Intelligent Information Management
Vol.6 No.4(2014), Article ID:47622,11 pages
DOI:10.4236/iim.2014.64021
Keystroke Authentication on Enhanced Needleman Alignment Algorithm
Seham Bamatraf1, Mohamed Bamatraf2, Osman Hegazy1
1Department of Information System, Faculty of Information and Computers, Cairo University, Giza, Egypt
2Department of Computer Science, Faculty of Science, Hadhramout University, Yemen
Email: mailtoseham@gmail.com, mbamatraf1@yahoo.com, o.hegazy@fci-cu.edu.eg
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/
![]()
![]()

Received 15 April 2014; revised 14 May 2014; accepted 13 June 2014
ABSTRACT
An important point for computer systems is the identification of users for authentication. One of these identification methods is keystroke dynamics. The keystroke dynamics is a biometric measurement in terms of keystroke press duration and keystroke latency. However, several problems are arisen like the similarity between users and identification accuracy. In this paper, we propose innovative model that can help to solve the problem of similar user by classifying user’s data based on a membership function. Next, we employ sequence alignment as a way of pattern discovery from the user’s typing behaviour. Experiments were conducted to evaluate accuracy of the proposed model. The results show high performance compared to standard classifiers in terms of accuracy and precision.
Keywords:Keystroke Dynamics, Authentication, Fuzzy Logic, Sequence Alignment, Classification

1. Introduction
Security has become an interesting and important challenge in many applications especially with the increasingly development of technology such as in business and governments applications. One of these security issues is the user authentication. The authorized user must be protected against the multiple attacks from the hackers or the illegal attempts of unauthorized users.
Recent literature has been concerned about improving security of financial and scientific data through computers in the world. Moreover, industry had already been used to provide explicit protection from piracy. Biometric industry is considered to be one of the most innovative techniques for authentication [1] .
Authentication is defined as the process of identifying someone or something by their characteristics. The characteristics describe uniquely features of a person or a thing. A lot of technologies have been presented for user identification so that he or she is granted to use system resources. Such technologies support varying degrees of security. User’s identity [2] can be verified through several ways such as password, smart card, and biometric authentication. The latter [3] , is used to verify users based on several measurements [4] such as physiological features (such as face [5] and palm [6] ) and behavioral features (voice [7] , handwriting [8] , signature [9] , keystroke dynamics [10] , etc.).
Keystroke dynamics is designed to recognize users based on the method of their typing features or number of attributes related to their keystroke duration, key hold time, and latency. These attributes may be taken individually or together.
Recently, machine learning techniques [1] have been presented to solve many problems of security since they proved a robust efficiency to distinguish between identities. Moreover, bioinformatics filed [11] is based on handling and maintaining biological data which is typically in increasingly for its amount and structures. Using machine learning techniques could improve the performance significantly when employed for such data. One of the main techniques utilized by the machine learning is the data mining where classification learning is one of the main objectives of the data mining. Essentially, classification predicts a target class of given collection items. The classification goal is to predict the best class that is more related to the classified pattern. Recent literature has showed best results in identifying data features using data mining techniques.
Sequence alignment aims to construct the best alignment between sequences. These
alignments are assessed by assigned scores which are calculated by the total of
substitution scores plus penalties of gaps. The substitution costs are computed
using
![]() matrix, where
matrix, where
![]() for DNA and
for DNA and
![]() for proteins. Several types of matrices are introduced like PAM
[12] and Blosum [13] . Practically, the
Blosum matrices are considered better. Although sequence alignment was used heavily
in the bioinformatics literature, it can be utilized for solving classification
problems when the corresponding data structures and learning process are directed
to certain areas. Needleman-Wunsch algorithm [14]
is one of the most important techniques used in sequence alignment. The Blosum matrix
is used to measure the converge degree between DNA sequences.
for proteins. Several types of matrices are introduced like PAM
[12] and Blosum [13] . Practically, the
Blosum matrices are considered better. Although sequence alignment was used heavily
in the bioinformatics literature, it can be utilized for solving classification
problems when the corresponding data structures and learning process are directed
to certain areas. Needleman-Wunsch algorithm [14]
is one of the most important techniques used in sequence alignment. The Blosum matrix
is used to measure the converge degree between DNA sequences.
