Paper Menu >>
Journal Menu >>
 American Journal of Plant Sciences, 2011, 2, 15-26 doi:10.4236/ajps.2011.21002 Published Online March 2011 (http://www.SciRP.org/journal/ajps) Copyright © 2011 SciRes. AJPS 15 Development of a Molecular Marker to Identify a Candidate Line of Turmeric (Curcuma longa L.) with a High Curcumin Content —Development of Molecular Marker of Turmeric Hiroshi Hayakawa1,2, Tetsuya Kobayashi2, Yukio Minaniya2,3, Katsu ra Ito4, Akira Miyazaki2, Tatsuya Fukuda2*, Yoshinori Yamamoto2 1United Graduate School of Agricultural Sciences, Ehime University, Monobe, Nankoku, Japan; 2Faculty of Agriculture, Kochi Uni- versity, Monobe, Nankoku, Japan; 3Graduate School of Integrated Arts and Sciences, Kochi University, Monobe, Nankoku, Japan; 4JST Innovation Satellite Kochi, Laboratory of Applied Entomology, Faculty of Agriculture, Kochi University, Monobe, Nankoku, Japan. Email: tfukuda@kochi-u.ac.jp Received October 13th, 2010; revised November 15th, 2010; accepted November 29th, 2010. ABSTRACT Dried and fresh rh izomes of the spice turmeric (Curcuma longa L.) are well kno wn in traditional medicine, and curcu- min is widely used in various geograph ic regions. Although there are differences in th e amount of curcumin within this species, identification of the ca ndidate lin e by rhizome is difficult beca use of the relative simplicity of its morp hologica l characteristics. To accura tely identify lines of C. longa with a high content of cu rcumin, we analysed several sequences of chloroplast DNA. First, to determine the appropriate outgroup taxa in which to conduct infra specific analyses of C. longa, we reconstructed the molecular phylogenetic tree of C. longa and its allied species. The results showed that C. aromatica and C. zedoa ria are closely related to C. longa. Next, to develop a molecular marker for identifying lines of C. longa with a high content of curcumin, a network analysis using chloroplast microsatellite regions was performed. Results showed that a unique haplotype within C. longa corresponds to the high curcumin content line. Therefore, the chloroplast microsatellite regions used for the analysis allowed us to determine the lines of this species with high cu r- cumin content. Keywords: Chloroplast DNA, Curcuma, Curcuma longa, Curcumin, Molecular Analysis 1. Introduction Traditional medicine is known to be fertile ground for the source of modern medicines [1]. One medicine in that category is curcumin, a yellow coloring agent present in the spice turmeric (Curcuma longa L.) that belongs to the ginger family (Zingiberaceae). Besides its use in cooking to add color and as a preservative, turmeric is used in Indian traditional medicine to treat various common ail- ments including stomach upset, flatulence, dysentery, ulcers, jaundice, arthritis, sprains, wound s, acne, and skin and eye infections [2]. In this way, curcumin is widely used in various geographic regions in which the mor- phologies of the dried or fresh rhizomes of Curcuma plants are highly similar, and it is therefore difficult to correctly identify individual Cur cu ma species. Recent molecular phylogenetic studies have revealed new aspects of the relationships among Curcuma species. For example, to identify the rhizomes of C. longa, Sasaki et al. [3] developed a molecular identification method fo r Curcuma species using an amplification-refractory muta- tion system analysis of 18S ribosomal ribonucleic acid (rRNA) and the trnK gene. Although this method can identify species in a line of samples with high accuracy, the process is complicated. This marker is sensitive to experimental conditions of the polymerase chain reaction (PCR) because judgment is based on the presence or ab- sence of PCR products. In addition, Minami et al. [4] reported that Curcuma species can b e identified using an intergenic spacer between trnS and trnfM of chloroplast  Development of a Molecular Marker to Identify a Candidate Line of Turmeric (Curcuma longa L.) with a High Curcumin Content Copyright © 2011 SciRes. AJPS 16 deoxyribonucleic acid (cpDNA). Although this method is useful for identifying C. longa from various Curcuma rhizomes, the study by Minami et al. [4] may require re-evaluation since the number of sample species used were few, and it did not employ appropriate outgroup taxa. The same can be said of the study by Sasaki et al. [3]. In our view, out-groups are important for assessing plesiomorphic or apomorphic states for each characteris- tic and for interpreting the phylogenetic relationships within C. longa. Alternatively, Aoi et al. [5] reported a difference in the amount of curcumin found among indi- viduals of C. longa. Therefore, it is necessary to use in- fraspecific genetic markers to determine the amount of curcumin in individuals of this species in order to acquire it efficiently. Large-scale sequencing of a predefined region of ap- proximately 1500 base pairs (bp) of the cpDNA matK has one main goal in monocotyledonous plants including C. longa: to clarify the phylogen etic position and closely related species in order to determine the outgroup taxa of unknown individuals [6]. Although the sequence of the matK gene plays an important role in identifying the out- group taxa of C. longa, it cannot be us ed to identify lin es in this species containing high amounts