Paper Menu >>
Journal Menu >>
 American Journal of Plant Sciences, 2011, 2, 56-62 doi:10.4236/ajps.2011.21008 Published Online March 2011 (http://www.SciRP.org/journal/ajps) Copyright © 2011 SciRes. AJPS Functional Analysis for Rolling Leaf of Somaclonal Mutants in Rice (Oryza sativa L.) Young-Hie Park1, Hyun-Suk Lee1, Gi-Hwan Yi2, Jae-Keun Sohn1, Kyung-Min Kim1* 1School of Plant Biosciences, Kyungpook National University, Daegu, Korea; 2Department of Functional Crop, National Institute of Crop Science, RDA, Milyang, Korea. Email: yhpark4026@hanmail.net, ghyi@rda.go.kr, {ddr3031, jhsohn, kkm}@knu.ac.kr Received December 28th, 2010; revised February 11th, 2010; accepted February 18th, 2011. ABSTRACT This study was carried out to facilitate the functional analysis of rice genes. Some 297 insertion plants (1.7%) of the entire lines with the endogenous retrotransposon Tos17 were produced. Phenotypes of these plants in the S2 generation were observed in the field acco rding to d ifferen t leaf typ es. Rolling lea f mutants sh owed thin ner sclerenchyma tou s cells, defective arrangement of vascular bundles, and well-formed bulliform cells as compared to the parental cultivar. Two new copies of Tos17 were detected in the rolling leaf type. In the new leaf type, the copy number and activation of Tos12, 15 did not appear as ‘Ilpum’. Flanking sequence tag (FST) analysis of Tos17 in the rolling leaf mutant indicated that new copies of Tos17 were transposed on chromosomes 11 and 12. Annotated homologues of the tagging genes on chromosome 11 were arabinoxylan rabino furanohydro lase isoenzym e AXAH-I and II. The tagg ing gene in chromoso me 12 was highly correlated with 6 kinds of genes including a transcript regu lated factor and a rough sheath 2-like protein. This rolling leaf and flanking sequence data will stimulate the functional analysis of rice genes. Keywords: Tissue Culture, Somaclonal Variation, Mutant, Opaque Endosperm, Tos Element 1. Introduction Tissue culture of plants is an important means to propa- gate genetically identical individuals asexually and to produce transgenic plants. However, undesired genetic and cytogenetic modifications are frequently generating genetic variability, somaclonal variations, induced during tissue culture. These tissue culture-induced mutations were reported in many plant species and seemed to be ubiquitous in plants [1,2]. Although tissue cu- ture-induced mutations were studied extensively as a source for plant improvement, little is known about their molecular causes. Specifically, somatic variations are becoming the limelight of breeders since they improve both the individual and species of crops [3,4]. Rice is used in genome study model of monocotyledonous plant because of the size of its genome (~430 Mb) being rela- tively small. Also, an international project had already gathered the functional analysis of gene from DNA se- quence [5]. There are various methods to analyze the function of the gene. One method used was a genome tilling array to a specific mutation group while handling chemical mutation materials such as N-methyl- N-nitrosourea (MNU) and ethyl methanesulfonate (EMS) or radiation. This method involved inserting foreign DNA fragments with labeled T-DNA, and using transition factor that exist in plants such as Ac/Ds and Tos derivatives which are a “Knock-out” or “Knock-down” of the gene