Paper Menu >>
Journal Menu >>
 American Journal of Plant Sciences, 2011, 2, 1-14 doi:10.4236/ajps.2011.21001 Published Online March 2011 (http://www.SciRP.org/journal/ajps) Copyright © 2011 SciRes. AJPS 1 Numerical Taxonomic Study of Some Tribes of Gramineae from Egypt Ahmed Osman1, Mohammed Zaki2, Sohar Hamed1, Nagwa Hussein1 1South Valley University, Qena, Egypt; 2Cairo University, Giza, Egypt. Email: ahmosman2000@yahoo.com Received October 10th, 2010; revised November 26th, 2010; accepted December 9th, 2010. ABSTRACT A systematic study of eleven tribes of Gramineae surveyed 34 characters including fruit morphology, fru it anatomy and palynology. The results were conducted to some numerical analysis aspects. On the basis of UPGMA (Unpaired Group Method of Average) clustering and PCA (Principal component analysis), the results show congruence between the UPGMA clustering and PCA method, in suggesting two major clad s/groups and five subclads. Keywords: Gr amineae, Numerical Taxonomy, UPGMA, Cladistic Tree 1. Introduction Poaceae (grasses) is one of the most species-rich flower- ing plant families and includes many economically im- portant crops. Parallel evolution of such features as the annual habit, C4 photosynthesis and several highly char- acteristic reproductive structures has facilitated a series of major radiations within Poaceae, culminating in the existing global distribution of about 10000 species and 700 genera [1,2]. A phylogeny of Poaceae was recently established using a combination of multiple data sets from both molecules and morphology [3], enabling im- proved understanding of relationships between basal and derived grasses. Poaceae tribes and genera are subject to different stud- ies in order to understand the phylogenetic relationships between taxa. Many attempts have been made to address phylogenetic relationships of Chloridoideae; synonym Eragrostoideae that comprises approximately 146 genera and 1360 species, whose adoption of efficient C4 photo- synthesis had led to its successful proliferation in the tropics and subtropics [1]; mainly based on the basis of morphological and molecular data [4,5], but general agreement is still lacking. The grass tribe Triticeae in- cludes some of the world’s most important cereals and a significant number of important forage grasses [6]. Due to its renownedly complicated evolutionary history and its economic importance there has been an increasing interest in producing molecular phylogenies for the Triticeae. Attempts to unravel the relationships in the group have been based on many different types of data e.g. isozymes [7], restriction site data [8,9] and sequence data from a number of different coding and/or non-coding regions, viz.5S RNA [10]. Among the mod- ern tools for plant taxonomy, reference [11] stated that increasing use has taken place of computers for data stor- age and analysis during the past twenty years. Data de- rived from all tools of taxonomical investigations has to be analyzed mathematically and cladistic trees have to be drawn. Despite of the criticism of using cladistic analysis in taxonomy, cladistic methods have become a most useful technical tool for clarifying intrafamilial relation- ships. Moreover; the advantages of using more rigorous techniques to elaborate natural classifications or evolu- tionary diagrams instead of those that have been used traditionally in botany have been well presented by [12]. A phylogenetic analysis of Triticeae was performed by means of numerical methods due to [13]. Five methods, each based on extreme assumptions of parameters so interpreted under [14] evolutionary model, were used. The most parsimonious tree obtained served as a base for subsequent elaboration of the final tree, taking into con- sideration genetic information primarily, and for the erection of the proposed phylogenetic classification of Triticeae. A key is provided for identification of the groupings in the tribe. The proposed classification is discussed in the light of previous classifications, even though none of them were phylogenetic in the sense of Hennig. Reference [15] have introduced a cladistic ana- lysis, primarily based on morphological data from 40  Numerical Taxonomic Study of Some Tribes of Gramineae from Egypt Copyright © 2011 SciRes. AJPS 2 taxa representing the 24 genera of the Triticeae. They used Bremer support as a measure of branch support. The trees based on morphology and on molecular data are largely incongruent. Also; [16] and [17]; in their study showed the relationships of graminid/restiid of poales in a cladistic tree. This report aims to apply numerical tax- onomy; UPGMA and Cluster Analysis; to reveal better the relationships between genera within a tribe and tribes within the family based on the data collected from the previous investigations for caryopses morphology and anatomy and pollen grains morphology. 