of curcumin be- cause of its low substitution rate [7]. Thus, determining more reliable genetic markers within C. longa requires more informative DNA regions. In recent years, cpDNA sequence data have been used frequently to reconstruct the phylogenetic relationships of a wide range of land plants [8]. Some regions were reported to have nucleo- tide substitution rates that are sufficiently high to facili- tate the elucidation of relationsh ips among closely related species [7,9]. Moreover, the utility of non-coding cpDNA regions within species-level phylogeny has been demon- strated in many taxonomic groups of land plants [10]. However, phylogenetic analysis using regions with high substitution rates, such as microsatellites, is often biased by multiple substitutions at a single site. In addition, be- cause algorithms of phylogenetic analysis can cause problems when data do not represen t a tree-like structure [11], we also analysed the cpDNA sequences with the statistical parsimony network approach. This approach reflects the genealogical relationships of the sequences used; that is, single-mutation steps separate adjacent haplotypes in the network, and older haplotypes are placed at internal branching points, whereas younger haplotypes occur towards the tip position. In the current study, we show that this analysis of th e infraspecific rela- tionships within C. longa can result in identification of clear relationships not only among young and rapidly speciating groups but also among old lineages reaching deep into the diverg ence of this group. Thus, the aim of our study was to develop infraspecific genetic primers, using a phylogenetic approach, for iden- tifying lines of C. longa with high curcumin content. 2. Materials and Methods 2.1. Cultivation and Measurements The experiments were carried out in the fields (sandy soil) of the Faculty of Agriculture, Kochi University, Japan for four years (2006 -2009). Ta b le 1 lists th e sample lines used for the cultivation experiments in this study: 2 C. aromatica Salisb., 12 C. longa L., and 1 C. zedoaria Rosc. Rows were constructed with a width of 70 cm and height of 20 cm. Rhizomes were transplanted 8 cm below the soil surface in one-row ridges, on hills separated by a distance of 30 cm, in late May for four years. For fertile- izer dressing, a total of 1.5 kg/a of N, 0.6 kg/a of P2O5, and 1.4 kg/a of K2O was applied over four years. In addi- tion, 200 kg/a of compost fertilizer, 15 kg/a of magnesia lime, and 30 kg/a of chicken droppings were applied (Chicken droppings were not used in 2006.). The ex- perimental plots were arranged in a randomized complete design with two replicates, which formed three rows. Due to a lack of seed rhizomes, some lines of C. longa were examined without replicate or two rows. Rows were covered by silver mulch. Irrigation was performed using a sprinkler, with observation of the amount of precipita- tion and the condition of th e plants. Fo ur or six h ills con- taining average-sized shoots per line were sampled in early December each year for four years. For curcumin analysis, the curcuminoid content of cur- cumin 1, curcumin 2, and curcumin 3 (curcumin, deme- thoxycurcumin, and bis-deme thoxy curcumin, respectively) of the primary branch rhizome was measured by high performance liquid chromatography (HPLC, HITACHI, L-2420), according to the method described by Sato et al. [12]. We extracted curcumin by 80% of ethanol from 0.3 g of the dried and gro und samples. H PLC was perfo rmed on a column of YMC-Pack Pro C18. Mixed acetonitrile with 0.1% of phosphoric acid (50:50) was used for the mobile phase. Current flowed at a rate of 1 mL per min. The amount of injection was 10 µL. 2.2. DNA Analysis Total DNA was isolated from fresh root with a Plant Genomic DNA Mini Kit (Viogene, Sunnyvale, CA, USA), according to the manufacturer’s protocol. The PCR mix contained approximately 100 to 200 ng of total DNA, 1 µM of each primer, 200 mM of each deoxynucleotide, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM of MgCl2, and 1.25 units o f Taq po lymerase. Doub le-stranded D NA was amplified, after incubation at 94˚C for 2 min, by 45  Development of a Molecular Marker to Identify a Candidate Line of Turmeric (Curcuma longa L.) with a High Curcumin Content Copyright © 2011 SciRes. AJPS 17 Table 1. List of sample of curcuma species used for cultivation experiments in this study. Nos. analysed hill1) Species Locality 2006 2007 20082009 Curcuma aromatica (Kochi) Kochi Prefecture, Japan 6 6 4 4 C. aromatica (Tanegashima) Tanegashima Island, Kagoshima Prefecture, Japan 6 6 6 4 C. longa (Kochi) Kochi Prefecture, Japan 6 6 6 4 C. longa (Tanegashima) Tanegashima Island , Kagoshima P r efecture, Japan 6 6 6 4 C. longa (Wakayama A) Wa k ayama Prefecture, Japan 6 6 4 4 C. longa (Wakayama B) Wakayama Prefecture, Japan 6 4 4 4 C. longa (Wakayama C) Wakayama Prefectur e, Japan ---2) 6 4 4 C. longa (Okinawa A) Okinawa Prefecture, Japan --- --- 4 4 C. longa (Okinawa B) Okinawa Prefecture, Japan --- --- 4 4 C. longa (Indonesia A) Bogol, West Java, Indone sia 4 4 4 4 C. longa (Indonesia B) Bogol, West Java, Indonesia --- 4 4 4 C. longa (Indonesia C) Bogol, West Java, Indonesia --- --- --- 4 C. longa (Vietnam A) Vietnam --- --- --- 4 C. longa (Vietnam B) Vietnam --- --- --- 4 C. zedoaria Unknown 4 4 4 