In this paper, we applied sequence alignment technique for improving user authentication based on keystroke behavior. However, a previous work [1] was presented in 2007 which also utilized bioinformatics techniques to improve user’s identification and verification. The work is dependent on the original Blosum of DNA sequence which may be affected the precise and accuracy of user’s authentication.
Here, we employed the sequence alignment to arrange the character sequences as DNA sequences to get its similarity based on Blosum matrix. We developed the Blosum to achieve the similarity based on the convergence degree of the behavioural attributes rather than letters’ sequences. Finally, we used Needleman-Wunsch algorithm as a sequence aligner tool with some modifications to efficiently authentication. Although Needleman-Wunsch algorithm was introduced in early 70’s, but it’s been used in several recent researches [15] .
Keystroke dynamics [16] -[18] could achieve security without any special hardware (keyboard only), if the generated patterns are strong enough. Thus, we depend only on the keyboard as the main factor to authenticate users. Additionally, data representation is one of the raised challenges. Data should meet the alignment requirements in terms of structure and semantic. Such challenges are solved in the proposed model in Section 4.
This paper is organized as follows: Section 2 presents a background about some related techniques and metrics. Section 3 describes Keystroke dynamics system. Section 4 presents the proposed method. Section 5 presents the experimental and results. Section 6 discusses the results and limitation of our approach, and Section 7 concludes the paper.
2. Background and Related Work
In the bioinformatics field, the sequence alignment aims to find the most common of motifs in sequences. Many researches addressed the sequence alignment issue and can be summarized in three types: global [14] , local [19] , and multiple [20] sequence alignment.
Global alignment of pairs of sequences means to make alignment over the entire length of both sequences. It is preferably when the pair has the same length and high similarity degree throughout. The well-known technique which used this global alignment and used in this paper is Needleman-Wunsch algorithm (NM&W) [14] . The goal of NM&W is to align the strings to achieve the closest similarity of both strings. Generally, the similarity of sequences is evaluated using a standardized substitution matrix that is based on Dayhoff [21] .
Several classification methods have been presented to identify the features of legal user. One of these methods was introduced by Kenneth [1] based on global alignment. The author normalized the time of user’s login data (include id and password) and divided them into the amino acid alphabet. The generated sequence after that is compared against the stored patterns. The comparison computed a score based on the typical global alignment technique.
Next approach, called Bio password [22] , was introduced which did not use any classification method. Two reasons prevented the authors from depend the assessment of classification methods. Firstly reason is due to the different parameters existing in the results. Secondly, most systems ignore the training and execution times. However, Bio password is considered one of the pioneers of the products that appeared in the commercial market for handling keystroke.
Gaines et al. [23] recorded the times of the successive keystrokes and authenticate users based on the probability distributions of such times.
Euclidean distance [24] was used between two vectors when typing characters: keystroke duration (i.e. total time to type a string) and pressure time. The vectors are stored as templates to be used latter in the classification process.
Another work presented in [25] , which was a simple classifier that can easily apply any distance measurement into the classification mechanism. Latter, Hidden Markov Model [26] was used as a classifier to classify the feature subsets generated from user typing behaviour.
Perceptron algorithm [27] was experimented using keystroke interval as input features for classification to provide linear decision functions to classify users and achieve low misclassification. Similarly, Bleha and Obaidat [28] used linear perception as their classifier to verify the identity of users.
In neural network, a new method called BPNN [10] was addressed which did not adhere to a certain number of inputs nodes. The method was used to differentiate between valid users from imposters based on their patterns that were extracted during typing their password keystroke.
Additionally, Belha at al [29] [30] applied real time measurement of keystroke duration and made use of several algorithms like Bayes classifier and Fisher’s linear Discriminate. Next, a minimum distance classifier was used to optimize results.
Obaidat [31] also used techniques like Potential Function, Bayes decision rule, K-means algorithm, and minimum distance algorithm and employed them for data classification.
Many other researchers focused on the combining various neural networks, pattern recognition, statistical measures, etc. In [32] , a suite of techniques for password authentication uses neural networks (3-layer feedforward network implementing with the back propagation algorithm), fuzzy logic (centre of gravity), and statistical methods (average and standard deviation) to solve the same problem.
