. http://dx.doi.org/10.1186/1755-8794-6-S1-S4 [7] K. D. Cole, H. J. He and L. Wang, “Breast Cancer Bio- marker Measurements and Standards,” PROTEOMICS- Clinical Applications, Vol. 7, No. 1-2, 2013, pp. 17-29. http://dx.doi.org/10.1002/prca.201200075 [8] A. Bedirian, et al., “Pleckstrin Homology and Phospho- tyrosine-Binding Domain-Dependent Membrane Asso- ciation and Tyrosine Phosphorylation of Dok-4, an In- hibitory Adapter Molecule Expressed in Epithelial Cells,” Journal of Biological Chemistry, Vol. 279, 2004, pp. 19335-19349. http://dx.doi.org/10.1074/jbc.M310689200 [9] A. H. Berger, et al., “Identification of DOK Genes as Lung Tumor Suppressors,” Nature Genetics, Vol. 42, 2010, pp. 216-223. http://dx.doi.org/10.1038/ng.527 [10] C. H. An, et al., “Mutational and Expressional Analysis of a Haploinsufficient Tumor Suppressor Gene DOK2 in Gastric and Colorectal Cancers,” APMIS, Vol. 119, No. 8, 2011, pp. 562-564. http://dx.doi.org/10.1111/j.1600-0463.2011.02749.x [11] H. Miyagaki, et al., “DOK2 as a Marker of Poor Progno- sis of Patients with Gastric Adenocarcinoma after Cura- tive Resection,” Annals of Surgical Oncology, Vol. 19, No. 5, 2012, pp. 1560-1567. http://dx.doi.org/10.1245/s10434-011-2157-6 [12] R. Mashima, et al., “Mice Lacking Dok-1, Dok-2, and Dok-3 Succumb to Aggressive Histiocytic Sarcoma,” Laboratory Investigation, Vol. 90, 2010, pp. 1357-1364. http://dx.doi.org/10.1038/labinvest.2010.121 [13] R. J. Crowder, et al., “Dok-6, a Novel p62 Dok Family Member, Promotes Ret -Mediated Neurite Outgrowth,” Journal of Biological Chemistry, Vol. 279, 2004, pp. 42072-42081. http://dx.doi.org/10.1074/jbc.M403726200 [14] J. Grimm, et al., “Novel p62dok Family Members, dok-4 and dok-5, Are Substrate s of the c-Ret Receptor Ty rosine OPEN ACCESS JCT  Evidence for Tumour Suppressor Function of DOK7 in Human Breast Cancer Kinase and Mediate Neuronal Differentiation,” Journal of Cell Biology, Vol. 154, No. 2, 2001, pp. 345-354. http://dx.doi.org/10.1083/jcb.200102032 [15] J. Cossins, et al., “The Spectrum of Mutations That Un- derlie the Neuromuscular Junction Synaptopathy in DOK7 Congenital Myasthenic Syndrome,” Human Molecular Genetics, Vol. 21, No. 17, 2012, pp. 3765-3775. http://dx.doi.org/10.1093/hmg/dds198 [16] E. Bergamin, et al., “The Cytoplasmic Adaptor Protein Dok7 Activates the Receptor Tyrosine Kinase MuSK via Dimerization,” Molecular Cell, Vol. 39, No. 1, 2010, pp. 100-109. http://dx.doi.org/10.1016/j.molcel.2010.06.007 [17] A. Inoue, et al., “Dok-7 Activates the Muscle Receptor Kinase MuSK and Shapes Synapse Formation,” Science Signaling, Vol. 2, No. 59, 2009, p. ra7. http://dx.doi.org/10.1126/scisignal.2000113 [18] A. Klein, et al., “DOK7 Congenital Myasthenic Syn- drome in Childhood: Early Diagnostic Clues in 23 Chil- dren,” Neuromuscular Disorders, Vol. 23, No. 11, 2013, pp. 883-891. http://dx.doi.org/10.1016/j.nmd.2013.06.002 [19] J. S. Muller, et al., “Phenotypical Spectrum of DOK7 Mutations in Congenital