4 1) Four or s i x hills conta ining average-sized shoots per line were harvested; 2) Not cultivated. cycles of incubation at 94˚C for 1.5 min, 48˚C for 2 min, and 72˚C for 3 min, with final extension at 72˚C for 15 min. We amplified four regions from cpDNA, namely matK, rp l16 intron 2, petB intron 1, and petB intron 2, with primers designed by Johnson and Soltis [13] and Nishizawa and Watano [9]. After amplification, reaction mixtures were subjected to electrophoresis in 1% low melting-temperature agarose gels for separation of spe- cific amplified products. We sequenced the purified PCR products using a Big Dye Terminator Cycle Sequencing Kit (ABI PRISM, Perkin Elmer Applied Biosystems, Foster City, CA, USA) and an ABI PRISM 3100-Avant Genetic Analyzer according to the manufacturer’s in- structions. The primers used for sequencing were the same as those used for amplification. To construct a phylogenetic tree based on matK se- quences of Curcuma and its allied species, the amplified regions were aligned using ClustalW [14] and were im- proved manually using MEGA 4 [15]. Phylogenetic rela- tionships were analysed using the neighbour-joining (NJ) method with PAUP* 4.08b [16]. The NJ analyses were performed using MEGA 4 with Kimura’s two-parameter model. For the NJ analyses, bootstrapping with 1000 pseudo-replicates was chosen to examine the robustness of the clades and their phylogenetic relationships. In ad- dition, a network tree of microsatellite reg ions of cpDNA was constructed with Network 1.4.1.2 (Fluxus Technol- ogy Ltd., Suffolk, UK) using the median-joining method [17]. 3. Results 3.1. Curcumin Content We mainly describe results of the cu ltivation experiments for 2009, since data for the previous years of the study (2006, 2007, and 2008) showed similar trends. Table 4 shows the curcumin content in the maturity period for the years 2006 through 2009. With respect to the curcumin content of primary branch rhizomes in the maturity period, total curcuminoid content, in order from highest to lowest, was as follows: South Asian C. longa (3198.4-2315.1 mg/100 g), with the exception of Indone- sia B > domestic C. longa (305.0-392.2 mg/100 g), with the exception of Wakayama B > Indonesia B (309.5 mg/100 g) > C. aromatica (121.9-126.9 mg/100 g). In- donesia A and B and Vietnam A and B showed a high content of accumulated curcuminoid in the rhizome. However, C. longa (Wakayama B) and C. zedoaria 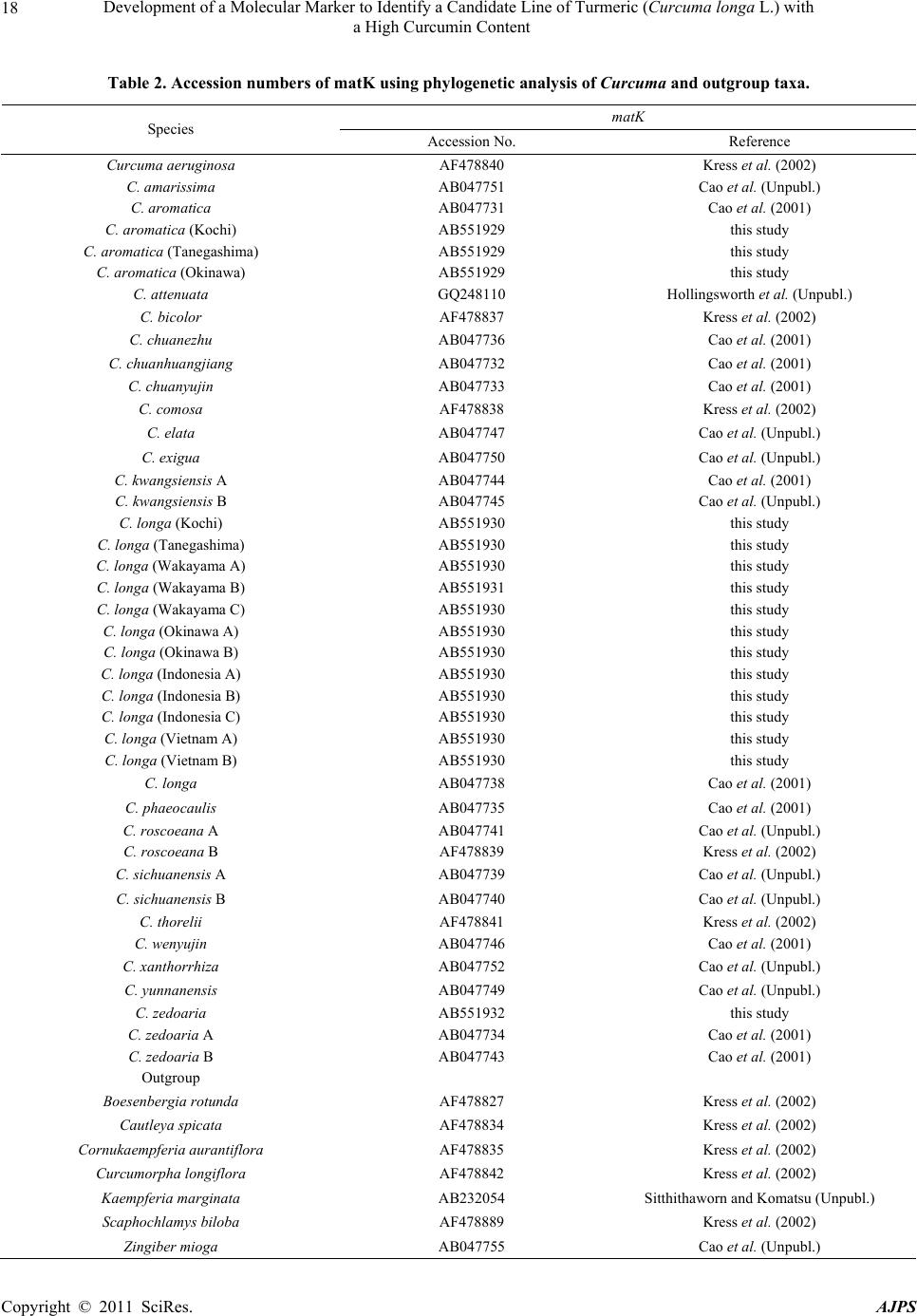 Development of a Molecular Marker to Identify a Candidate Line of Turmeric (Curcuma longa L.) with a High Curcumin Content Copyright © 2011 SciRes. AJPS 18 Table 2. Accession numbers of matK using phy logenetic analy sis of Curcuma and outgroup taxa. matK Species Accession No. Reference Curcuma aeruginosa AF478840 Kress et al. (2002) C. amarissima AB047751 Cao et al. (Unpubl.) C. aromatica AB047731 Cao et al. (2001) C. aromatica (Kochi) AB551929 this study C. aromatica (Tanegashima) AB551929 this study C. aromatica (Okinawa) AB551929 this study C. attenuata GQ248110 Hollingsworth et al. (Unpubl.) C. bicolor AF478837 Kress et al. (2002) C. chuanezhu AB047736 Cao et al. (2001) C. chuanhuangjiang AB047732 Cao et al. (2001) C. chuanyujin AB047733 Cao et al. (2001) C. comosa AF478838 Kress et al. (2002) C. elata AB047747 Cao et al. (Unpubl.) C. exigua AB047750 Cao et al. (Unpubl.) C. kwangsiensis A AB047744 Cao et al. (2001) C. kwangsiensis B AB047745 Cao et al. (Unpubl.) C. longa (Kochi) AB551930 this study C. longa (Tanegashima) AB551930 this study C. longa (Wakayama A) AB551930 this study C. longa (Wakayama B) AB551931 this study C. longa (Wakayama C) AB551930 this study C. longa (Okinawa A) AB551930 this study C. longa (Okinawa B) AB551930 this study C. longa (Indonesia A) AB551930 this study C. longa (Indonesia B) AB551930 this study C. longa (Indonesia C) AB551930 this study C. longa (Vietnam A) AB551930 this study C. longa (Vietnam B) AB551930 this study C. longa AB047738 Cao et al. (2001) C. phaeocaulis AB047735 Cao et al. (2001) C. roscoeana A AB047741 Cao et al. (Unpubl.) C. roscoeana B AF478839 Kress et al. (2002) C. sichuanensis A AB047739 Cao et al. (Unpubl.) C. sichuanensis B AB047740 Cao et al. (Unpubl.) C. thorelii AF478841 Kress et al. (2002) C. wenyujin AB047746 Cao et al. (2001) C. xanthorrhiz a AB047752 Cao et al. (Unpubl.) C. yunnanensis AB047749 Cao et al. (Unpubl.) C. zedoaria AB551932 this study C. zedoaria A AB047734 Cao et al. (2001) C. zedoaria B AB047743 Cao et al. (2001) Outgroup Boesenbergia rotunda AF478827 Kress et al. (2002) Cautleya spicata AF478834 Kress et al. (2002) Cornukaempferia auran tiflora AF478835 Kress et al. (2002) Curcumorpha longiflora AF478842 Kress et al. (2002) Kaempferia marginata AB232054 Sitthithaworn and Komatsu (Unpubl.) Scaphochlamys biloba AF478889 Kress et al. (2002) Zingiber mioga AB047755 Cao et al. (Unpubl.)  