All the previously mentioned researches used various types of classification techniques directly without any customization neither enhancements to sense the type of data and the reality problem domain. We think, the sense factor must be considered to improve the performance, stability and applicability of the proposed systems.
However, considering all valuable efforts, we try to prove the applicability of alignment techniques to represented numerical data. The state of art can be concluded in enhancing the Needleman-Wunsch alignment algorithm to as a score of similarity measure between keystroke dynamics (numerical data) which implies implementation of a customized score matrix. The enhanced algorithm can be later applied to any other similar data in any other domain.
3. Keystroke Dynamics’ Overview
This section provides an overview about keystroke dynamics including process, variations, and evaluation metrics. Keystroke dynamics is a biometric identifier used to discriminate valid users and imposters by analyzing their typing behaviour. Keystroke analysis is a measurement of the typing of the individual users including two main attributes: keystroke duration, and keystroke latency as shown in Figure 1.
Duration (Td) measures the period (Dwell time) starting when the key is pressed down until it is released. On the other hand, latency or flight time (Tf) measures the elapsed time between keystrokes, i.e. the interval between releasing one key and pressed the other [33] .
For example, they measure the keystrokes in three groups: from the first time the key is pressed and ends when the pressure is released from the key. Using statistical functions in the analysis process give accurate results.
The keystroke dynamics model is depicted in Figure 2. Generally, the model is divided into two stages: enrolment and testing. The enrolment stage involves collecting the data of users during login process (Id/password) and generating a unique signature reference which acts as a template to authenticate user. The templates are stored in database to be used latter in the testing stage.
At the testing stage, the user login data (id/password) has been extracted to be compared with the signature references to authenticate users when the template meets the user data. Otherwise, the workstation is closed (or does anything to prevent the user from entering the system).
Practically, there are two basic types of errors to measure the performance of biometric systems [34] . The first is called False Acceptance Rate (FAR) and the second is called False Reject Rate (FRR).
While FAR represents the percentage of incorrectly acceptance of imposters, FRR represents the percentage of incorrectly rejection of authorized users. However, there is a trade-off between the two measures, i.e. the system is either strictly in authentication and rejecting every attempts to login (leads to high FRR) or flexible leading to access the imposters to the system resources (leads to high FAR).
Moreover, bioinformatics and machine learning use two major factors to measure the performance of the learning classification: precision and accuracy [35] [36] . In summary, the testing stage of classification model produces either the known class of the instance (called positive class) otherwise it is called negative class. There are four options for the instance: true positive (TP) means that the instance has true condition and positive predicted class. True negative (TN) means that the instance has false condition and predict negative class. False positive (FP) means that the instance has false condition but predict positive class. False negative (FN) means the instance has true condition but predict negative class. Precision and accuracy are computed as follows equations:
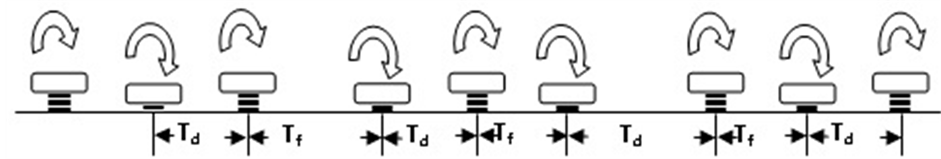
Figure 1. Keystroke features: dwell time (Td) and flight time (Tf).
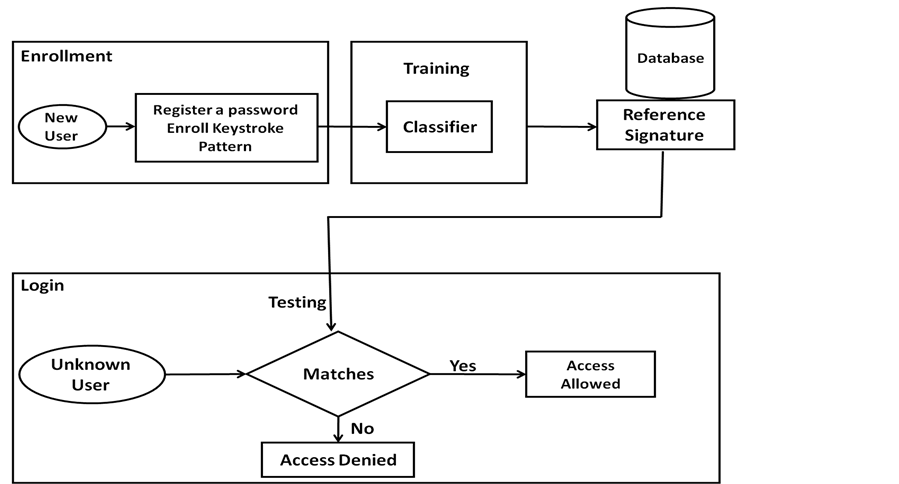
Figure 2. User authentication process using keystroke dynamics method.