or activation [6-8]. T-DNA or Ac/Ds gene trap sys- tems needed a lot of time and effort because it involved breeding large-scale mutation populations through the process of transformation to introduce a marked-gene on a rice genome. On the other hand, retrotransposon such as Tos can easily get a somaclonal variant through tissue culture without processing the mutagen. This method was widely used for the rice gene function analysis in Japan’s NIAS (National Institute of Agrobiological Sci- ence, http://Tos.nias.affrc.go.jp), France’s CIRAD (Cen- tre de Coopération Internationale en Recherche Agrono- mique pour le Développement, http://urgi.versailles. inra. fr/OryzaTagline), and Korea’s POSTECH (Pohang Uni- versity of Science and Technology, http://www.postech. ac.kr/life/pfg) [1]. Meanwhile, there are about 1,000 re- trotransposons with 32 classes in the rice genome that existed [6] and, LTR (long terminal repeats) retrotrans- poson with a minimum of 59 classed which composed  Functional Analysis for Rolling Leaf of Somaclonal Mutants in Rice 1 (Oryza sativa L.) Copyright © 2011 SciRes. AJPS 57 17% of the rice entire genome, but research about the evolution or introduced time is insufficient [3]. The re- verse transcription modulator (RTs; Reverse Transcrip- tase) of Tos family with 20 classes caused somaclonal variation because it was activated at the tissue culture which were only found in the japonica variety ‘Nippon- bare’ [9-18]. However, very little is known about the eco- type and copy number of the rice species or kind of Tos. Hence, japonica variety, ‘Ilpum’ was bred domestic- cally for rice gene function analysis. S1 and S2 progeny populations were bred from the cultured seed, and the incidence of variant in main agronomic characteristics was investigated. The “Rolling leaf” mutation was ana- lyzed to search for the kind of Tos and difference of copy number in rice, and the retrotransposon present in Tos family. 2. Materials and Methods 2.1. Plant Materials Growing Conditions Calli of the japonica variety, ‘Ilpum’ of Oryza sativa were induced in germinated dehulled seeds on a Mura- shige-Skoog solid medium [19] containing 2,4-20 di- chlorophenoxyacetic acid at 2 mg·L-1, casein hydrolysate (2 g·L-1), sucrose (30 g·L-1), 21 and gelrite (5 g·L-1). De-hulled seeds were incubated for 4 weeks at 26 ± 1˚C in permanent darkness. After incubation for 4 weeks, calli on the selection medium with somatic embryo-like structures were transferred to MS medium supplemented with 30 g·L-1 sucrose, 1 mg·L-1 1-naphthaleneacetic acid, 5 mg·L-1 kinetin, and 5 g·L-1 gelrite and exposed to in- tense light (approx. 45-5 mEm-2 s-1) at 26 ± 1˚C. After 3 weeks, the regenerated shoots were transferred to a bottle (6 × 16 cm) containing a fresh medium. When the shoots reached the top of the box, the plantlets were transferred to soil. 2.2. Measurement of Agronomic Traits Progeny populations (S1, S2) grown from the regenerated seed-culture (S0) were planted 8 at 30 × 15 cm planting distance on the practice field in Kyungpook National University, and the main agronomic characters were in- vestigated. Brown rice’s protein local, and starch content were analyzed by NIRS (Near-Infrared Spectroscopy, Foss 6500), and the amylose content used 3 repeated mean values provided by colorimetry of Juliano et al. [9]. 