Myasthenic Syndromes,” Brain, Vol. 130, No. 6, 2007, pp. 1497-1506. http://dx.doi.org/10.1093/brain/awm068 [20] K. Okada, et al., “The Muscle Protein Dok-7 Is Essential for Neuromuscular Synaptogenesis,” Science, Vol. 312, No. 5781, 2006, pp. 1802-1805. http://dx.doi.org/10.1126/science.1127142 [21] T. Cheusova, et al., “Casein Kinase 2-Dependent Serine Phosphorylation of MuSK Regulates Acetylcholine Re- ceptor Aggregation at the Neuromuscular Junction,” Genes & Development, Vol. 20, 2006, pp. 1800-1816. http://dx.doi.org/10.1101/gad.375206 [22] D. J. Glass, et al., “Agrin Acts via a MuSK Receptor Complex,” Cell, Vol. 85, No. 4, 1996, pp. 513-523. http://dx.doi.org/10.1016/S0092-8674(00)81252-0 [23] S. Klein-Scory, et al., “Immunoscreening of the Extracel- lular Proteome of Colorectal Cancer Cells,” BMC Cancer, Vol. 10, 2010, p. 70. http://dx.doi.org/10.1186/1471-2407-10-70 [24] D. Drygin, et al., “Protein Kinase CK2 Modulates IL-6 Expression in Inflammatory Breast Cancer,” Biochemical and Biophysical Research Communications, Vol. 415, No. 1, 2011, pp. 163-167. http://dx.doi.org/10.1016/j.bbrc.2011.10.046 [25] H. Heyn, et al., “DNA Methylation Profiling in Breast Cancer Discordant Identical Twins Identifies DOK7 as Novel Epigenetic Biomarker,” Carcinogenesis, Vol. 34, No. 1, 2013, pp. 102-108. http://dx.doi.org/10.1093/carcin/bgs321 [26] M. Esteller, “Epigenetics in Cancer,” New England Jour- nal of Medicine, Vol. 358, 2008, pp. 1148-1159. http://dx.doi.org/10.1056/NEJMra072067 [27] Y. Huang, et al., “Epigenetics in Breast Cancer: What’s New?” Breast Cancer Research, Vol. 13, 2011, p. 225. http://dx.doi.org/10.1186/bcr2925 [28] S. Sharma, T. K. Kelly and P. A. Jones, “Epigenetics in Cancer,” Carcinogenesis, Vol. 31, No. 1, 2010, pp. 27-36. http://dx.doi.org/10.1093/carcin/bgp220 [29] J. F. Costello, et al., “Aberrant CpG-Island Methylation Has Non-Random and Tumour-Type-Specific Patterns,” Nature Genetics, Vol. 24, 2000, pp. 132-138. http://dx.doi.org/10.1038/72785 [30] J. Hamuro, et al., “The Transcription Factor Sp1 Plays a Crucial Role in dok-7 Gene Expression,” Biochemical and Biophysical Research Communications, Vol. 408, No. 2, 2011, pp. 293-299. http://dx.doi.org/10.1016/j.bbrc.2011.04.020 [31] S. Lu and M. C. Archer, “Sp1 Coordinately Regulates de Novo Lipogenesis and Proliferation in Cancer Cells,” In- ternational Journal of Cancer, Vol. 126, No . 2, 2010, pp. 416-425. http://dx.doi.org/10.1002/ijc.24761 [32] S. U. Mertens-Talcott, et al., “Betulinic Acid Decreases ER-Negative Breast Cancer Cell Growth in Vitro and in Vivo: Role of Sp Transcription Factors and microRNA- 27a:ZBTB10,” Molecular Carcinogenesis, Vol. 52, No. 8, 2013, pp. 591-602. http://dx.doi.org/10.1002/mc.21893 [33] X. B. Wang, et al., “Expression and Prognostic Value of Transcriptional Factor sp1 in Breast Cancer,” Chinese Journal of Cancer, Vol. 26, No. 9, 2007, pp. 996-1000. [34] M. Wei, et al., “Stat6 Cooperates with Sp1 in Controlling Breast Cancer Cell Proliferation by Modulating the Ex- pression of p21 (Cip1/WAF1) and p27 (Kip1),” Cellular Oncology, Vol. 36, No. 1, 2013, pp. 79-93. http://dx.doi.org/10.1007/s13402-012-0115-3 [35] X. Yang, et al., “HSF1 and Sp1 Are Involved in the Regulation of FUT4 Gene Expression and Cell Prolifera- tion in Breast Cancer Cells,” Journal of Cellular Bio- chemistry, Vol . 