2. Materials and Methods 2.1. Plant Material The study dealt with 34 species belonging to 25 genera of 11 tribes of Gramineae; Andropogoneae, Aristideae, Arundineae, Aveneae, Brachypodieae, Bromeae, Er- agrostideae, Paniceae, Poeae, Stipeae and Triticeae. The study based on herbarial specimens dried and kept in the QNA Herbarium (in South Valley University, Qena, Botany Department) and some species received on loan from CAI Herbarium for the palynological study (Table 1). In the following analysis, species and genera consti- tuted the OTUs (Operational Taxonomic Units). In order to broadly sample variation, the OUTs consist of a num- ber of collections from different localities in Egypt, illus- trated in Table 2. 2.2. Characters Observations Table 3 shows the characters and character states scored for fruit anatomy, fruit morphology and pollen morphol- ogy, averaged for each OUT. A total of 37 characters were measured, comprising 22 qualitative and 15 quanti- tative characters. For recording the total characters; a main using of different microscopic techniques; light, scanning electron and stereomicroscope were used for investigating different samples and recording data col- lected. Table 4 shows the data matrix used for analysis of taxa studied. For some of the OTUs, some characters’ observations were lacked and these omissions were coded as missing data (−0.999). 2.3. Data Analysis Two types of analysis were performed with STATIS- TICA version 5.0 computer software. Firstly, the total data coded were analyzed by the Unpired Group Method of Average (UPGMA) clustering. Construction of the tree illustrating the relationships between the studied species was performed using Arthimetic Average (UP- GMA) proposed by [18]. Secondly, factor analysis and factor loadings were applied to determine the major and specific characters that aid in separation using the same program. A principal component analysis (PCA) was also performed according to [19]. 3. Results Figure 1 shows the UPGMA cladistic tree comprising all OTUs in the present study. The tree separated into two major clads at 100 dissimilarity distance. The first major clad at 53 dissimilarity distance, comprised only two species of the total number; Panicum turgidum and Arundo donax; while the second major clad at 93 dis- similarity comprised the rest 32 species. The second major clad separated into two branches, the first branch includes five subclads: 1) A subclad at 86 dissimilarity distance with five species; Lamarckia aurea, Oryzopsis miliacae, Polypogon monspeliensis, Eragrostis cilinensis and Stipagrostis ciliata. 2) A subclad at 84 dis- similarity distance with Aegilops kotshyi. 3) A subclad at 80 dissimilarity distance comprises six species; Aegilops ventricosa, Hordeum murinum ssp. leporinum, Lolium perenne, Bromus scoparius, Brachypodium distachyum and Avena fatua. 4) A subclad at 74 dissimilarity dis- tance includes nine species; Stipa capensis, Dactylis glomerata, Stipa lagascae, Bromus rubens, Echinocoloa colona, Coelachryum bervifolium, Schismus arabicus, Stipa parviflora and Aristida funiculata and 5) A subclad at 54 dissimilarity distance with nine species, Poa annua, Polypogon maritimus, Eragrostis minor, Phalaris minor, Avena barbata, Aristida mutabilis, Cenchrus ciliaris, Leptochloa fusca and Aristida adscensionis. The second branch of the second major clad comprises two species; Dactylochtenium aegyptium and Sorghum variegatum; at 66 dissimilarity distance. Factor analysis using Principal Component Analysis (PCA) showed that the most intrinsic characters en- hanced separation of the total OTUs are fruit shape, color type and fruit surface sculpture of the morphological characters, of the fruit anatomical characters; section outline shape, hull cells type, aleurone cells shape and orientation, scutellum shape and thickness and en- dosperm thickness are intrinsic characters for separation. Meanwhile, all the pollen characters are good data for separating of taxa; pollen class, shape, size, surface sculpture, annulus thickness, pore diameter, pollen wall thickness, sexine and nexine thickness. The characters of separation are of high factor loadings ≥ (± 0.7) Table 5. These represented by a percentage of the total variation as 24.01% from the three factors extracted as; factor 1 is responsible for 16.54% of the variation, factor 2 is re- sponsible for 4.34% of the variation and factor 3 is re- sponsible for the minimum value of the total variation; is 3.13%. The plot of 34 OTUs on the first two factors ex- tracted in the PCA method is shown in (Figure 2). Plot of factor 1/2 shows two groups. 