2.3. Genomic DNA Extraction, PCR, Southern, and RT-PCR Analysis DNA from the mutants was extracted using the CTAB method [20-25]. Ten-microgram aliquots of DNA were first digested with Hae III and then electrophoresed onto a 0.8% 18 agarose gel. PCR was performed in 50 volume of [1.5 mM MgCl2, Taq DNA polymerase, and a corresponding buffer from GibCo (Invitrogen, Carlsbad, Calif., http://www.invitrogen.com). Primer made amino acid sequences [8] of Tos 4 ~ Tos 20 by DNAsis 3.0 Pro- grams (Hitachi, Japan) using GenBank’s data base con- firmed the relationship existence of retrotransposon ele- ments of ‘Ilpum’ (Table 1). The DNA was denatured at 93˚C for 5 min followed by 35 cycles of amplification (1 min at 93˚C; 2 min at 40˚C; 2 min at 72˚C); the final in- cubation at 72˚C was extended to 5 min, and the reaction was cooled and kept at 4˚C. PCR were performed in a PTC-200 thermal cycler (MJ Research). RT-PCR (re- verse transcriptase-PCR) was used to analyze the mutants containing the retrotransposon elements as determined by the PCR and Southern analysis [2]. The total RNAs that were from the putative rice plants (leaves) using gua- nidine thiocyanate were isolated [11]. 2.4. Histology and Microscopy Observation Fresh hand-cut sections (~20 mm) of rice leaf were fixed in FAA solution [(10 ml Formalin, 5 ml Acetic acid (gla- cial), 95 ml 50% Ethylalcohol), (16 to 48 h, 48˚C)] and dehydrated through a graded ethanol series. Treated samples were transferred into water. The sections were microscopically examined (Automatic plant microtome, Mt-3, NK system) and photographed. The samples were embedded in paraffin (Fluka) and polymerized at 56~ 58˚C. Sections (50 μm) were cut and dropped with filtered paraffin, microscopically examined (Olympus, BX50), and photographed. Area measurements of the vascular elements were performed using an Olympus analySIS TS Lite software. 2.5. Amplification of Sequences Flanking Transposed Tos l7 Sequences flanking the transposed Tos 17 (target-site sequences) were amplified by an adaptor-PCR. Target- site sequences were amplified using the rice total DNA, respectively, as described [24] except that the total DNA was digested with Hae III. Two sets of primers were used for the two-step PCR: adaptor primer-1, GCGTAATAC- GACTCACTATAGCAATTAACC and Tos l7 primer-1, TGCTCTCCACTATGTGCCCTCCGAGCTA were used for the first PCR; the adaptor primer-2, GACTCACTA- TAGCAATTAAC and To s 17 primer-2, ACAAGTCG- CTGATTTCTTCAC were used for the second reaction. PCR was performed in a PTC-200 thermal cycler (MJ Research) and conducted as follows: first, the denature- tion step at 95˚C for 5 minutes, 20 cycles of 30 seconds at 94˚C and 1 minutes at 72˚C, followed by a final elon- gation step at 72˚C for 10 minutes; second the PCR was conducted with 5ul of the first PCR product under the following conditions: denaturation step at 94˚C for 5 min,  Functional Analysis for Rolling Leaf of Somaclonal Mutants in Rice 1 (Oryza sativa L.) Copyright © 2011 SciRes. AJPS 58 Table 1. The primer sequence of Tos families for PCR. Primer sequence Tos families Forward (5' → 3') Reverse (3' → 5') Tos 4 ACATTTTTACATggAgAgCTTgAg ATTCTACATCCTCATCagTAC Tos 5 ATATgCATCTCCTgATgCAgACTA CAAgCACAAAATAACTCCCTC Tos 6 gCACACTATgATTTAgAgTTgCAT AgTgTTCCTgTCCTggATTgC Tos 7 gACAgATATAAAgCTAgATTggTT AgTTTCTCAgTTggTgACAgA Tos 8 gCTATgTTACAAgCgAggACATAC CTTTTgCAACTAAgCgAgCTT Tos 9 gCTTACTCAgCTTCgCAAgCTCgA AgTCTTgCCTTgTgCCTAATT