Development of a Molecular Marker to Identify a Candidate Line of Turmeric (Curcuma longa L.) with a High Curcumin Content Copyright © 2011 SciRes. AJPS 19 Table 3. Accession numbers bas ed on network analysis of Curcuma longa and allied species. Type of chloroplast DNA Species rps16 intron 2 petB intron 1 petB intron 2 Haplotype Curcuma zedoaria a1) e g A C. aromatica (Kochi) b f i B C. aromatica (Tanegashima) b f i B C. longa (Kochi) c e j D C. longa (Tanegashima) c e j D C. longa (Wakayama A) c e j D C. longa (Wakayama B) c e j D C. longa (Wakayama C) c e j D C. longa (Okinawa A) c e j D C. longa (Okinawa B) c e j D C. longa (Indonesia A) d f h C C. longa (Indonesia B) c e j D C. longa (Indonesia C) d f h C C. longa (Vietnam A) d f h C C. longa (Vietnam B) d f h C 1) Type of chloroplast DNA. Accession numbers; a: AB557651, b: AB557648, c: AB557650, d: AB557649, e: AB557653. f: AB557652, g: AB557657, h: AB557655, i: AB557654, j: AB5576 5 6. Haploty pe is corresponded to Figure 3. showed little curcuminoid content. The inside color of the rhizome in C. longa (Wakayama B) and C. zedoaria was white and purple, respectively, whereas C. longa, which showed an accumulation of curcuminoid, had a yellow-colored rhizo me (Figure 1). In order from highest to lowest of each content of curcumin 1, 2, and 3 showed a similar trend with the or- der of total curcuminoid con tent. With respect to th e ratio of its curcumin 1, 2, and 3 in domestic C. longa (with the exception of Wakayama B) was 69%-71%, 21%-23%, and 8%-9%, respectively. However, the ratio of curcu- minoid content in South Asian C. longa (with the excep- tion of Indonesia B) was 46%-52% (curcumin 1), 26%-28% (curcumin 2), and 22%-26% (curcumin 3), indicating a low percentage of curcuminoid in curcumin 1 and a high percentage of curcuminoid in curcumin 3, compared with domestic C. longa. The ratio of curcu min 1, 2, and 3 in Indonesia B differed from that in Indonesia A and C and more closely resembled domestic C. longa, despite the fact that Indonesia A, B, and C originated in the same country. In C. aromatica, the ratio of curcumin 1, 2, and 3 was 47%-53%, 47%-52%, and 0-1%, respect- tively. C. aromatica also had a high percentage of cur- cumin 2. The relative order of curcuminoid content in the pri- mary branch rhizomes was similar throughout the four years of the study. Morishita et al. [18] reported that year-to-year variation in curcuminoid content is more vigorous than that of the varietal differences in turmeric. In the present study, the difference of curcuminoid con- tent in Curcuma species had not only varietal difference but also year-to-year variation. 3.2. DNA Analysis To construct the molecular phylo genetic tree o f Curcuma and its allied species, we determined sequences of the matK gene of Curcuma cpDNA and seven outgroups of Boesenbergia rotunda, Cautleya spicata, Cornukaemp- feria aurantiflora, Curcumorpha longiflora, Hedychium greenei, Scaphochlamys biloba , and Zingiber mioga. The lengths of the matK gene of Curcuma species varied from 1831 bp (C. longa (Wakayama B)) to 1846 bp (C. thorelii). The results of the phylogenetic analysis indicated that Curcuma species consists of a monophyletic group (Fig- ure 2), with the genus Curcu ma primarily divided into two monophyletic groups: clade 1 and clade 2. Clade 1, located at the most basal monophyletic position of Cur - 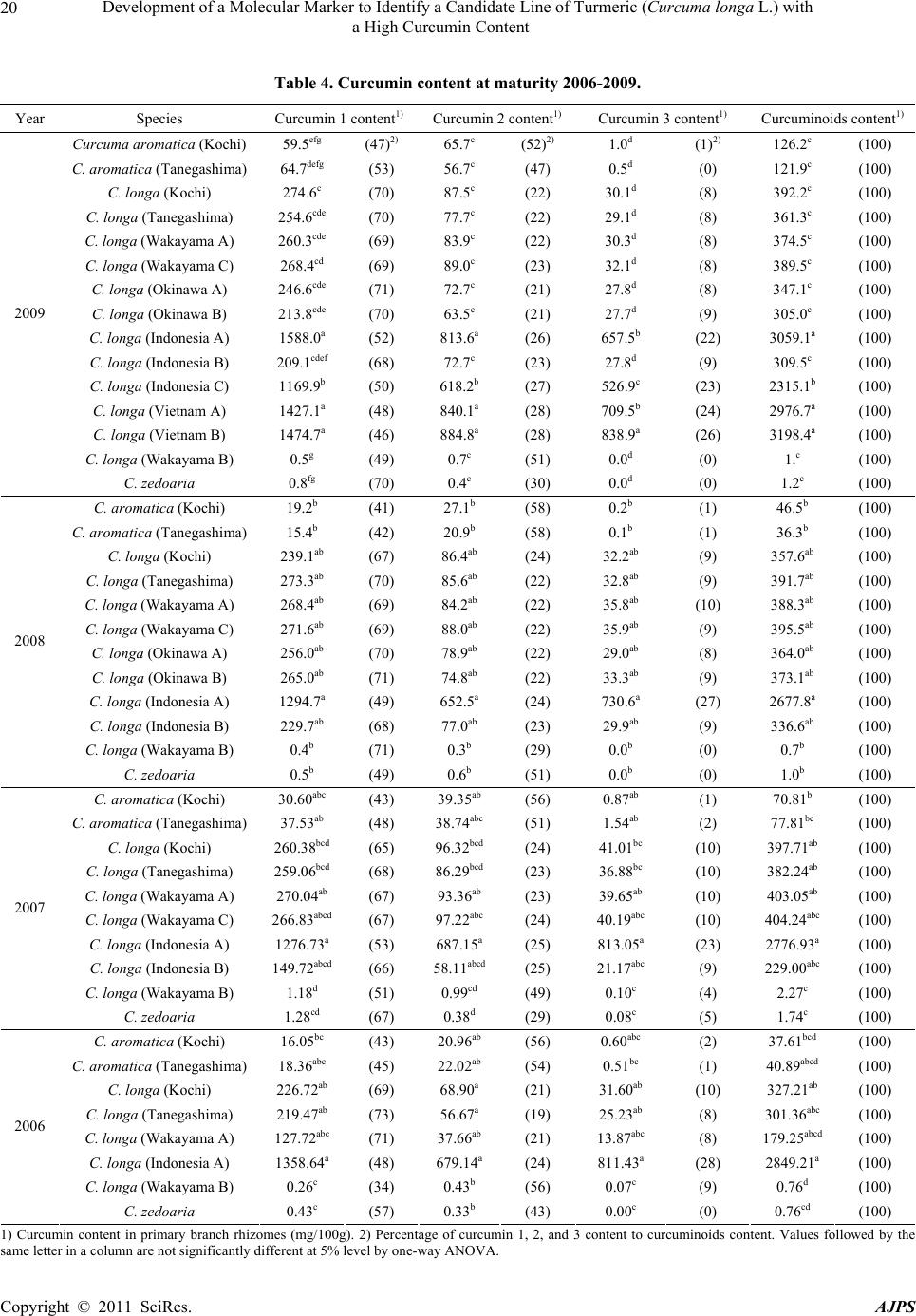 Development of a Molecular Marker to Identify a Candidate Line of Turmeric (Curcuma longa L.) with a High Curcumin Content Copyright © 2011 SciRes. AJPS 20 Table 4. Curcumin content at maturity 2006-2009. Year Species Curcumin 1 content1) Curcumin 2 content1) Cu rcumin 3 content1) Curcuminoids content1) Curcuma aromatica (Kochi) 59.5efg (47)2) 65.7c (52)2) 1.0d (1)2) 126.2c (100) C. aromatica (Tanegashima) 64.7defg (53) 56.7c (47) 0.5d (0) 121.9c (100) C. longa (Kochi) 274.6c (70) 87.5c (22) 30.1d (8) 392.2c (100) C. longa (Tanegashima) 254.6cde (70) 77.7c (22) 29.1d (8) 361.3c (100) C. longa (Wakayama A) 260.3cde (69) 83.9c (22) 30.3d (8) 374.5c (100) C. longa (Wakayama C) 268.4cd (69) 89.0c (23) 32.1d (8) 389.5c (100) C. longa (Okinawa A) 246.6cde (71) 72.7c (21) 27.8d (8) 347.1c (100) C. longa (Okinawa B) 213.8cde (70) 63.5c (21) 27.7d (9) 305.0c (100) C. longa (Indonesia A) 1588.0a (52) 813.6a (26) 657.5b (22) 3059.1a (100) C. longa (Indonesia B) 209.1cdef (68) 72.7c (23) 27.8d (9) 309.5c (100) C. longa (Indonesia C) 1169.9b (50) 618.2b (27) 526.9c (23) 2315.1b (100) C. longa (Vietnam A) 1427.1a (48) 840.1a (28) 709.5b (24) 2976.7a (100) C. longa (Vietnam B) 1474.7a (46) 884.8a (28) 838.9a (26) 3198.4a (100) C. longa (Wakayama B) 0.5g (49) 0.7c (51) 0.0d (0) 1.c (100) 2009 C. zedoaria 0.8fg (70) 0.4c (30) 0.0d (0) 1.2c (100) C. aromatica (Kochi) 19.2b (41) 27.1b (58) 0.2b (1) 46.5b (100) C. aromatica (Tanegashima) 15.4b (42) 20.9b (58) 0.1b (1) 36.3b (100) C. longa (Kochi) 239.1ab (67) 86.4ab (24) 32.2ab (9) 357.6ab (100) C. longa (Tanegashima) 273.3ab (70) 85.6ab (22) 32.8ab (9) 391.7ab (100) C. longa (Wakayama A) 268.4ab (69) 84.2ab (22) 35.8ab (10) 388.3ab (100) C. longa (Wakayama C) 271.6ab (69) 88.0ab (22) 35.9ab (9) 395.5ab (100) C. longa (Okinawa A) 256.0ab (70) 78.9ab (22) 29.0ab (8) 364.0ab (100) C. longa (Okinawa B) 265.0ab (71) 74.8ab (22) 33.3ab (9) 373.1ab (100) C. longa (Indonesia A) 1294.7a (49) 652.5a (24) 730.6a (27) 2677.8a (100) C. longa (Indonesia B) 229.7ab (68) 77.0ab (23) 29.9ab (9) 336.6ab (100) C. longa (Wakayama B) 0.4b (71) 0.3b (29) 0.0b (0) 0.7b (100) 2008 C. zedoaria 0.5b (49) 0.6b (51) 0.0b (0) 1.0b (100) C. aromatica (Kochi) 30.60abc (43) 39.35ab (56) 0.87ab (1) 70.81b (100) C. aromatica (Tanegashima) 37.53ab (48) 38.74abc (51) 1.54ab (2) 77.81bc (100) C. longa (Kochi) 260.38bcd (65) 96.32bcd (24) 41.01bc (10) 397.71ab (100) C. longa (Tanegashima) 259.06bcd (68) 86.29bcd (23) 36.88bc (10) 382.24ab (100) C. longa (Wakayama A) 270.04ab (67) 93.36ab (23) 39.65ab (10) 403.05ab (100) C. longa (Wakayama C) 266.83abcd (67) 97.22abc (24) 40.19abc (10) 404.24abc (100) C. longa (Indonesia A) 1276.73a (53) 687.15a (25) 813.05a (23) 2776.93a (100) C. longa (Indonesia B) 149.72abcd (66) 58.11abcd(25) 21.17abc (9) 229.00abc (100) C. longa (Wakayama B) 1.18d (51) 0.99cd (49) 0.10c (4) 2.27c (100) 2007 C. zedoaria 1.28cd (67) 0.38d (29) 0.08c (5) 1.74c (100) C. aromatica (Kochi) 16.05bc (43) 20.96ab (56) 0.60abc (2) 37.61bcd (100) C. aromatica (Tanegashima) 18.36abc (45) 22.02ab (54) 0.51bc (1) 40.89abcd (100) C. longa (Kochi) 226.72ab (69) 68.90a (21) 31.60ab (10) 327.21ab (100) C. longa (Tanegashima) 219.47ab (73) 56.67a (19) 25.23ab (8) 301.36abc (100) C. longa (Wakayama A) 127.72abc (71) 37.66ab (21) 13.87abc (8) 179.25abcd (100) C. longa (Indonesia A) 1358.64a (48) 679.14a (24) 811.43a (28) 2849.21a (100) C. longa (Wakayama B) 0.26c (34) 0.43b (56) 0.07c (9) 0.76d (100) 2006 C. zedoaria 0.43c (57) 0.33b (43) 0.00c (0) 0.76cd (100) 1) Curcumin content in primary branch rhizomes (mg/100g). 2) Percentage of curcumin 1, 2, and 3 content to curcuminoids content. Values followed by the same letter in a column are not significantly different at 5% level by one-way ANOVA. 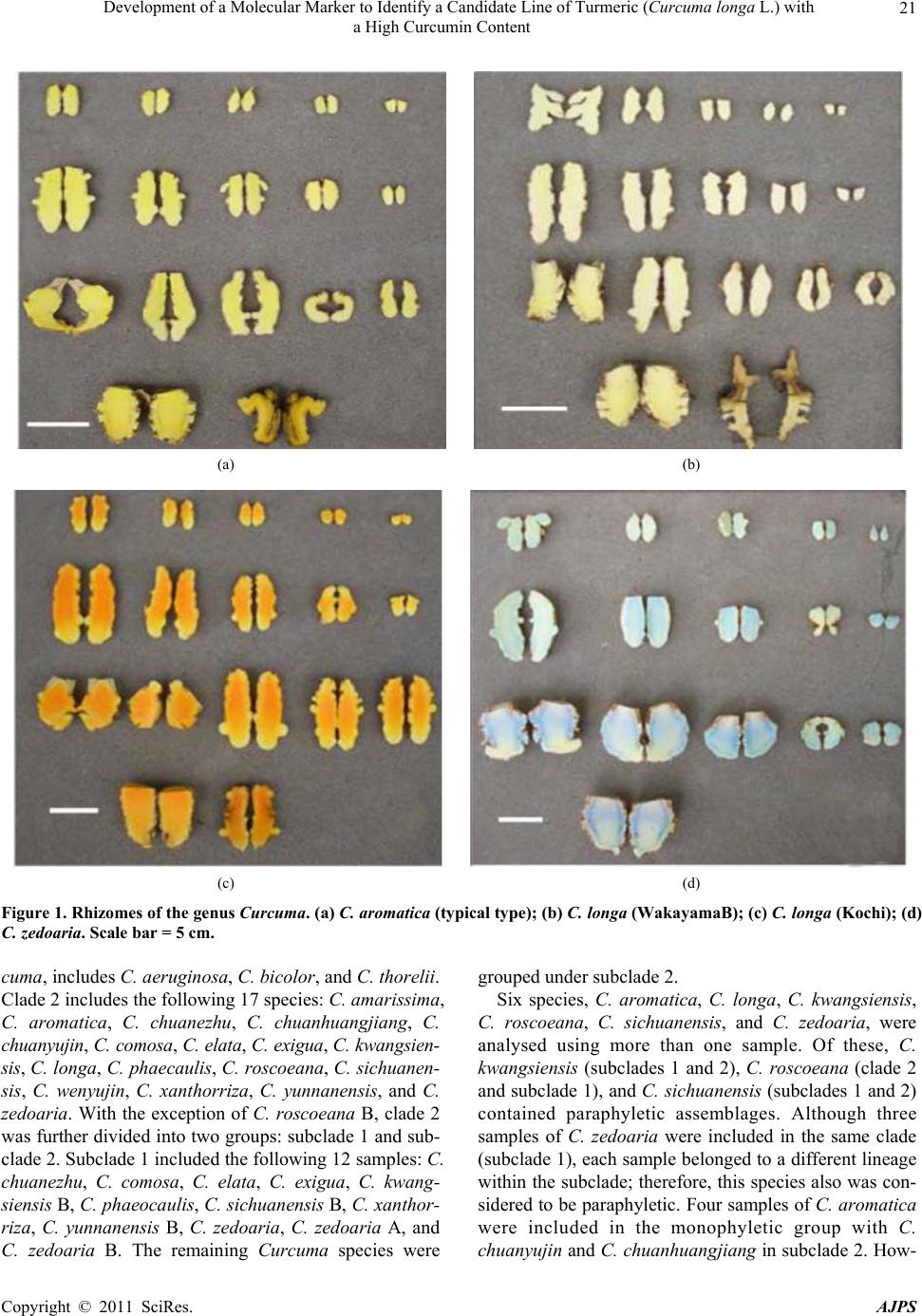 Development of a Molecular Marker to Identify a Candidate Line of Turmeric (Curcuma longa L.) with a High Curcumin Content Copyright © 2011 SciRes. AJPS 21 (a) (b) (c) (d) Figure 1. Rhizomes of the genus Curcuma. (a) C. aromatica (typical type); (b) C. longa (WakayamaB); (c) C. longa (Kochi); (d) C. zedoaria. Scale bar = 5 cm. cuma, includes C. aeruginosa, C. bicolor, and C. thorelii. Clade 2 includes the following 17 species: C. amarissima, C. aromatica, C. chuanezhu, C. chuanhuangjiang, C. chuanyujin, C. comosa, C. elata, C. exigua, C. kwangsien- sis, C. longa, C. phaecaulis, C. roscoeana, C. sichuanen- sis, C. wenyujin, C. xanthorriza, C. yunnanensis, and C. zedoaria. With the exception of C. roscoeana B, clade 2 was further divided into two groups: subclade 1 and sub- clade 2. Subclade 1 included the following 12 samples: C. chuanezhu, C. comosa, C. elata, C. exigua, C. kwang- siensis B, C. phaeocaulis, C. sichuanensis B, C. xantho r- riza, C. yunnanensis B, C. zedoaria, C. zedoaria A, and C. zedoaria B. The remaining Curcuma species were grouped under subclade 2. Six species, C. aromatica, C. longa, C. kwangsiensis, C. roscoeana, C. sichuanensis, and C. zedoaria, were analysed using more than one sample. Of these, C. kwangsiensis (subclades 1 and 2), C. roscoeana (clade 2 and subclade 1), and C. sichuanensis (subclades 1 and 2) contained paraphyletic assemblages. Although three samples of C. zedoaria were included in the same clade (subclade 1), each sample belonged to a different lineage within the subclade; therefore, th is species also was con- sidered to be paraphyletic. Four samples of C. aromatica were included in the monophyletic group with C. chuanyujin and C. chuanhuan gjiang in subclade 2. How-  Development of a Molecular Marker to Identify a Candidate Line of Turmeric (Curcuma longa L.) with a High Curcumin Content Copyright © 2011 SciRes. AJPS 22 Figure 2. Phylogenetic tree of Curcuma and its allied species using the neighbour-joining (NJ) method. The numbers below the branches indicate the bootstrap value. Black squares indicate high curcumin cont ent. Black circles indicate medium cur- cumin content. White circles indicate low curcumin content. ever, because there was no synapomorphic character in this group, we were unable to determine whether C. aromatica was monophyletic or paraph yletic. In add itio n, 13 samples of C. longa did not share a synapomorphic 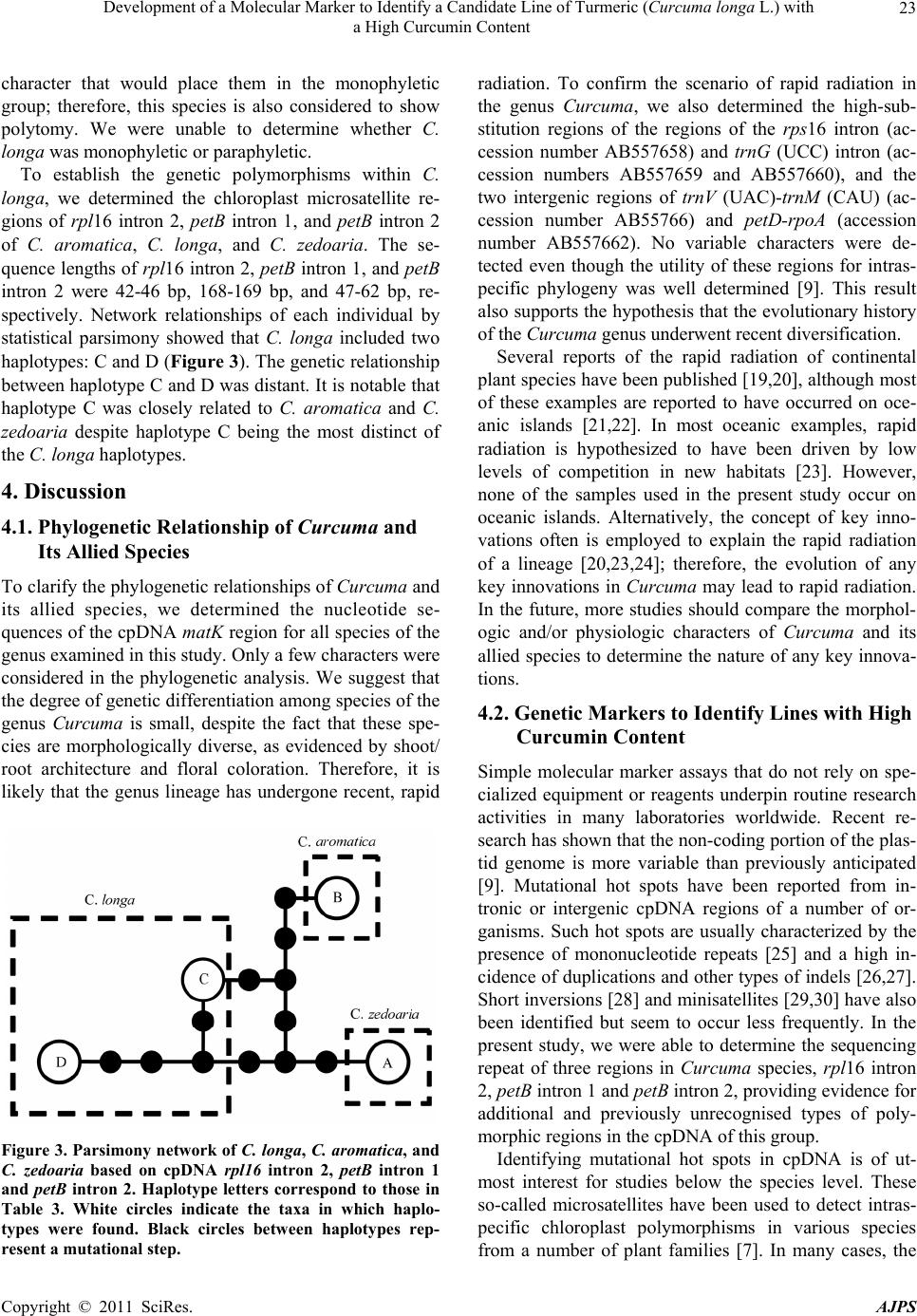 Development of a Molecular Marker to Identify a Candidate Line of Turmeric (Curcuma longa L.) with a High Curcumin Content Copyright © 2011 SciRes. AJPS 23 character that would place them in the monophyletic group; therefore, this species is also considered to show polytomy. We were unable to determine whether C. longa was monophyletic or paraphyletic. To establish the genetic polymorphisms within C. longa, we determined the chloroplast microsatellite re- gions of rp l16 intron 2, petB intron 1, and petB intron 2 of C. aromatica, C. longa, and C. zedoaria. The se- quence lengths of rpl16 intron 2, petB intron 1, and petB intron 2 were 42-46 bp, 168-169 bp, and 47-62 bp, re- spectively. Network relationships of each individual by statistical parsimony showed that C. longa included two haplotypes: C and D (Figure 3). The genetic relationship between haplotype C and D was distant. It is notable that haplotype C was closely related to C. aromatica and C. zedoaria despite haplotype C being the most distinct of the C. longa haplotypes. 