1) Group of 6. Arundo 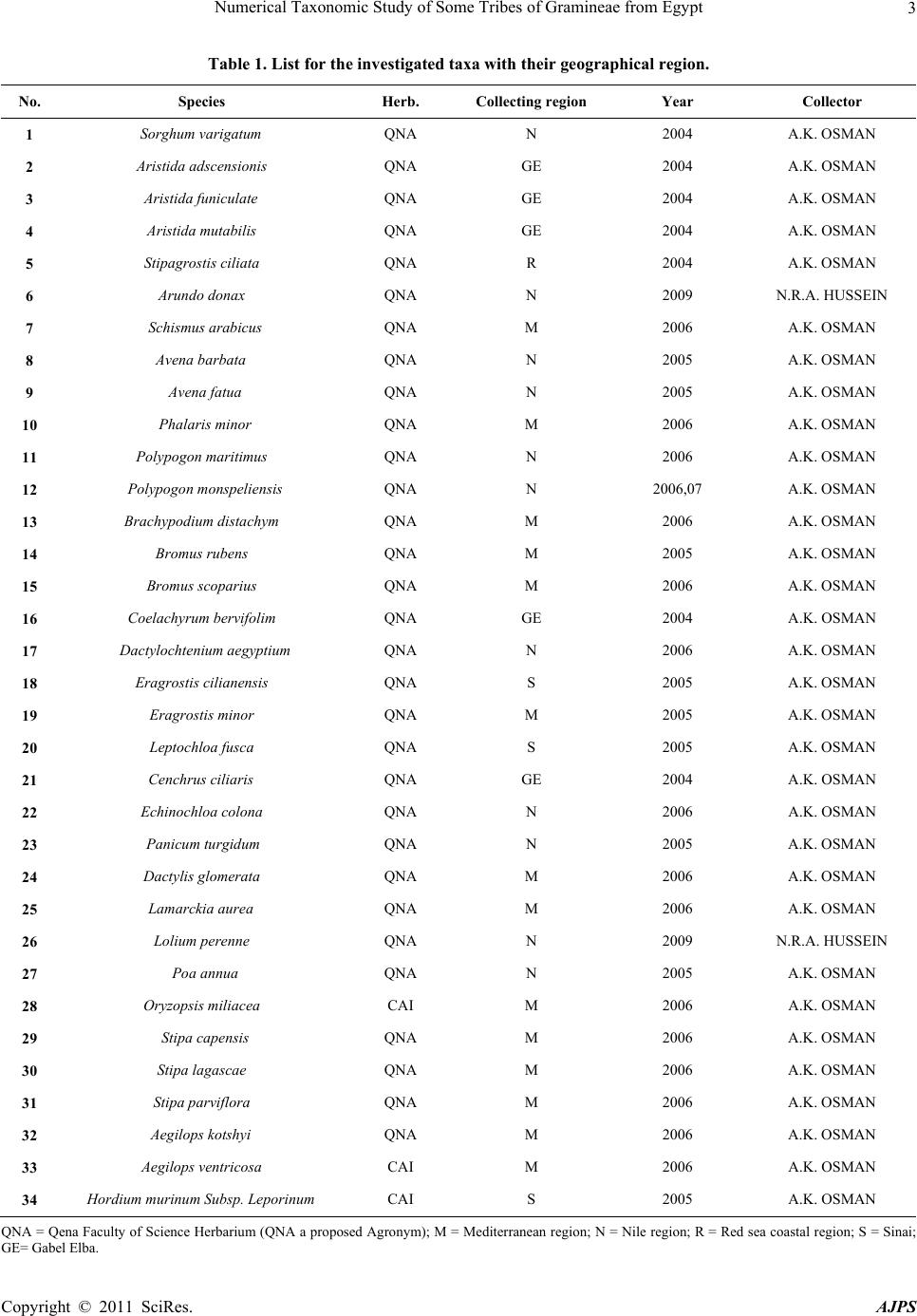 Numerical Taxonomic Study of Some Tribes of Gramineae from Egypt Copyright © 2011 SciRes. AJPS 3 Table 1. List for the investigated taxa with their geographical region. No. Species Herb. Collecting region Year Collector 1 Sorghum varigatum QNA N 2004 A.K. OSMAN 2 Aristida adscensionis QNA GE 2004 A.K. OSMAN 3 Aristida funiculate QNA GE 2004 A.K. OSMAN 4 Aristida mutabilis QNA GE 2004 A.K. OSMAN 5 Stipagrostis ciliata QNA R 2004 A.K. OSMAN 6 Arundo donax QNA N 2009 N.R.A. HUSSEIN 7 Schismus arabicus QNA M 2006 A.K. OSMAN 8 Aven a b a r b a t a QNA N 2005 A.K. OSMAN 9 Avena fatua QNA N 2005 A.K. OSMAN 10 Phalaris minor QNA M 2006 A.K. OSMAN 11 Polypogon maritimus QNA N 2006 A.K. OSMAN 12 Polypogon monspeliensis QNA N 2006,07 A.K. OSMAN 13 Brachypodium distachym QNA M 2006 A.K. OSMAN 14 Bromus rubens QNA M 2005 A.K. OSMAN 15 Bromus scoparius QNA M 2006 A.K. OSMAN 16 Coelachyrum bervifolim QNA GE 2004 A.K. OSMAN 17 Dactylochtenium aegyptium QNA N 2006 A.K. OSMAN 18 Eragrostis cilianensis QNA S 2005 A.K. OSMAN 19 Eragrostis minor QNA M 2005 A.K. OSMAN 20 Leptochloa fusca QNA S 2005 A.K. OSMAN 21 Cenchrus ciliaris QNA GE 2004 A.K. OSMAN 22 Echinochloa colona QNA N 2006 A.K. OSMAN 23 Panicum turgidum QNA N 2005 A.K. OSMAN 24 Dactylis glomerata QNA M 2006 A.K. OSMAN 25 Lamarckia aurea QNA M 2006 A.K. OSMAN 26 Lolium perenne QNA N 2009 N.R.A. HUSSEIN 27 Poa annua QNA N 2005 A.K. OSMAN 28 Oryzopsis miliacea CAI M 2006 A.K. OSMAN 29 Stipa capensis QNA M 2006 A.K. OSMAN 30 Stipa lagascae QNA M 2006 A.K. OSMAN 31 Stipa parviflora QNA M 2006 A.K. OSMAN 32 Aegilops kotshyi QNA M 2006 A.K. OSMAN 33 Aegilops ventricosa CAI M 2006 A.K. OSMAN 34 Hordium murinum Subsp. Leporinum CAI S 2005 A.K. OSMAN QNA = Qena Faculty of Science Herbarium (QNA a proposed Agronym); M = Mediterranean region; N = Nile region; R = Red sea coastal region; S = Sinai; GE= Gabel Elba.  Numerical Taxonomic Study of Some Tribes of Gramineae from Egypt Copyright © 2011 SciRes. AJPS 4 Table 2. List for the names of total OTUs studied and their corresponding numbers and tribe names. OTUs no. Species name Tribes OTUs no. Species name Tribes Sp.1 Sorghum varigatum Andro Sp.18 Eragrostis cilianensis Eragro Sp.2 Aristida adscensionis Arist Sp.19 Eragrostis minor Eragro Sp.3 Aristida funiculate Arist Sp.20 Leptochloa fusca Eragro Sp.4 Aristida mutabilis Arist Sp.21 Cenchrus ciliaris Panic Sp.5 Stipagrostis ciliate Arist Sp.22 Echinochloa colona Panic Sp.6 Arundo donax Arund Sp.23 Panicum turgidum Panic Sp.7 Schismus arabicus Arund Sp.24 Dactylis glomerata Poeae Sp.8 Avena barbata Aven Sp.25 Lamarckia aurea Poeae Sp.9 Avena fatua Aven Sp.26 Lolium perenne Poeae Sp.10 Phalaris minor Aven Sp.27 Poa annua Poeae Sp.11 Polypogon maritimus Aven Sp.28 Oryzopsis miliacea Stipeae Sp.12 Polypogon monspeliensis Aven Sp.29 Stipa capensis Stipeae Sp.13 Brachypodium distachym Brach Sp.30 Stipa lagascae Stipeae Sp.14 Bromus rubens Brom Sp.31 Stipa parviflora Stipeae Sp.15 Bromus scoparius Brom Sp.32 Aegilops kotshyi Triti Sp.16 Coelachyrum bervifolim Eragro Sp.33 Aegilops ventricosa Triti Sp.17 Dactylochtenium aegyptium Eragro Sp.34 Hordium murinum Subsp. Leporinum Triti Andro= Andropogoneae, Arist= Aristideae, Arund= Arundineae, Aven= Aveneae, Brach = Brachypodieae, Brom= Bromeae, Eragro= Eragrostideae, Panic= Paniceae,Triti= Triticeae. Figure 1. Cladogram of 34 species studied by UPGMA method.  