Tos 10 gCAAAATCCTAATggAAAggTggA AgTTATgTCAggTCgTgTAtg Tos 11 gCTAgTAgTgATgTCAgTCTACTg gTTgTgAgTgTACATTACTgC Tos 12 gCATTTTTACATggggAgTTAggA gAAgATAATCTgAAATgTgCA Tos 13 gCgTTTTTgCATggAgAgCTTgAg ACTgAgCAgCTgACAACTTAA Tos 14 TACATTgTATACCTCgTggATgAC TAAgCACAAgATCACCCCTTC Tos 15 gACggTACTATTgAAAAgTACAag AgTTgATTCCTAgCAATTCTC Tos 16 CTCAACCTCATAATggACgTgATA CATggTTTgTATCTCATgAgC Tos 17 gCTTTTCTTCATggTgATCTTCAT gAgTAACAACAAAgTAcgACC Tos 18 gCATTTTTACATggggAgTTAgAA AgTCAggACgAgAACATACCA Tos 19 gTTACTTCTTgggAATTgAggTTT ggATCATTAgCAATCTgTAtg Tos 20 ACAgTTTgCCCTCAgAACATgTTC AgTCTTCACATCTAACTgCTC 40 cycles of 30 seconds at 94˚C, 30 seconds at 60˚C, and 1 minute at 72˚C, followed by a final elongation step at 72˚C for 10min. Amplified products were loaded on a 1% agarose gel. The PCR products were purified by Hi- Yield™ Gel/PCR DNA Extraction Kit (RBC) and se- quenced by ABI3730XL (using Tos 17 primer-2). 2.6. Sequence Analysis and Database Search Handling of primary sequences and multiple sequence alignment were carried out using the DNAsis 3.0 soft- ware (Hitachi, Japan). Computer-based amino acid simi- larity searches of the Protein Identification Research, Genome annotation DB (Gramene, http://www.gramene. org/) and NCBI (http://www.ncbi.nlm.nih.gov/) were carried out with the FST (Flanking Sequence Tag). 3. Results There were 4 lines (1.3%) among the entire 297 lines that showed evidence of “rolling 19 leaf” mutant that oc- curred at the progeny (S1) using the brown rice culture of japonica type ‘Ilpum’. Compared with the “rolling leaf” mutant mutant, the donor plant ‘’Ilpum’, showed normal leaf with equal small veins, protoplasm formations, and conductive tissue of right and left in the midrib (Figure 1). The mutant had no equal arrangement of midrib and small veins. Because the bulliform cell was well-develo- ped, it ranged extensively, and the conductive tissue of the midrib was thick. Also, the sclerenchyma was thin (Figures 1(c) & (d)). Though the mutant “rolling leaf” plant high and the panicle number was less than the donor plant (Table 2), it was believed that the growth change with morphologi- cal characteristicsof leaf was secondary. These agro- nomic characteristics should be studied further to under- stand deeply the mechanism of such leaves. The retrotransposon’s activation known as one of the main causes of the conversion that occurred in tissue culture of rice, specifically for the 17 set oligonucleotide primers from conserved domain of Tos 4 ~ 20 reported in the ‘Nipponbare’ were analyzed 13 (Table 1). The PCR that the primers used for 3 classes (Tos 12, 15, 17) of the Tos 14 element was amplified in the ‘Ilpum’ of the mu- tants (Figure 2(a)). Tos element’s that appeared in the ‘Ilpum’ were different from the ‘Nipponbare’ [8]. These differences were believed to be from a genotype differ- ence caused by the cultivar which was used in PCR am- 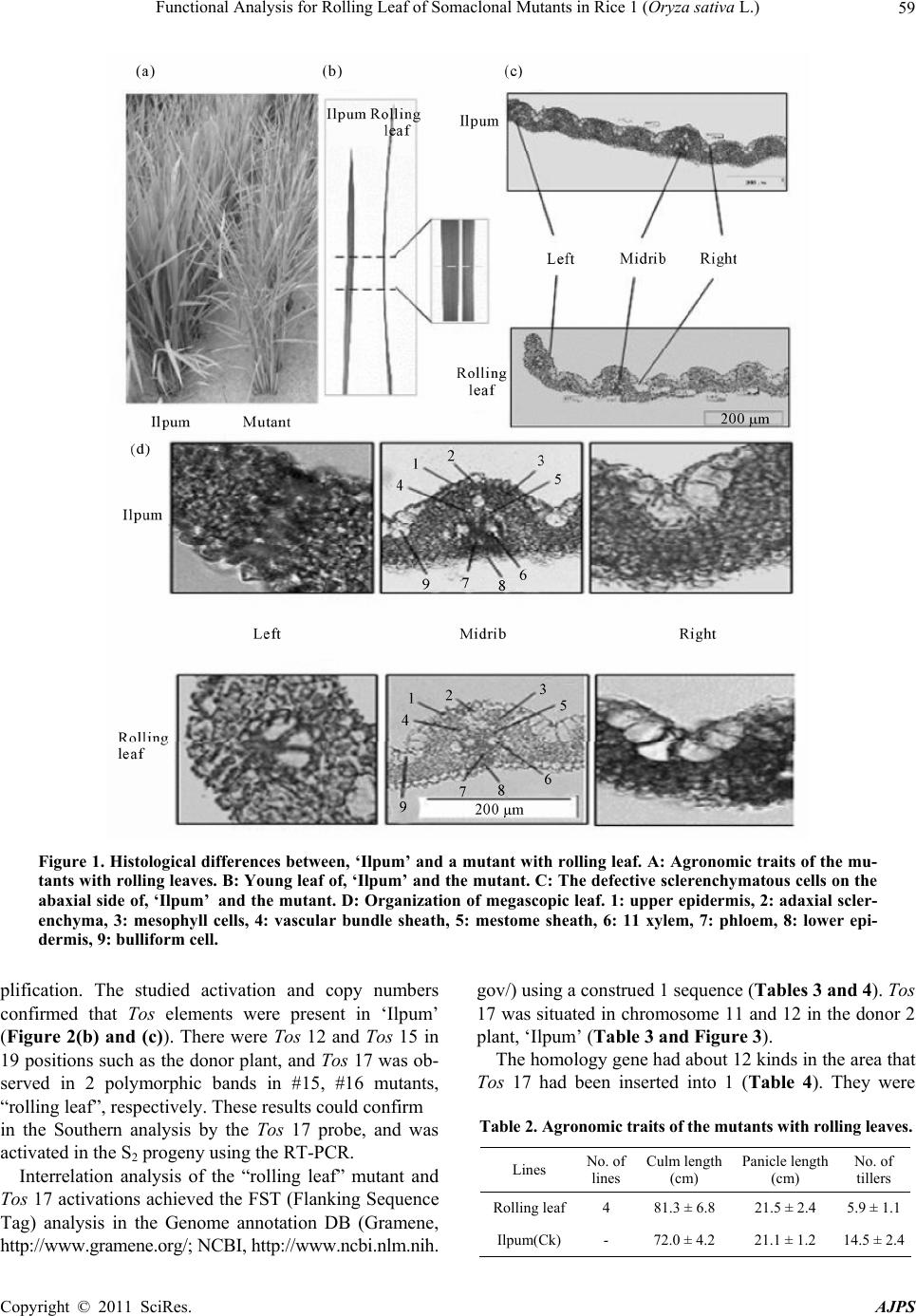 Functional Analysis for Rolling Leaf of Somaclonal Mutants in Rice 1 (Oryza sativa L.) Copyright © 2011 SciRes. AJPS 59 Figure 1. Histological differences between, ‘Ilpum’ and a mutant with rolling leaf. A: Agronomic traits of the mu- tants with rolling leaves. B: Young leaf of, ‘Ilpum’ and the mutant. C: The defective sclerenchymatous cells on the abaxial side of, ‘Ilpum’ and the mutant. D: Organization of megascopic leaf. 1: upper epidermis, 2: adaxial scler- enchyma, 3: mesophyll cells, 4: vascular bundle sheath, 5: mestome sheath, 6: 11 xylem, 7: phloem, 8: lower epi- dermis, 9: bulliform cell. plification. The studied activation and copy numbers confirmed that Tos elements were present in ‘Ilpum’ (Figure 2(b) and (c)). There were Tos 12 and Tos 15 in 19 positions such as the donor plant, and Tos 17 was ob- served in 2 polymorphic bands in #15, #16 mutants, “rolling leaf”, respectively. These results could confirm in the Southern analysis by the Tos 17 probe, and was activated in the S2 progeny using the RT-PCR. Interrelation analysis of the “rolling leaf” mutant and Tos 17 activations achieved the FST (Flanking Sequence Tag) analysis in the Genome annotation DB (Gramene, http://www.gramene.org/; NCBI, http://www.ncbi.nlm.nih. gov/) using a construed 1 sequence (Tables 3 and 4). Tos 17 was situated in chromosome 11 and 12 in the donor 2 plant, ‘Ilpum’ (Table 3 and Figure 3). The homology gene had about 12 kinds in the area that Tos 17 had been inserted into 1 (Table 4). They were Table 2. Agronomic traits of the mutants with rolling leaves. Lines No. of lines Culm length (cm) Panicle length (cm) No. of tillers Rolling leaf4 81.3 ± 6.8 21.5 ± 2.4 5.9 ± 1.1 Ilpum(Ck) - 72.0 ± 4.2 21.1 ± 1.2 14.5 ± 2.4  Functional Analysis for Rolling Leaf of Somaclonal Mutants in Rice 1 (Oryza sativa L.) Copyright © 2011 SciRes. AJPS 60 Figure 2. Detection of retrotransposon elements in the mu- tants and a donor cultivar ‘Ilpum’ using genomic DNA analysis. A: Detection of retrotransposon Tos 17 primer by PCR. m; size marker (λ/HindIII). c; ‘Ilpum’. #15~#17; mu- tants. B: DNA gel blot analysis by S1 progeny and RT-PCR by S2 progeny of the mutants, ‘rolling leaf’. C: The second PCR products derived from PCR amplification between the first PCR products and adaptor primer-1 in somaclonal mutants and a donor cultivar ‘Ilpum’. v; The bands were isolated from agarose gel, and sequenced (pass). ; The ☆ bands were isolated from agarose gel, and sequenced (fail). c; Ilpum, #15~16; mutants of rolling leaf. arabinoxylan arabinofuranohydrolase isoenzyme AXAH- II, cytochrome cd1-nitrite reductase-like, RNA-binding region RNP-1, metallothionein-like protein 1, and Cyto- chrome P450. Also, transcription factor rough sheath 2 like proteins were analyzed and showed high interrela- tionship in morphogenesis of “rolling leaf”. 4. Discussions The mutant “rolling leaf (rl)” was reported in 11 different kinds of the transposon element which affected the roll- ing leaf until the present. Also the rl1~rl6 alelle, reces- sive gene of 6 kinds, were situated within each chromo- some 1, 4, 12, 3, 7 thru formal marker analysis [12,13]. The rl7~rl10 alelle mapping in the chromosome 5 [16,17, 21,26,27], Rl(t), had an incomplete recessive gene, map- ping between the long arms (137 kb area) of chromosome 2 [22]. Yet there was also no report that there was any di- rect gene cloning. OsAGO7, the “rolling leaf” related gene, was reported to have an upward leaf curling in the over- expressing transformants [23]. Zhang et al. [28] re- ported that the Shallot-like 1 (SLL 1), MYB transcription control factor, belonging to the KANADI family was caused by the leaf-rolling cause on the chromosome 9 in the rice. The plant height of the mutant “rolling leaf” was higher than the donor plant. It was reported that there was a photosynthesis