4. Discussion 4.1. Phylogenetic Relationship of Curcuma and Its Allied Species To clarify the phylogenetic relationships of Curcuma and its allied species, we determined the nucleotide se- quences of the cpDNA matK region for all species of the genus examined in this study. Only a few characters were considered in the phylogenetic analysis. We suggest that the degree of genetic differentiation among species of the genus Curcuma is small, despite the fact that these spe- cies are morphologically diverse, as evidenced by shoot/ root architecture and floral coloration. Therefore, it is likely that the genus lineage has undergone recent, rapid Figure 3. Parsimony network of C. longa, C. aromatica, and C. zedoaria based on cpDNA rpl16 intron 2, petB intron 1 and petB intron 2. Haplotype letters correspond to those in Table 3. White circles indicate the taxa in which haplo- types were found. Black circles between haplotypes rep- resent a mutational step. radiation. To confirm the scenario of rapid radiation in the genus Curcu ma, we also determined the high-sub- stitution regions of the regions of the rps16 intron (ac- cession number AB557658) and trnG (UCC) intron (ac- cession numbers AB557659 and AB557660), and the two intergenic regions of trnV (UAC)-trnM (CAU) (ac- cession number AB55766) and petD-rpoA (accession number AB557662). No variable characters were de- tected even though the utility of these regions for intras- pecific phylogeny was well determined [9]. This result also supports the hypothesis that the evolutionary history of the Curcum a genus underwent re c ent divers ificati o n. Several reports of the rapid radiation of continental plant species have been published [19,20], although most of these examples are reported to have occurred on oce- anic islands [21,22]. In most oceanic examples, rapid radiation is hypothesized to have been driven by low levels of competition in new habitats [23]. However, none of the samples used in the present study occur on oceanic islands. Alternatively, the concept of key inno- vations often is employed to explain the rapid radiation of a lineage [20,23,24]; therefore, the evolution of any key innovations in Curcuma may lead to rapid radiation. In the future, more studies should compare the morphol- ogic and/or physiologic characters of Curcuma and its allied species to determine the nature of any key innova- tions. 4.2. Genetic Markers to Identify Lines with High Curcumin Content Simple molecular marker assays that do not rely on spe- cialized equipment or reagents underpin routine research activities in many laboratories worldwide. Recent re- search has shown that the non-coding portion of the plas- tid genome is more variable than previously anticipated [9]. Mutational hot spots have been reported from in- tronic or intergenic cpDNA regions of a number of or- ganisms. Such hot spots are usually characterized by the presence of mononucleotide repeats [25] and a high in- cidence of duplications and other types of indels [26,27]. Short inversions [28] and minisatellites [29,30 ] have also been identified but seem to occur less frequently. In the present study, we were able to determine the sequencing repeat of three regions in Curcuma species, rpl16 intron 2, petB intron 1 and petB intron 2, providing ev idence fo r additional and previously unrecognised types of poly- morphic regions in the cpDNA of this group. Identifying mutational hot spots in cpDNA is of ut- most interest for studies below the species level. These so-called microsatellites have been used to detect intras- pecific chloroplast polymorphisms in various species from a number of plant families [7]. In many cases, the  Development of a Molecular Marker to Identify a Candidate Line of Turmeric (Curcuma longa L.) with a High Curcumin Content Copyright © 2011 SciRes. AJPS 24 amplified fragments also contained size-variable sites in the target species. Thus, microsatellites may provide reasonable genetic markers to amplify the polymorphic chloroplast regions from any grasses, crops, fruits, and vegetables of interest. As the position of mutational hot spots in general is usually not conserved across species, these primer pairs will have to be tested in any new taxon on the basis of trial and error. However, the present re- port could extend th e application of microsatellites to the Curcuma species. In fact, we observed three instances of haplotype sharing within C. longa, together with a con- siderable degree of intraspecific variation. Moreover, it is interesting that C. longa exhibits unique chloroplast hap- lotypes (e.g. haplotype C) that are involved in lines with high curcumin content (Figure 3). This result indicates that the microsatellite regions are useful for identifying C. longa candidate lines with high curcumin content. Fur- ther studies will determine whether more comprehensive sampling and addition al evidence will support the results of the present study regarding the identification of C. longa lines with high curcumin content. In this study, we showed that the cpDNA of C. longa can be divided into two groups, expecting to expose a cryptic species with the appropriate biologic characteris- tics. Cryptic species are closely related species that differ critically in genetic, physiologic, behavioral, and eco- logical traits [31], and the abundance of morphologically cryptic or unrecognized species, even in well-known taxa, suggests that there are more species than are currently recognized or estimated [32]. Recent studies have re- ported that cryptic species seem to be especially common in certain taxa [33-36], and similar evidence for cryptic species was found in our results. Therefore, in order to verify this hypothesis with respect to C. longa, further