115, No . 1 , 2014, pp. 168-178. [36] L. Yue, et al., “The Oncoprotein HBXIP Activates Tran- scriptional Coregulatory Protein LMO4 via Sp1 to Pro- mote Proliferation of Breast Cancer Cells,” Carcinogene- sis, Vol. 34, No. 4, 2013, pp. 927-935. http://dx.doi.org/10.1093/carcin/bgs399 [37] Y. Zhang, et al., “The Oncoprotein HBXIP Upregulates PDGFB via Activating Transcription Factor Sp1 to Pr o- mote the Proliferation of Breast Cancer Cells,” Bio- chemical and Biophysical Research Communications, Vol. 434, No. 2, 2013, pp. 305-310. http://dx.doi.org/10.1016/j.bbrc.2013.02.123 [38] W. Sasi, et al., “Higher Expression Levels of SOCS 1,3,4,7 Are Associated with Earlier Tumour Stage and Better Clinical Outcome in Human Breast Cancer,” BMC Cancer, Vol. 10, 2010, p. 178. http://dx.doi.org/10.1186/1471-2407-10-178 [39] W. Al Sarakbi, et al., “The mRNA Expression of SETD2 in Human Breast Cancer: Correlation with Clinico- Pathological Parameters,” BMC Cancer, Vol. 9, 2009, p. 290. http://dx.doi.org/10.1186/1471-2407-9-290 [40] W. G. Jiang, et al ., “Prognostic Value of rho GTPases and rho Guanine Nucleotide Dissociation Inhibitors in Human Breast Cancers,” Clinical Cancer Research, Vol. 9, No. 17, 2003, pp. 6432-6440. [41] T. Nakagawa, et al., “Decreased Expression of SOCS-3 mRNA in Breast Cancer with Lymph Node Metastasis,” Oncology Reports, Vol. 19, No. 1, 2008, pp. 33-39. OPEN ACCESS JCT  Evidence for Tumour Suppressor Function of DOK7 in Human Breast Cancer [42] Z. Dogusan, et al., “Expression of SOCS Genes in Nor- mal and Leukemic Human Leukocytes Stimulated by Prolactin, Growth Hormone and Cytokines,” Journal of Neuroimmunology, Vol. 109, No. 1, 2000, pp. 34-39. http://dx.doi.org/10.1016/S0165-5728(00)00300-3 [43] A. Saulnier, et al., “Inactivation of the Putative Suppres- sor Gene DOK1 by Promoter Hypermethylation in Pri- mary Human Cancers,” International Journal of Cancer, Vol. 130, No. 11, 2012, pp. 2484-2494. http://dx.doi.org/10.1002/ijc.26299 [44] A. Ben Ammar, et al., “A Mutation Causes MuSK Re- duced Sensitivity to Agrin and Congenital Myasthenia,” PLoS One, Vol. 8, 2013, Article ID: e53826. http://dx.doi.org/10.1371/annotation/3ff2b918-c83c-4c 6f- a2e2-4d91294ec92f [45] W. C. Chang and J. J. Hung, “Functional Role of Post- Translational Modifications of Sp1 in Tumorigenesis,” Journal of Biomedical Science, Vol. 19, 2012, p. 94. http://dx.doi.org/10.1186/1423-0127-19-94 Abbreviations NPI: Nott i ng ham Prog nostic Index DF: Disease Free LR: Local Reoccurrence DR: Distant Reoccurrenc e TNM: TNM Classification of Malignant Tumours OPEN ACCESS JCT
|