difference because of the carbon fixing or the gas exchange according to the three-dimen- sional structure of leaf [5]. Though the mutant “rolling leaf” was the transposon of the Tos 17, it occurred in chromosome 11 and 12, each differed from the studies of Shi et al. [23] and Zhang et al. [28] and the findings in this research (Table 3 and Figure 3). This result maybe Figure 3. Genes disrupted by insertion of Tos 17. Amino acid sequences deduced from target-site sequence (rolling leaf) are aligned with sequenced of GenBank data (‘Nipponbare’). Red letters: amino acid sequence of Tos 17, Rectangles: no homology.  Functional Analysis for Rolling Leaf of Somaclonal Mutants in Rice 1 (Oryza sativa L.) Copyright © 2011 SciRes. AJPS 61 Table 3. Chromosome locations of Tos 17 in a rice mutant with rolling leaf. Sequenced flanking region of Tos-DNA Matched region with rice chromosomes Tos- DNA end Adaptor start Reading length from to length Chr. from to Tos- DNA end Adaptor start Reading length 195 306 112 11 1492553 1492664 194 307 340 195 372 178 12 23812926 23812749 194 373 406 Table 4. Annotated genes of Tos 17 inserted flanking regions on chromosome in a rice 4 mutant with “rolling leaf”. Chromosome No. Accession No. Protein Origin NM_001072196.1 Arabinoxylan arabinofuranohydrolase isoenzyme AXAH-II Oryza sativa L. NM_001072197.1 Cytochrome cd1-nitrite reductase-like Oryza sativa L. NM_001072198.1 Protein kinase family protein Oryza sativa L. NM_001072200.1 TPR-like domain containing protein Oryza sativa L. NM_001072201.1 Resistance protein candidate (Fragment) Oryza sativa L. 11 NM_001072202.1 RNA-binding region RNP-1 Oryza sativa L. FAA00315.1 TPA: metallothionein-like protein Oryza sativa L. ABR25982.1 Metallothionein-like protein 1 Oryza sativa L. ABR25363.1 Mitochondrial import inner membrane translocase subunit tim9 Oryza sativa L. BAB61618.1 Transcription factor rough sheath 2 like protein Oryza sativa L. BAC78592.1 pre-mRNA splicing factor Oryza sativa L. 12 AAY44413.1 Cytochrome P450 Oryza sativa L. brought about by the mixing of a lot of gene numbers that caused the “rolling leaf” mutant. The FST analysis of the mutant “rolling leaf” and the cause of the muta- tion should be studied continuously via genetic and mo- lecular breeding of the S3 progeny to determine the rela- tion of gene function. 5. Acknowledgements We are grateful to Dr. Baek Hie Nahm (bhnahm@ gmail.com) for the technological study and critical read- ing of the manuscript. Sylvia T. Lapitan (stlapitan@yahoo. com) for critical proofreading of the manuX-X-SCRIPT. Flanking DNA sequencing was supported by GreenGene Biotech Inc.This work was supported by a grant from the Next Biogreen 21 R&D program, Rural Development Administration, Republic of Korea. REFERENCES [1] K. Arjun, E. Guiderdoni, G. An, Y. C. Hsing, C. D. Han, M. C. Lee, S. M. Yu, N. Upadhyaya, S. Ramachandran and Q. Zhang, “Mutant Resources in Rice for Functional Genomics of the Grasses,” Plant physiology, Vol. 149, No. 1, 2009, pp. 165-170. doi:10.1104/pp.108.128918 [2] H. G. Chin, M. S. Choe, S. H. Lee, S. H. Park, S. H. Park, J. C. Koo, N. Y. Kim, J. J. Lee, B. G. Oh, G. Yi, S. C. Kim, H. C. Choi, M. J. Cho, C. D. Han, “Molecular Analysis of Rice Plants Harboring an Ac/Ds Transposable Elementmediated Gene Trapping System,” The Plant Journal, Vol. 19, No. 5, 1999, pp. 615-623. [3] L. Gao, E. M. McCarthy, E. W. Ganko