reciprocal crosses among the haplotypes discussed in the present study may lead to a conclusion of cryptic diver- sity. In the future, additional research will be needed to determine if the various cryptic species differ in their ecological requirements and tolerance to potential envi- ronmental stresses. There is also the need to re-examine the alpha taxonomy of the four species showing cryptic diversity. Without this basic taxonomic research, these cryptic species will remain in taxonomic crypsis [37]. Our limited data set does not allow us to decide the most appropriate genetic marker at the present time. To accurately map the distribution of C. longa, and to monitor phylogeographic patterns within this species in more detail, we are currently applying the chloroplast markers developed in the present study to a large number of specimens sampled throughout the distributional range of this species. In addition, genetic and phylogenetic studies in Curcuma species have lagged behind those of other crop species. The network analysis presented here, along with the polymorphic information presented, pro- vide valuable genetic resources for future studies of C. longa and related species. 5. Acknowledgements We wish to thank R. Arakawa and J. Yokoyama, T. Yo- shida, A. Matsuzawa, A. Hirata, Y. Muramatsu, M. Saito, R. Ueda, K. Ohga, N. Yokoyama, M. Muroi, and K. Ma- tsuyama for providing much help. This study was partly supported by a grant-in-aid for scientific research from the Ministry of Education, Science, and Culture of Japan (to TF). REFERENCES [1] T. W. Corson and C. M. Crews, “Molecular Understand- ing and Modern Application of Traditional Medicines: Triumphs and Trials,” Cell, Vol. 130, No. 5, 2007, pp. 769-774. doi:10.1016/j.cell.2007.08.021 [2] S. Singh, “From Exotic Spice to Modern Drug?” Cell, Vol. 130, No. 5, 2007, pp. 765-768. doi:10.1016/j.cell.2007.08.024 [3] Y. Sasaki, H. Fushimi, H. Cao, S. Q. Cai and K. Komatsu, “Sequence Analysis of Chinese and Japanese Curcuma Drugs on the 18S rRNA Gene and trnK Gene and the Ap- plication of Amplification-Refractory Mutation System Analysis for Their Authentication,” Biological & Phar- maceutical Bulletin, Vol. 25, No. 12, 2002, pp. 1593-1599. doi:10.1248/bpb.25.1593 [4] M. Minami, K. Nishio, Y. Ajioka, H. Kyushima, K. Shi- geki, K. Kinjo, K. Yamada, M. Nagai, K. Satoh and Y. Sakurai, “Identification of Curcuma Plants and Curcumin Content Level by DNA Polymorphisms in the trnS-trnfM intergenic Spacer in Chloroplast DNA,” Journal of Natu- ral Medicines, Vol. 63, No. 1, 2009, pp. 75-79. doi:10.1007/s11418-008-0283-7 [5] K. Aoi, K. Kaburagi, T. Seki, T. Tobata, M. Sarak and M. Kuroyanagi, “Studies on the Cultivation of Turmeric (Curcuma longa L.) I: Varietal Differences in Rhizome Yield and Curcuminoid Content,” Bulletin of National In- stitute of Hygienic Sciences, Vol. 104, 1986, 124-128. [6] M. N. Tamura, J. Yamashita , S. Fuse and M. Hamaguchi, “Molecular Phylogeny of Monocotyledons Inferred from Combined Analysis of Plastid matK and rbcL Gene Se- quences,” Journal of Plant Research, Vol. 117, No. 2, 2004, pp. 109-120. doi:10.1007/s10265-003-0133-3 [7] D. E. Soltis and P. S. Soltis, “Choosing an Approach and an Appropriate Gene for Phylogenetic Analysis,” In: D. E. Soltis, P. S. Soltis and J. J. Doyle, Eds. Molecular Sys- tematics of Plants II. Kluwer Academic Publishers, Bos- ton, 1998, pp. 1-42. [8] R. G. Olmstead and P. A. Reeves, “Evidence for the Polyphyly of the Scrophulariaceae Based on Chloroplast rbcL and ndhF Sequences,” Annals of the Missouri Bo-  Development of a Molecular Marker to Identify a Candidate Line of Turmeric (Curcuma longa L.) with a High Curcumin Content Copyright © 2011 SciRes. AJPS 25 tanical Garden, Vol. 82, No. 2, 1995, pp. 176-193. doi:10.2307/2399876 [9] T. Nishizawa and Y. Watano, “Primer Pairs Suitable for PCR-SSCP Analysis of Chloroplast DNA in Angio- sperms,” Journal of Phytogeography and Taxonomy, Vol. 48, No. 1, 2000, pp. 67-70. [10] S. A. Kelchner, “The Evolution of Non-Coding Chloro- plast DNA and Its Application in Plant Systematics,” Ann. Annals of the Missouri Botanical Garden, Vol. 87, No. 4, 2000, pp. 482-498. doi:10.2307/2666142 [11] D. Posada and K. A. Crandall, “Intraspecific Gene Gene- Alogies: Trees Grafting into Networks,” Trends in Ecol- ogy & Evolution, Vol. 16, No. 1, 2001, pp. 37-45. doi:10.1016/S0169-5347(00)02026-7 [12] M. Sato, K. Shimura and K. Hashizume, “Quality Valua- Tion of Turmeric Powders on Market,” Mie Prefectural Health and Environment Research Institution, Vol. 6, 2004, pp. 52-54. [13] L. A. Johnson and D. E. Soltis, “matK DNA Sequences and Phylogenetic Reconstruction in Saxifragaceae s.str.,” Systematic Botany, Vol. 19, No. 1, 1994, pp. 143-156. doi:10.2307/2419718 [14] J. D. Thompson, D. G. Higgins and T. J. Gibson, “CLUSTAL W: Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weight- ing, Positions-Specific Gap Penalties and Weight Matrix Choice,” Nucleic Acids Research, Vol. 22, No. 22, 1994, pp. 4673-4680. doi:10.1093/nar/22.22.4673 [15] K. Tamura, J. Dudley, M. Nei and S. Kumar, MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software. Version 4.0,” Molecular Biology and Evolution, Vol. 24, No. 8, 2007, pp. 1596-1599. doi:10.1093/molbev/msm092 [16] D. L. Swofford, “paup*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4,” Sinauer