Numerical Taxonomic Study of Some Tribes of Gramineae from Egypt Copyright © 2011 SciRes. AJPS 5 Table 3. Characters and character states used in the analysis of Gramineae tribes. Characters Character states Code Fruits Morphological characters 1. Fruit shape Elliptic Rectangular Cordate Oblong Linear Narrow cordate Cordate with hollow part Circular Oval Oval with acute protrusion (tall) oblong with tapered ends Elliptic with tapered ends (acute elliptic) 1 2 3 4 5 6 7 8 9 10 11 12 2. Fruit coloring Onecoloured (uniformly coloured) Bicoloured 1 2 3. Color type Light brown Dark brown with light yellowish sheath Brown Beage Dark red Light beage Light beage and light green strips Brown with pale beage sheath Beage with light violet sheath Brown and light orange Beage and brown protrusions Shiny beage and light brown small spots Dark beage Beage and brown black spots Light brown and dark brown ends Light brown with light green sheath Brown with light green sheath Gradient beage with light green and brown spot at grain top 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 Trichomes on grains 4. Trichomes type Simple hairs 1 5. Trichome presence Absent (glabrous) Present Present on grain sheath 1 2 3 6. Hair length Short Long With different lengths 1 2 3 7. Hair coloring Shiny transparent (colorless) Colored 1 2 8. Position o f attaching Around all grain surface At grain edges Around all surface, condensed at the top At the top At the base Few around all surface, condensed at the base On the side margins of the grain sheath 1 2 3 4 5 6 7 9. Fruit surface sculpture Reticulate with straight cell wall Striate Scabrate 1 2 3  Numerical Taxonomic Study of Some Tribes of Gramineae from Egypt Copyright © 2011 SciRes. AJPS 6 Reticulate with undulate granulate cell wall Compound reticulate with foveolate Reticulate with undulate cell wall Striate to scabrate Scalariform Striate at the intermediate and reticulate with straight cell wall next to hilum Compound reticulate with tubrculate Smooth Rugose Reticulate with straight to undulate cell wall Ribbed pattern cell wall Compound reticulate with granulate Scaly surface 4 5 6 7 8 9 10 11 12 13 14 15 16 Fruits Morph metrical characters 10. Fruit weight ( mg) =(0.03-2.12) =(2.12-4.21) =(4.21-6.3) =(6.3-8.39) =(8.39-10.48) =(10.48-12.57) =(12.57-14.66) 1 2 3 4 5 6 7 Fruit size 11. Wide (µm) =(29-112) =(112-195) =(195-279) 1 2 3 12. Length (µm) =(1.9-52.9) =(52.9-103.9) =(103.9-154.9) =(154.9-205.9) =(205.9-256.9) =(256.9-307.94) =(307.94-358.9) 1 2 3 4 5 6 7 Fruit anatomy 13. Section outline shape Circular Circular to cordate Circular to oval Semi-circular Oval Oval to cordate Cordate Oval to rectangular with curved corners Oval to rectangular C- shaped Circular to triangular with obtused corners Triangular with obtused corners Triangular Rectangular with obtused corners Rectangular with obtused corners to oval Rectangular with acuted corners Rectangular Polygonal Linear with folded ends Circled linear 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 14. Hull cells type Parenchyma Epithelial Not observed 1 2 3  Numerical Taxonomic Study of Some Tribes of Gramineae from Egypt Copyright © 2011 SciRes. AJPS 7 15. Aleurone cells shape Rectangular Rectangular and quadrate Quadrate Rectangular and polygonal Not observed 1 2 3 4 5 16. Aleurone cells oreintation Horizontal Vertical Horizontal and vertical Not observed 1 2 3 4 17. Scutellum cells shape Strip of cells Elliptic mass of cells Rectangular to cordate mass of cells Quinqangular mass of cells Oval mass and strip of cells Triangular mass and strip of cells Not observed 1 2 3 4 5 6 7 18. Endosperm differentiation Differentiated Differentiated and Undifferentiated 1 2 19. Type of endosper m Starchy Starchy and fluidy 1 2 20. Section wide (µm) =(5.33-31.66) =(31.66-57.99) =(57.99-84.32) =(84.32-110.65) =110.65-136.98) =(136.98-163.31) =(163.31-189.64) 1 2 3 4 5 6 7 21. Section length (µm) =(33.33-71.94) =(71.94-110.55) =(110.55-149.16) =(149.16-187.77) =(187.77-226.38) =(226.38-264.99) =(264.99-303.6) 1 2 3 4 5 6 7 22. Hull cells thickness (µm) Not ovserved =(9.39-14.79) =(14.79-20.19) =(20.19-25.59) =(25.59-30.99) =(30.99-36.39) 0 1 2 3 4 5 23. Seed coat thickness (µm) =(2.43-8.63) =(8.63-14.83) =(14.83-21.03) =(21.03-27.23) =(27.23-33.43) =(33.43-39.63) 1 2 3 4 5 6 24. Aleurone layer thickness (µm) Not observed =(1.92-9.42) =(9.42-16.92) =(19.92-24.42) =(24.42-31.92) =(31.92-39.42) =(39.42-46.92) 0 1 2 3 4 5 6 25. Scutellum thickness ( µm) Not observed =(5.54-85.94) =(85.94-166.43) =(166.34-246.74) 0 1 2 3  Numerical Taxonomic Study of Some Tribes of Gramineae from Egypt Copyright © 2011 SciRes. AJPS 8 =(246.74-327.14) =(327.14-407.54) =(407.54-487.94) Thicknesses in the range codes 1&4 Thicknesses in the range codes 1&3 4 5 6 7 8 26. Endosperm thickness (µm) =(21.52-125.02) =(125.02-228.52) =(228.52-332.02) =(332.02-435.52) =(435.52-539.02) =(539.02-642.52) Two different thicknesses in range of codes 1&2 1 2 3 4 5 6 7 Pollen grains Morphological characters 27. Pollen class Monoporate Diporate 1 2 28. Annulus Pollen annulate 1 29. OperculumPollen operculate1 30. Pollen size Pollen small Pollen medium 1 2 