and J. F. McDon- ald, “Evolutionary History of Oryza sativa LTR Retro- Transposons: A Preliminary Survey of the Rice Genome Sequences,” BMC Genomics, Vol. 5, No. 1, 2004, pp. 1-18. doi:10.1186/1471-2164-5-18 [4] G. Gavazzi, C. Tonelli, G. Todesco, E. arreghini, F. Raf- faldi, F. Vecchio, G. Barbuzzi, M. Biasini and F. Sala, “Somaclonal Variation Versus Chemically Induced Muta- Genesis in Tomato(Lycopersicon esculentum L.),” Theo- retical and Appled Genetics, Vol. 74, No. 6, 1987, pp. 733-738. [5] Y. M. Govaerts, S. Jacquemoud, M. M. Verstraete and S. L. Ustin, “Three-10 Dimensional Radiation Transfer Modeling in a Dicotyledon Leaf,” Applied Optics, Vol. 35, No. 33, 1996, pp. 6585-6598. doi:10.1364/AO.35.006585 [6] H. Hirochika, A. Fukuchi and F. Kikuchi, “Retrotranspo- son Families in Rice,” Molecular and General Genetics, Vol. 233, No. 1-2, 1992, pp. 209-216. doi:10.1007/BF00587581  Functional Analysis for Rolling Leaf of Somaclonal Mutants in Rice 1 (Oryza sativa L.) Copyright © 2011 SciRes. AJPS 62 [7] H. Hirochika, H. Otsuki, M. Yoshikawa, Y. Otsuki, K. Sugimoto and S. Takeda, “Autonomous Transposition of the Tobacco Retrotransposon Tto 1 in rice,” The Plant Cell, Vol. 8, No. 4, 1996a, pp. 725-734. [8] H. Hirochika, K. Sugimoto, Y. Otski, H. Tsugawa and M .Kanda, “Retrotransposones of Rice Involved in Muta- tions Induced by Tissue Culture,” Proceedings of Na- tional Acadenny of Sciences of the United States of America, Vol. 93, No. 15, 1996b, pp. 7783-7788. doi:10.1073/pnas.93.15.7783 [9] B. O. Juliano, C. M. Perez, A. B. Blakeney, T. Castillo, N. Ongseree, B. Laignelet, E. T. Lapis, V. V. S. Murty, C. M. Paule and B. D. Webb, “International Cooperative Test- ing on the Amylose Content of Milled Rice,” Starch, Vol. 33, No. 5, 1981, pp. 157-162. [10] Y. U. Jun, S. Hu, J. Wang, S. Li, K. S. G. Wong, B. Liu, Y. Deng, L. Dai, Y. Zhou, X. Zhang, M. Cao, J. Liu, J. Sun, J. Tang, Y. Chen, X. Huang, W. Lin, C. Ye, W. Tong, L. Cong, J. Geng, Y. Han, L. Li, W. Li, G. Hu, X. Huang, W. Wi, J. Li, Z. Liu, L. Li, J. Liu, Q . Qi, J. Liu, L. Li, X .Wang, H. Lu, T. Wu, M. Zhu, P. Ni, W. Dong, X. Ren, X. Feng, P. Cui, X. Li, H. Wang, X. Xu, W. Zhai, Z. Xu, J. Zhang, S .He, J. Zhang, J. Xu, K. Zhang, X. Zheng, J. Dong, W. Zeng, L. Tao, X. Chen, J. He, D. Liu, W. Tian, H .Xia, G. Li, H. Gao, P. Li, W. Chen, X. Wang, Y. Zhang, J. Hu, J. Wang, S. Liu, J. Yang, G. Zhang, Z. Li, C. Zhou, R. Chen, B. Hao, W. Zheng, S. Chen, W. Guo, G. Li, S. Liu, G. Hunag, M. Tao, J. Wang, L. Zhu, L. Yuan and H. Yang, “A Draft Sequence of the Rice (Oryza sativa ssp. indica) Genome,” Chinese Science Bulletin, Vol. 46, No. 23, 2001, pp. 1937-1942. doi:10.1007/BF02901901 [11] M. Kawai, S. Kidou, A. Kato, H. Uchimiya, “Molecular Characterization of cDNA Encoding for Adenylate Kinase of Rice (Oryza sativa L.),” The Plant Journal, Vol. 2, No. 6, 1992, pp. 845-854. [12] G. S. Khush and T. Kinoshita, “Rice Karyotype, Marker Genes, and Linkage Group,” Rice Biotechnology, CAB International, Wallingford, 1991, pp. 83-107. [13] T. Kinoshita, “Gene Analysis and Linkage Map,” Biology of Rice, JSSP/Elsevier, Tokyo, 1984 pp. 187-274. [14] P. J. Larkin, P. M. Bank, R. Bhati, R. I. S. Brettelli, P. A. Davies, S. A. Ryan, W. R. Scowcroft, L. H. Spindle and G. T. Tanner, “From Somatic Variant to Variant Plants: Mechanisms and Applications,” Genomes, Vol. 31, No. 2, 1989, pp. 