 (1)
(1)
 (2)
(2)
4. The Proposed Model
The model consists of several phases, starts with data representation using fuzzy sets, and ends with identification, passing through a set of steps based on enhanced NM&W algorithm. The whole process is described in the following subsections.
4.1. Data Representation
Data is entered by users presents delays between sequential pair of characters for a password string (typing behavior). It is recorded as features (duration of a keystroke or key hold time, latency of keystrokes), cleaned from outlier statistically using median and standard deviation.
Fuzzy-logic is then used to range features and assign it into fuzzy sets to categorize the typing speed of the user into classes based on the membership function as described below:
The collected keystroke sequences will be used later to generate unique patterns of typing for each user representing the reference signature as shown in Figure 3.
Using fuzzy unit sequences are converted into characters as shown in Figure 4 which is as explained in this example:
The ranges of typing times are assigned to fuzzy sets (e.g., the times in the range of 2 - 3 milliseconds are part of a set named “very fast”) and can be represented as letter A and so on, where the time intervals are assigned according to the alphabetic order. However, if the time is between 2 - 8 msec, it can be assigned into 6 fuzzy sets (A-F) as in Equation (3).
The measure value of each character depends on the gap between every pair of characters, in other words, when an “A” is compared with a “B”, indicating minor difference compared to a “D”, such consideration could make a significant difference while computing the penalty and the score as well in the next phase.
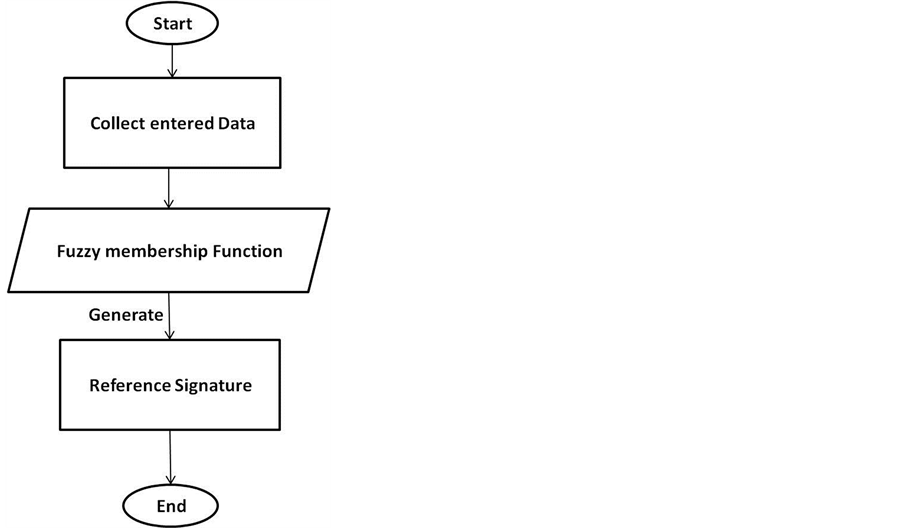
Figure 3. Phases of proposed model.

Figure 4. Pseudo code for membership function.
4.2. Classification
This section describes how to enhance the NM&W algorithm to predict classes of individuals.
Typically, NM&W algorithm is used to calculate the scores for aligned characters based on a substitution matrix. Instead of the conventional Blosum used in DNA sequences comparison, a custom similarity matrix is introduced. This matrix consists of the arrangement for alphabet letters and the degree of similarity according to the alphabet arrangement (considering difference measures).