31. Pollen shape Pollen Oblate-spheroidal Pollen Suboblate Pollen Prolate-spheroidal Pollen spheroidal 1 2 3 4 32. Surface sculpturing Areolate Granulate Scabrate verrucate 1 2 3 4 Pollen grains Morph metrical characters 33. Annulus thickness (µm) =(1.16-1.52) =(1.52-1.88) =(1.88-2.24) =(2.24-2.6) =(2.6-2.96) =(2.96-3.32) 1 2 3 4 5 6 34. Pore diameter (µm) =(1.57-2.17) =(2.17-2.77) =(2.77-3.37) =(3.37-3.97) =(3.97-4.57) 1 2 3 4 5 35. Pollen wall thickness (µm) =(0.698-0.848) =(0.848-0.998) =(0.998-1.148) =(1.148-1.298) =(1.298-1.448) 1 2 3 4 5 36. Sexine thickness (µm) =(0.434-0.454) =(0.454-0.474) =(0.474-0.494) =(0.494-0.514) =(0.514-0.534) =(0.534-0.554) 1 2 3 4 5 6 37. Nexine thickness (µm) =(0.314-0.354) =(0.354-0.394) =(0.394-0.434) =(0.434-0.474) =(0.474-0.514) =(0.514-0.554) 1 2 3 4 5 6 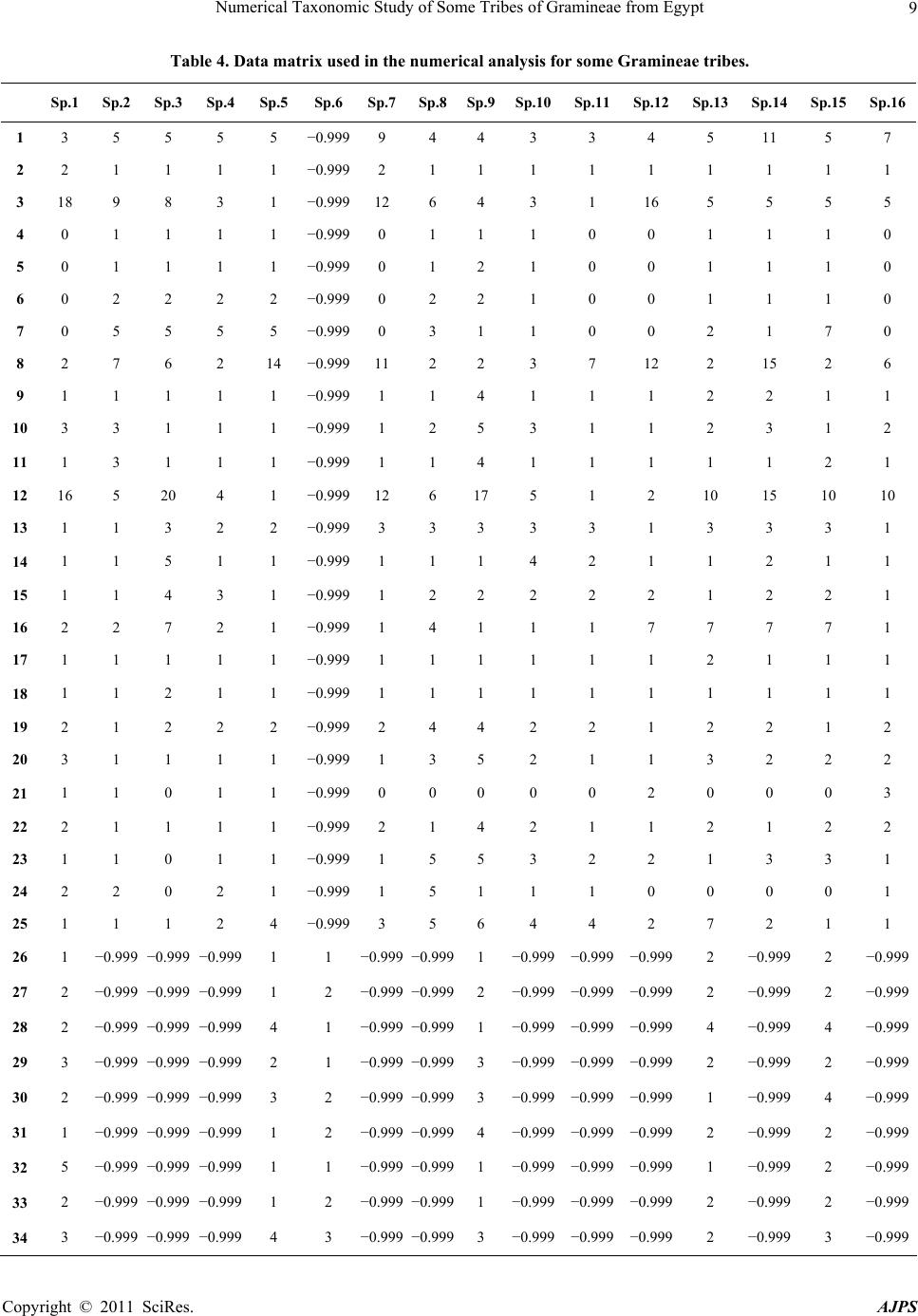 Numerical Taxonomic Study of Some Tribes of Gramineae from Egypt Copyright © 2011 SciRes. AJPS 9 Table 4. Data matrix used in the numerical analysis for some Gramineae tribes. Sp.1 Sp.2 Sp.3 Sp.4Sp.5 Sp.6 Sp.7Sp.8Sp.9Sp.10Sp.11Sp.12Sp.13 Sp.14 Sp.15Sp.16 1 3 5 5 5 5 −0.999 9 4 4 3 3 4 5 11 5 7 2 2 1 1 1 1 −0.999 2 1 1 1 1 1 1 1 1 1 3 18 9 8 3 1 −0.999 12 6 4 3 1 16 5 5 5 5 4 0 1 1 1 1 −0.999 0 1 1 1 0 0 1 1 1 0 5 0 1 1 1 1 −0.999 0 1 2 1 0 0 1 1 1 0 6 0 2 2 2 2 −0.999 0 2 2 1 0 0 1 1 1 0 7 0 5 5 5 5 −0.999 0 3 1 1 0 0 2 1 7 0 8 2 7 6 2 14 −0.999 11 2 2 3 7 12 2 15 2 6 9 1 1 1 1 1 −0.999 1 1 4 1 1 1 2 2 1 1 10 3 3 1 1 1 −0.999 1 2 5 3 1 1 2 3 1 2 11 1 3 1 1 1 −0.999 1 1 4 1 1 1 1 1 2 1 12 16 5 20 4 1 −0.999 12 6 17 5 1 2 10 15 10 10 13 1 1 3 2 2 −0.999 3 3 3 3 3 1 3 3 3 1 14 1 1 5 1 1 −0.999 1 1 1 4 2 1 1 2 1 1 15 1 1 4 3 1 −0.999 1 2 2 2 2 2 1 2 2 1 16 2 2 7 2 1 −0.999 1 4 1 1 1 7 7 7 7 1 17 1 1 1 1 1 −0.999 1 1 1 1 1 1 2 1 1 1 18 1 1 2 1 1 −0.999 1 1 1 1 1 1 1 1 1 1 19 2 1 2 2 2 −0.999 2 4 4 2 2 1 2 2 1 2 20 3 1 1 1 1 −0.999 1 3 5 2 1 1 3 2 2 2 21 1 1 0 1 1 −0.999 0 0 0 0 0 2 0 0 0 3 22 2 1 1 1 1 −0.999 2 1 4 2 1 1 2 1 2 2 23 1 1 0 1 1 −0.999 1 5 5 3 2 2 1 3 3 1 24 2 2 0 2 1 −0.999 1 5 1 1 1 0 0 0 0 1 25 1 1 1 2 4 −0.999 3 5 6 4 4 2 7 2 1 1 26 1 −0.999 −0.999 −0.9991 1 −0.999 −0.9991 −0.999 −0.999 −0.9992 −0.999 2 −0.999 27 2 −0.999 −0.999 −0.9991 2 −0.999 −0.9992 −0.999 −0.999 −0.9992 −0.999 2 −0.999 28 2 −0.999 −0.999 −0.9994 1 −0.999 −0.9991 −0.999 −0.999 −0.9994 −0.999 4 −0.999 29 3 −0.999 −0.999 −0.9992 1 −0.999 −0.9993 −0.999 −0.999 −0.9992 −0.999 2 −0.999 30 2 −0.999 −0.999 −0.9993 2 −0.999 −0.9993 −0.999 −0.999 −0.9991 −0.999 4 −0.999 31 1 −0.999 −0.999 −0.9991 2 −0.999 −0.9994 −0.999 −0.999 −0.9992 −0.999 2 −0.999 32 5 −0.999 −0.999 −0.9991 1 −0.999 −0.9991 −0.999 −0.999 −0.9991 −0.999 2 −0.999 33 2 −0.999 −0.999 −0.9991 2 −0.999 −0.9991 −0.999 −0.999 −0.9992 −0.999 2 −0.999 34 3 −0.999 −0.999 −0.9994 3 −0.999 −0.9993 −0.999 −0.999 −0.9992 −0.999 3 −0.999  Numerical Taxonomic Study of Some Tribes of Gramineae from Egypt Copyright © 2011 SciRes. AJPS 10 Sp.17 Sp. 18 Sp. 19 Sp. 20Sp. 21 Sp. 22 Sp. 23Sp. 24Sp. 25Sp. 26Sp. 27Sp. 28Sp. 29Sp. 30 Sp. 31 Sp. 32 Sp. 33Sp. 34 1 9 8 1 1 3 3 6 10 12 2 1 9 4 5 5 4 2 4 2 2 1 1 2 2 2 1 1 2 1 1 1 1 1 1 1 1 1 3 11 3 1 10 14 7 4 3 15 1 3 13 1 3 4 17 1 4 4 0 0 0 0 0 0 0 0 1 0 1 1 2 1 1 1 1 0 5 0 0 0 0 0 0 0 0 2 0 1 1 3 1 1 1 1 0 6 0 0 0 0 0 0 0 0 2 0 2 2 2 2 1 2 2 0 7 0 0 0 0 0 0 0 0 5 0 4 4 6 1 5 4 4 0 8 10 16 1 1 6 5 1 6 6 2 8 12 9 13 2 2 2 4 9 1 1 1 1 1 1 1 1 1 2 1 1 1 1 2 3 5 2 10 1 1 1 1 2 3 5 1 2 3 1 2 1 1 2 4 5 2 11 1 1 1 1 1 1 1 1 1 1 1 1 1 1 5 1 1 1 12 18 1 2 1 5 13 −0.99911 −0.9998 3 1 1414 19 7 9 8 13 3 3 3 3 3 1 −0.9993 −0.9993 3 3 3 3 3 3 3 3 14 1 1 5 1 1 1 −0.9995 −0.9991 3 3 1 2 5 1 2 1 15 1 1 4 1 1 1 −0.9994 −0.9992 1 1 1 1 4 1 2 2 16 6 5 1 1 1 1 −0.9997 -0.9997 7 7 1 7 7 3 7 7 17 1 1 1 1 1 1 −0.9991 −0.9991 1 1 1 1 1 1 1 1 18 1 1 1 1 1 1 −0.9991 −0.9991 1 1 1 1 1 1 1 1 19 2 2 2 1 2 2 −0.9993 −0.9993 2 2 1 2 1 6 5 6 20 1 1 1 1 3 1 −0.9991 −0.9993 1 1 1 1 2 5 6 4 21 0 0 0 0 0 5 −0.9990 −0.9990 0 0 0 0 0 0 0 0 22 2 1 2 1 1 1 −0.9991 −0.9994 6 6 1 2 2 3 6 5 23 1 1 0 1 1 2 −0.9990 −0.9995 2 2 1 1 0 5 4 6 24 7 8 1 1 1 1 −0.9990 −0.9990 0 0 1 0 0 6 0 0 25 2 3 4 3 4 4 −0.9994 −0.9993 3 3 5 3 1 5 6 3 26 2 −0.999 −0.999 −0.999 −0.999 −0.999 1 −0.999 −0.9991 −0.9991 −0.999−0.999 −0.999 −0.999 2 2 27 2 −0.999 −0.999 −0.999 −0.999 −0.999 2 −0.999 −0.9992 −0.9992 −0.999−0.999 −0.999 −0.999 2 2 28 3 −0.999 −0.999 −0.999 −0.999 −0.999 1 −0.999 −0.9994 −0.9991 −0.999−0.999 −0.999 −0.999 4 1 29 2 −0.999 −0.999 −0.999 −0.999 −0.999 1 −0.999 −0.9994 −0.9992 −0.999−0.999 −0.999 −0.999 1 2 30 2 −0.999 −0.999 −0.999 −0.999 −0.999 4 −0.999 −0.9993 −0.9991 −0.999−0.999 −0.999 −0.999 5 3 31 1 −0.999 −0.999 −0.999 −0.999 −0.999 1 −0.999 −0.9994 −0.9992 −0.999−0.999 −0.999 −0.999 3 2 32 3 −0.999 −0.999 −0.999 −0.999 −0.999 2 −0.999 −0.9993 −0.9992 −0.999−0.999 −0.999 −0.999 4 1 33 2 −0.999 −0.999 −0.999 −0.999 −0.999 2 −0.999 −0.9992 −0.9992 −0.999−0.999 −0.999 −0.999 2 2 34 3 −0.999 −0.999 −0.999 −0.999 −0.999 1 −0.999 −0.9993 −0.9993 −0.999−0.999 −0.999 −0.999 3 2  Numerical Taxonomic Study of Some Tribes of Gramineae from Egypt Copyright © 2011 SciRes. AJPS 11 Figure 2. Scatter-plot of 34 studied taxa plotted against the first factor by the second factor. donax and 23. Panicum turgidum. 2) Group including the rest 32 species. There are some characters; character of trichomes type, annulus of the pollen and also the oper- culum of the pollen grain (Table 3); which not been fit- ted into the data matrix because they are of only one code, so they were excluded in the analysis because they had no variation in the matrix. 4. Discussion In the present study a large number of grains macro- and micro-morphological, anatomical and pollen grains char- acters were scored and numerical methods (UPGMA and PCA) were applied to study the relationship among eleven Poaceae tribes and estimate the level of variation within and among these tribes. UPGMA gives insight into degree of similarity among the studied species and whether they form groups // clusters and gives an indica- tion of the level of variation within and between tribes. PCA reflects which characters are important on the axes, and indicates the significant characters based on the highest factor score (Table 5). Therefore it becomes clear which characters cause the separation between groups and can be useful to distinguish taxa. Pollen grains showed the most powerful significant characters, whereas all characters have been recorded are of high factor scores. Generally, our results show congruence between the UPGMA clustering and PCA analysis in suggesting two main groups and five subgroups which included the distribution of eleven tribes studied. Our UPGMA results show that the tribe Andropo- goneae is separated in one branch of the cladistic tree, the tribe Aristidieae is separated in three branches of the tree through three different subclads // subgroups. The tribe Arundineae with two species is separated in two branches of different clads in the tree. The tribe Aveneae is separated in five branches of the tree while the tribe Brachypodieae is separated in one branch and the Bromeae is separated in two branches within two differ- ent subclads. The Eragrostideae separated in four branches and the Paniceae separated in three branches. The Stipeae separated in four branches in only two clads while the Triticeae separated in three branches in also two clads. All the mentioned species, tribes, major clades and subclads are arranged as the following: The first main clads // groups of two species 6. Arundo donax (Tribe: Arundineae) and 23. Panicum turgidum (Tribe: Paniceae). While the second main group includes a large variety of taxa from different tribes; 32 species of tribes: Andropogoneae, Aristideae, Arundineae, Aveneae, Brachypodieae, Bromeae, Eragrostideae, Paniceae, Sti- peae and Triticeae. These tribes are separated through five distinct subgroups: 1) Sub-clad of species 25. La- marckia aurea, 28. Oryzopsis miliacea, 12. Polypogon monspeliensis, 18. Eragrostis cilianensis and 5. Stipa- grostis ciliata belonging to Poeae, Stipeae, Aveneae, Er- agrostideae, and Aristideae. 2) Sub-clad only of species  Numerical Taxonomic Study of Some Tribes of Gramineae from Egypt Copyright © 2011 SciRes. AJPS 12 Table 5. Factor loadings showed the most intrinsic characters enhanced separations of the studied species. Factor Loadings Rotation: Un-rotated Extraction: Principal components Characters Factor 1 Factor 2 Factor 3 1. Fruit shape 1.578348 0.871665 −1.24653 2. Coloring mode −0.23281 0.691848 0.173174 3. Color type 2.27393 2.051546 −3.32845 4. Trichome presence −0.54117 0.571291 0.467989 5. Hair length −0.50498 0.545862 0.444604 6. Hair coloring −0.36187 0.617713 0.489408 7. Position of attaching 0.194314 0.461728 0.273855 8. Fruit surface sculpture 2.058189 2.095991 −0.0891 9. Fruit weight (mg) −0.15868 −0.01955 0.347091 10. Fruit wide (mm) 0.125955 0.069094 −0.25377 11. Fruit length (mm) −0.18849 0.39183 0.305645 12. Section outline shape 3.173544 −3.89842 −0.03563 13. Hull cells type 0.443799 0.150657 1.117598 14. Aleurone