705-711. [15] P. J. Larkin and W. R. Scowcroft, “Somaclonal Varia- tion—A Novel Source of Variability from Cell Cultures for Plant Improvement,” Theoretical and Applied Genet- ics, Vol. 60, No. 4, 1981, pp. 197-214. doi:10.1007/BF02342540 [16] S. G. Li, Y. Q. Ma, P. He, H. Y. Li, Y. Chen, K. D. Zhou and L. H. Zhu, “Genetics Analysis and Mapping the Flag Leaf Roll in Rice (Oryza. sativa. L.),” Journal Sichuan Ag- riculture University, Vol. 16, 1998, pp. 391-393. [17] Z. Luo, Z. Yang, B. Zhong, Y. Li, R. Xie, F. Zhao, Y. Ling and G. He, “Genetic Analysis and Fine Mapping of a Dynamic Rolled Leaf Gene, RL 10(t), in Rice (Oryza. sativa. L.),” Genome, Vol. 50, No. 9, 2007, pp. 811-817. doi:10.1139/G07-064 [18] A. Miyao, Y. Iwasaki, H. Kitano, J. I. Itoh, M. Maekawa, K. Murata, O. Yatou, Y. Nagato and H. Hirochika, “A Large-Scale Collection of Phenotypic Data Describing an Insertional Mutant Population to Facilitate Functional Analysis of Rice Genes,” Plant Molecular Biology , Vol. 63, No. 5, 2007, pp. 625-635. doi:10.1007/s11103-006-9118-7 [19] T. Murashige and F. Skoog, “A revised Medium for Rapid Growth and Bioassays with Tobacco Tissue Cul- tures,” Physioogial Plantarum, Vol. 15, No. 3, 16 1962, pp. 473-497. [20] K. M. Nori, K. Miyoshi, K. I. Nonomura, Y. Yamazaki and Y. Ito, “Rice Mutants and Genes Related to Organ Development, Morphogenesis and Physiological Traits,” Plant Cell Physiology, Vol. 46, No. 1, 2005, pp. 48-62. doi:10.1093/pcp/pci506 [21] Y. j. Shao, Z. X. Chen, Y. F. Zhang, E. H. Chen, D. C. Qi, J. Miao and X. B. Pan, “One Major QTL Mapping and Physical Map Construction for Rolled Leaf in Rice,” Acta Genetica Sinica (in Chinese), Vol. 32, No. 5, 2005a, pp. 501-506. [22] Y. J. Shao, C. H. Pan, Z. X. Chen, S. M. Zuo, Y. F. Zhang and X. B. Pan, “Fine Mapping of an Incomplete Recessive Gene for Leaf Rolling in Rice(Oryza. sativa. L.),” Chiness Science Bulletin (in Chinese), Vol. 50, No. 21, 2005b, pp. 2466-2472. [23] Z. Y. Shi, J. Wang, X. S. Wan, G. Z. Shen, X. Q. Wang and J. l. Zhang, “Over-Expression of Rice OsAGO 7 Gene Induces Upward Curling of the Leaf Blade that Enhanced Erect-Leaf Habit,” Planta, Vol. 226, No. 1, 2007, pp. 99-108. doi:10.1007/s00425-006-0472-0 [24] K. Sugimoto, Y. Otsuki, S. Saji and H. Hirochika, “Transposition of the Maize Ds Element from a Viral Vector to the Rice Genome,” The Plant Journal, Vol. 5, No. 6, 1994, pp. 863-871. doi:10.1046/j.1365-313X.1994.5060863.x [25] B. Winnepenninckx, T. Backeljau and R. De Wachter, “Extraction of high molecular weight DNA from mol- lusks,” Trends in Genetics, Vol. 9, No. 12, 1993, pp. 407. doi:10.1016/0168-9525(93)90102-N [26] Q. J. Xie, M. C. Ruch and J. H. Oard, “Homozygous Variation in Rice Somaclones: Non Random Variation In- stead of Mitotic Recombination,” Crop Science, Vol. 35, No. 4, 1995, pp. 954-957. doi:10.2135/cropsci1995.0011183X003500040002x [27] C. J. Yan, S. Yan, Z. Q. Zhang, G. H. Liang, J. F. Lu and M. H. Gu, “Genetic Analysis and Gene Fine Mapping for a Rice Novel Mutant rl 9(t) with Rolling Leaf Character,” Chi- ness Science Bulletin (in Chinese), Vol. 51, No. 1, 2006, pp. 63-69. [28] G. H. Zhang, Q. Xu, X. D. Zhu, Q. Qian and H. W. Xue, “Shallot-Like is a KANDI Transcription Factor that Modulates Rice Leaf Rolling by Regulating Leaf Abaxial Cell Development,” The Plant Cell Prev iew, Vol. 21, No. 3, 2009, pp. 719-735. doi:10.1105/tpc.108.061457 |