The similarity degree between letters A and A takes the largest similarity degree, and the similarity degree between letters A and B will be less than (A, A), however the degree of similarity for letters that are far then their similarity will be less. The similarity degree between letters is computed according to the constructed Blosum matrix. Every pair of characters is applied to the similarity matrix in order and the score decision is recoded according to the steps described below.
Let n = number of characters, med = median position of characters (n + 1)/2.
For example, let’s the alphabet composed of 5 letters: A, B, C, D, and E. n = 5 and med = 3.
The Blosum matrix will be composed by![]() . The letters are arranged according to its order
and the computation for the degree of similarity is started from the median column
(the third column). The median column is initialized to zero and incrementally increased
by 1 with each row and decreased when reach the max value of similarity (2 in this
case). Meanwhile, for columns, the value is increased when proceeding to the left
and decreased when proceeding to the right as shown in
Figure 5.
. The letters are arranged according to its order
and the computation for the degree of similarity is started from the median column
(the third column). The median column is initialized to zero and incrementally increased
by 1 with each row and decreased when reach the max value of similarity (2 in this
case). Meanwhile, for columns, the value is increased when proceeding to the left
and decreased when proceeding to the right as shown in
Figure 5.
Consider the two strings keystroke fuzzy sequences to be globally aligned are:
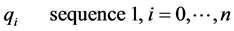
![]()
Each sequence consists of a set of alphabet characters. The following 3 steps show how scores are calculated using classical NM&W algorithm with slight modification.
Step I: Initialization
Create a score matrix
![]() with initial values set to the first row and column by multiplication of each cell
by the value of the gap penalty (
with initial values set to the first row and column by multiplication of each cell
by the value of the gap penalty (![]() in this case) as:
in this case) as:
![]()
![]()
Step II: Scoring
The rest of the cells of the score matrix are filled iteratively, cell by cell,
starting from the cell![]() , the score of every cell
, the score of every cell
![]() is
is
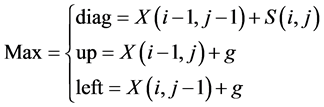
where
![]() is the similarity value of letters at
is the similarity value of letters at
![]() and
and![]() , and
, and
![]() is the gap penalty.
is the gap penalty.
In other words, the value of the cell
![]() is assigned based on one of three values in the three adjacent cells: up, left,
or diagonally as shown in Figure 6.
is assigned based on one of three values in the three adjacent cells: up, left,
or diagonally as shown in Figure 6.
Once the calculation of cells in the
![]() matrix is finished, the cell entry
matrix is finished, the cell entry
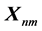 contains the highest score for all possible alignments.
contains the highest score for all possible alignments.
Step III: Trace Back (Alignment)
At this step, the alignment of two sequences is computed as shown in Figure 7. The alignment starts at the bottom right cell of the matrix, moving back in three directions: left, up, or diagonally. Trace back step is completed when accessing the first, top-left cell of the matrix.
The argument is based on the way sequence alignment can identify or extract similar and un-similar motifs in DNA sequences. Generally, DNA sequence proteins are similar except for few regions. Such regions can differentiate between the presence of the protein sequence, which could be a disease or any other motive responsible for the presence or the absences of a feature in the DNA source.
The same can be shown in keystroke similar sequences. The sequence(s), covers/classifies all user trials based on highest score obtained is/are considered as the user signature. The generated set is stored as the representative of the user class. It can be later used as the basis for comparison with any new user trial.
The similarity between users occurs due to global judgment over the sequence. In the proposed model the gap penalty while back tracing can successfully identify the set of character pairs where similar users differ from each other.

Figure 5. The proposed Blosum for five letters.

Figure 6. Calculation of X(i, j) value.

Figure 7. Tracback phase.
5. Experiments and Results
The used keystroke data benchmark for experimental purpose is downloaded from [37] . It is a set of timestampsof the typed passwords which consist of 10-characters composed of letters, number and punctuation. The generated password “.tie5Roanl” is chosen as it represents a strong one.
As mentioned on the web site the data are collected from about fifty volunteers (users) who are asked to write the same password in eight sessions each of which has 3 minute in average. Totally, the users had typed 400 passwords. The data is extracted involving the times of keydown-keydown (DD), keyup-keydown (UD), and hold (H) for all eleventh keys of the password (including the enter key).