cells shape 0.12138 0.430593 1.567286 15. Aleurone cells orientation 0.052685 0.232461 1.360735 16. Scutellum shape 1.010615 −1.21997 0.271807 17. Endosperm differentiation −0.33176 0.452823 0.673013 18. Endosperm ty pe −0.33784 0.490211 0.698834 19. Section wide (µm) 0.296755 −0.18378 0.934019 20. Section length (µm) 0.097103 −0.48171 0.569057 21. Hull cells thickness (µm) −0.60307 0.690619 0.471997 22. Seed coat thickness (µm) 0.18211 −0.36414 0.73636 23. Aleurone layer thickness (µm) 0.130375 −0.39784 0.654374 24. Scutellum cells thickness (µm) −0.18054 1.09338 0.69615 25. Endosper m thickness (µm) 0.764397 −0.05966 1.460169 26. Pollen class −1.07075 −0.2744 −0.64054 27. Pollen siz e −1.05331 −0.44013 −1.01514 28. Pollen shap e −0.92265 −0.79847 −0.89728 29. Pollen surface sculpture −0.98552 −0.54043 −0.84148 30. Annulus thickness (µm) −0.96595 −0.83463 −1.30983 31. Pore diameter (µm) −1.02089 −0.78764 −0.8329 32. Pollen wall thickness (µm) −1.00316 −0.51699 −0.97543 33. Sexine thickness (µm) −1.06557 −0.39242 −1.01249 34. Nexine thickness (µm) −0.97452 −0.69914 −1.23458 Percentage per PCA 16.54 4.34 3.13 Percentage for total variation for the three factors extracted 24.01 % *PCA: Principal Component Analysis  Numerical Taxonomic Study of Some Tribes of Gramineae from Egypt Copyright © 2011 SciRes. AJPS 13 32. Aegilops kotshyi belongs to Triticeae. 3) Sub-clad of 33. Aegilops ventricosa, 34. Hordium murinum Subsp. Leporinum, 26. Lolium perenne, 15. Bromus scoparius, 13. Brachypodium distachym and 9. Avena fatua be- longing to Triticeae, Poeae, Bromeae, Brachypodieae and Aveneae. 4) Sub-clad of species 29. Stipa capensis, 24. Dactylis glomerata, 30. Stipa lagascae, 14. Bromus rubens, 22. Echinochloa colona, 16. Coelachyrum bervi- folium, 7. Schismus arabicus, 31. Stipa parviflora and 3. Aristida funiculata belonging to Stipeae, Poeae, Bromeae, Paniceae, Eragrostideae, Arundineae and Aristideae. 5) Sub-clad of species 27. Poa annua, 11. Polypogon mari- timus, 19. Eragrostis minor, 10. Phalaris minor, 8. Avena barbata, 4. Aristida mutabilis, 21. Cenchrus ciliaris, 20. Leptochloa fusca and Aristida adscensionis within tribes: Poeae, Aveneae, Eragrostideae, Paniceae and Aristideae. Several various monophyletic species which regarded as sister-groups are distinct within five subclads men- tioned. Firstly, in the tree (Figure 1) Paniceae and Arun- dineae are a two-species sister-group to the rest whole cluster of the tree, on the other hand, Eragrostideae and Andropogoneae are another two-species sister-group within the second branch of the second major clad in the tree. Moreover, other species within Paniceae, Er- agrostideae and Arundineae are separated through some different subclads; thus Andropogoneae alone can be conspicuously differentiated from other tribes by means of its characteristic features for the fruit morphology, fruit anatomy and pollen grains morphology. Secondly, different tribes consume sister-grouping within each of the five subclads distinguished. Tribe Poeae conform a monophyletic sister-group in subclad (1) in a cluster of Stipeae, Aveneae, Eragrostideae and Aristideae and in subclad (5) in a cluster of Stipeae, Eragrostideae, Aris- tideae, Bromeae, Paniceae and Arundineae. Therefore, Poeae is preferably separated from these tribes depending on its own marked pollen grains characters. Moreover, the Triticeae shows a distinct variation that can aid the comparison of the relationships between Triticeae, Bromeae and Brachypodieae revealed by [16], where they suggested that the Brachypodieae is the sister group of the Triticeae while the Bromeae is the sister group of the Brachypodieae plus the Triticeae. Brachy- podium is the sister-group of a clad including both Bro- mus and the Triticeae. While, [5] illustrated that the rela- tionships between Bromus and the Triticeae is unre- solved, so there is a possibility that the Triticeae is a non-monophyletic group. Meanwhile, in our results, the Triticeae is a monophyletic sister-group to the neighbor- ing clad of Triticeae, Poeae, Bromeae, Brachypodieae and Aveneae (sub-clad 3). This clad which can be sepa- rated conspicuously through the Aveneae which is a monophyletic branch through Avena fatua, in addition to the separation of Poeae among tribes of subclads 1 and 5. Thus the Triticeae, Bromeae and Brachypodieae are closely related as confirmed by their palynological simi- larity, in addition to the compatibility of the fruit mor- phological (Table 5) that enhanced the understanding of the degree of