Associates, Sunderland, MA. 2001. [17] H. J. Bandelt, P. Forster and A. Röhl, “Median-Joining Networks for Inferring Intraspecific Phylogenies,” Mo- lecular Biology and Evolution, Vol. 16, No. 1, 1999, pp. 37-48. [18] T. Morishita, H. Yamaguchi, K. Degi, T. Shimizu and H. Nakagawa, “Varietal Difference and Year-to-Year Varia- tion of the Contents of Curcuminoids in Turmeric,” Japanese Journal of Crop Sciences, Vol. 75, 2006, pp. 130-131. doi:10.1626/jcs.75.335 [19] A. B. Utelli, B. A. Roy and M. Baltisberger, “Molecular and morphological analysis of European Aconitum spe- cies (Ranunculaceae),” Plant Systematics and Evolution, Vol. 224, No. 3-4, 2000, pp. 195-212. doi:10.1007/BF00986343 [20] S. T. Malcomber, “Phylogeny of Gaertinera Lam. (Lu- biaceae) Based on Multiple DNA Markers: Evidence of Rapid Radiation in a Widespread, Morphologically Di- verse Genus,” Evolution, Vol. 56, No. 1, 2002, pp. 42-57. [21] B. G. Baldwin, “Adaptive Radiation of Hawaiian Silvers- Word Alliance: Congruence and Conflict of Phylogenetic Evidence from Molecular and Non-Molecular Investiga- Tions,” In: T. J. Givnish and K. J. Sytsma, Eds., Molecu- lar Evolution and Adaptive Radiation, Cambridge Uni- versity Press, Cambridge, 1997, pp. 103-128. [22] J. Francisco-Ortega, D. J. Crawford, A. S. Guerra and R. K. Jansen, “Origin and Evolution of Argyranthmum (As- teraceae: Anthemideae) in Macaronesia,” In: T. J. Givnish and K. J. Sytsma, Eds., Molecular Evolution and Adap- tive Radiation, Cambridge University Press, Cambridge, 1997, pp. 407-431. [23] K. F. Liem, “Key Evolutionary Innovations, Differential Diversity, and Symecomorphosis,” In: M. Nitecki, Ed., Evolutionary Innovations, University of Chicago Press, Chicago, 1990, pp. 147-170. [24] S. A. Hodges and M. L. Arnold, “Columbines: A Geo- graphically Widespread Species Flock,” Proceedings of the National Academy of Sciences of the USA, Vol. 91, No. 11, 1994, pp. 5129-5132. doi:10.1073/pnas.91.11.5129 [25] J. Provan, N. Soranzo, N. J. Wilson, J. W. McNicol, M. Morgante and W. Powell, “The Use of Uniparentally In- herited Simple Sequence Repeat Markers in Plant Popula- tion Studies and Systematics,” In: P. M. Hollingsworth, R. M. Bateman an d R. J. Gornall, Eds., Molecular Systemat- ics and Plant Evolution, Taylor & Francis, London, 1999, pp. 1-19. [26] B. R. Morton and M. T.Clegg, “A Chloroplast DNA Mu- tational Hotspot and Gene Conversion in a Noncoding Region near rbcL in the Grass Family (Poaceae),” Cur- rent Genetics, Vol. 24, No. 4, 1993, pp. 357-365. doi:10.1007/BF00336789 [27] D. A. Johnson and J. Hattori, “Analysis of a Hotspot for Deletion Formation within the Intron of the Chloroplast trnI Gene,” Genom e, Vol. 39, No. 5, 1996, pp. 999-1005. doi:10.1139/g96-124 [28] S. A. Kelchner and J. F. Wendel, “Hairpins Create Minute Inversions in Non-Coding Regions of Chloroplast DNA,” Current Genetics, Vol. 30, No. 3, 1996, pp. 259-262. doi:10.1007/s002940050130 [29] D. Cafasso, G. Pellegrino, A. Musacchio, A. Widmer and S. Cozzolino, “Characterization of a Minisatellite Repeat Locus in the Chloroplast Genome of Orchis palustris (Or- chidaceae),” Current Genetic s, Vol. 39, No. 5-6, 2001, pp. 394-398. doi:10.1007/s002940100226 [30] R. A. King and C. Ferris, “A Variable Minisatellite Se- quence in the Chloroplast Genome of Sorbus L. (Rosaceae: Maloideae),” Genome, Vol. 45, No. 3, 2002, pp. 570-576. doi:10.1139/g02-018 [31] D. H. Darling and J. H. Werren, “Biosystematics of Na- sonia (Hymenoptera: Pteromalidae): Two New Species Reared from Birds’ Nests in North America,” Annals of the Entomological Society of America, Vol. 83, No. 3, 1990, pp. 352-370. [32] E. M. Barratt, R. Deaville, T. M. Burl, M. W. Bruford, G.  Development of a Molecular Marker to Identify a Candidate Line of Turmeric (Curcuma longa L.) with a High Curcumin Content Copyright © 2011 SciRes. AJPS 26 Jones, P. A. Racey a nd R. K. Wayne, “DNA Answer s the Call for Pipistrelle Bat Species,” Nature, Vol. 387, 1997, pp. 138-139. doi:10.1038/387138b0 [33] I. D. Hogg, M. I. Stevens, K. E. Schnabel and M. A. Chapman, “Deeply Divergent Lineages of the Wide- spread New Zealand Amphipod Paracalliope Fluviatilis Revealed Using Allozyme and Mitochondrial DNA Analyses,” Freshwater Biology, Vol. 51, No. 2, 2006, pp. 236-248. doi:10.1111/j.1365-2427.2005.01491.x [34] T. Tully, C. A. D'Haese, M. Richard and R. Ferriere, “Two Major Evolutionary Lineages Revealed by Mo- lecular Phylogeny in the Parthenogenetic Collembola Species Folsomia Candida,” Pedobiologia, Vol. 50, No. 2, 2006, pp. 95-104. doi:10.1016/j.pedobi.2005.11.003 [35] D. Bickford, D. J. Lohman and N. S. Sodhi, “Cryptic Species as a Window on Diversity and Conservation,” Trends in Ecology & Evolution, Vol. 22, No. 3, 2007, pp.148-155. doi:10.1016/j.tree.2006.11.004 [36] T. L. Finston, M. S. Johnson, W. F. Humphreys, S. M. Eberhard and S. A. Halse, “Cryptic Speciation in Two Widespread Subterranean Amphipod Genera Reflects Historical Drainage Patterns in an Ancient Landscape,” Molecular Ecology, Vol. 16, No. 2, 2007, pp. 355-365. doi:10.1111/j.1365-294X.2006.03123.x [37] B. C. Schlick-Steiner, B. Seifert and C. Stauffer, “With- out Morphology, Cryptic Species Stay in Taxonomic Crypsis Following Discovery,” Trends in Ecology & Evolution, Vol. 22, No. 8, 2007, pp. 391-392. doi:10.1016/j.tree.2007.05.004 |