similarity between taxa of these tribes. The Stipeae is a sister-group of the sub-clad (4), with exclud- ing tribes Poeae, Bromeae, Paniceae, Arundineae and Eragrostideae from this sub-clad, thus the Stipeae is separated from the Aristideae and also the similarity de- gree between them can be conducted to the characters of the fruit morphology and pollen grain morphology illus- trated in Table 5. Therefore, the applied methods of UPGMA and PCA can be used to study the variation within the tribe and the tribes in the family to determine the relationships between genera and tribes. Our results revealed there is a much separation between tribes An- dropogoneae, Arundineae, Aristideae, Stipeae, Poeae and Eragrostideae. However, tribes Triticeae, Bromeae and Brachypodieae showed much closer relationships. In addition to the consideration of those tribes Aveneae, Eragrostideae and Stipeae are the most heterogeneous tribes because the taxa of these tribes found to be inter- spersed with taxa from tribes Poeae, Paniceae and Aris- tideae. REFERENCES [1] W. D. Clayton and S. A. Renvoize, “Grasses of the World,” Genera Graminum, Her Majesty’s Stationary Of- fice, London, 1986. [2] H. P. Linder and P. J. Rudall, “The Evolutionary History of Poales,” Annual Reviews in Ecology and Systematics, Vol. 36, No. 1, 2005, pp. 107-124. doi:10.1146/annurev.ecolsys.36.102403.135635 [3] Grass Phylogeny Working Group, “Phylogeny and Sub- familial Classification of the Grasses (Poaceae),” Annals of the Missouri Botanical Garden, Vol. 88, No. 3, 2001, pp. 373-457. doi:10.2307/3298585 [4] K. W. Hilu and K. Wright, “Systematics of Gramineae: A Cluster Analysis Study,” Taxon, Vol. 31, No. 1, 1982, pp. 9-36. doi:10.2307/1220585 [5] K. W. Hilu and L. A. Alice, “A Phylogeny of Chlori- doideae (Poaceae) Based on matK Sequences,” System- atic Botany, Vol. 26, No. 2, 2001, pp. 386-405. [6] O. Seberg and S. Frederiksen, “A Phylogenetic Analysis of the Monogenomic Triticeae (Poaceae) Based on Mor- phology,” Botanical Journal of the Linnean Society, Vol. 136, No. 1, 2001, pp. 75-97. doi:10.1111/j.1095-8339.2001.tb00557.x [7] C. L. McIntyre, “Variation in Isozyme Loci in Triticeae,” Plant Systematics and Evolution, Vol. 160, No. 1-2, 1988, pp. 123-142. doi:10.1007/BF00936714 [8] J. V. Monte, C. L. McIntyre and J. P. Gustafson, “Analy- sis of Phylogenetic Relationships Using RFLPs,” Theo-  Numerical Taxonomic Study of Some Tribes of Gramineae from Egypt Copyright © 2011 SciRes. AJPS 14 retical and Applied Genetics, Vol. 86, No. 5, 1993, pp. 649-655. doi:10.1007/BF00838722 [9] R. J. Mason-Gamer, E. A. Kellogg, “Chloroplast DNA Analysis of the Monogenomic Triticeae: Phylogenetic Implications and Genome-Specific Markers,” In: J. J. Jau- har, Ed., Methods of genome analysis in plants. Boca Raton, CRC Press, Florida, 1996, pp. 301-325. [10] E. A. Kellogg and R. Apple, “Intraspecific and Inter- specific Variation in 5S RNA Genes are Decoupled in Diploid Wheat Relatives,” Genetics, Vol. 140, No. 1, 1995, pp. 325-343. [11] W. K. Taia, “Modern Trends in Plant Taxonomy,” Asian Journal of Plant Sciences, Vol. 4, No. 2, pp. 2005, 184- 202. [12] L. R. Parenti, “A Phylogenetic Analysis of the Land Plants,” Biological Journal of the Linnean Society, Vol. 13, No. 3, 1980, pp. 225-242. doi:10.1111/j.1095-8312.1980.tb00084.x [13] B. R. Baum, “A Phylogenetic Analysis of the Tribe Triticeae (Poaceae) Based on Morphological Characters of the Genera,” Canadian Journal of Botany, Vol. 61, No. 2, 1983, pp. 518-535. [14] J. Felsenstein, “Alternative Methods of Phylogenetic Inference and Their Interrelationship,” Systematic Zool- ogy, Vol. 28, No. 1, 1979, pp. 49-62. doi:10.2307/2412998 [15] K. Bremer, “Gondwanan Evolution of the Grass Alliance of Families (Poales),” Evolution, Vol. 56, No. 7, 2002, pp. 1374-1387. [16] R. R. Sokal and C. D. Michener, “A Statistical Method for Evaluating Systematic Relationships,” University of Kansas Scientific Bulletin, Vol. 28, 1958, pp. 1409-1438. [17] A. K. Osman, “Numerical Taxonomic Study of Some Tribes of Compositae (Subfamily Asteroideae) from Egypt,” Pakistan Journal of Botany, Vol. 43, No. 1, 2010, pp. 171-180. [18] O. Seberg, S. Frederiksen, C. Baden and I. Linde-Laursen, “Peridictyon, a New Genus from the Balkan Peninsula, and Its Relationship with Festucopsis (Poaceae),” Will- denowia, Vol. 21, No. 1-2, 1991, 87-104. [19] S. Frederiksen and O. Seberg, “Phylogenetic Analysis of the Triticeae (Poeae),” Hereditas, Vol. 116, No. 1-2, 1992, pp. 15-19. doi:10.1111/j.1601-5223.1992.tb00198.x |