The data are organized in a table of![]() . Each row represents the timing information extracted
when typing the password for a single user in a single repetition within one session.
The first three columns indicate: user id (1-50), session index (1-8) and repetition
number within the session (1-50).
. Each row represents the timing information extracted
when typing the password for a single user in a single repetition within one session.
The first three columns indicate: user id (1-50), session index (1-8) and repetition
number within the session (1-50).
Next columns include the timing information of the password. Columns are named with codes [1] indicating the timing information type as shown in Figure 8.
Consider the following one-line as an example of the table contents
As said previously, the data sets consist of 50 reads of each user of 50 users. About 90 percent of data reads of each user was applied in the training phase, while the remaining 10 per cent was utilized in the testing phase using random folds. These data are represented as a character sequences to be applied to the proposed system.
The same data set is applied to the Weka [38] experimental package. Various classifiers from different categories (trees, decision tables, decision rules, ANN, etc.) were experimented, in order to be compared with the proposed method under identical conditions.
Table 1 illustrates the results in terms of accuracy and precision computed as given in equations (1) and (2) respectively. The same results are also depicted in Figure 9 to be more visible.
In general, the experimental results indicate that the performance of the proposed system (Enhanced NM-W) is more efficient in terms of accuracy and precision compared to the other classifiers. The accuracy of our pro-

Figure 8. Timing information types.
Table 1. Classifier results using Weka tool vs enhanced approach.
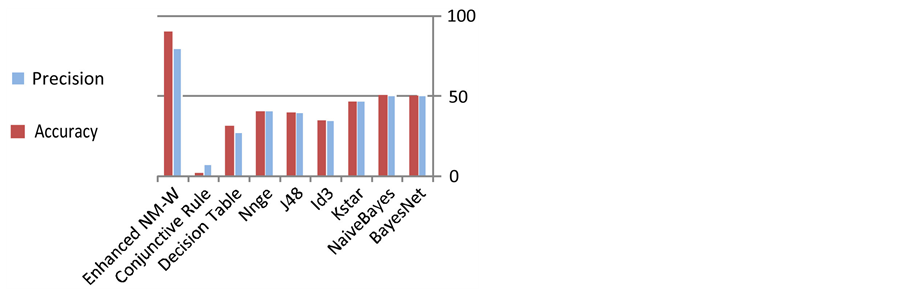
Figure 9. Classifier results of Weka and our enhanced approach.
posed system is better at approximately 30% over the nearest accuracy which is achieved by BayesNet. Similarity, the precision of our proposed system is better by about 40% over the nearest precision which is also achieved by BayesNet.
6. Discussion and Limitations
The main reason behind the far better results of the proposed model over other classifiers is due to several reasons. One reason is the nature of the data in terms of quantity and the problem domain, in some classifiers that are tree, table, and rule based when entropy is calculated for data with more than 40 attributes and about 50 exemplars and set as the base for the tree root or decision it leads in most of the cases into unbalanced judgment as there is high similarity between several users if most of attributed are used in the construction of the model, leading to high misclassification. One more reason is with nominal values such classifiers performance is lower compared to continuous data, when the raw data is applied to the classifiers it showed closed results to the proposed model. Moreover we used Weka classifier that deals with nominal data; it may be possible some other classifiers (out of our scope or knowledge) can generate similar results. Any how the proposed model results evidently proves the applicability of the model in similar domains, more datasets can also be experimented in the future with the proposed model.
However, there are some limitations for our approach must be considered. One limitation with the proposed model is the nature of data the technique can deal with; it can't be applied directly to continuous data. Another limitation lies in the nature of such classification problem as the relation between attributes where in some cases some keystrokes must be ignored in some users and kept for the rest sequence alignment skips such cases with penalties not effecting the judgment of relating such sequence to a user, where other classifiers usually considers the selection process to the whole data. Even though such feature is an advantage in the other hand it is a limitation for other type of nominal data. Moreover a problem lies with nature of the Needleman alignment regarding the local-minima trap.
7. Conclusion
This work handles the problem of how to authenticate users efficiently based on their keystroke behaviour. The method creates a unique signature for each user using a membership function as a sequence of letters. Hence, we utilize the sequence alignment Needleman-Wunsch algorithm to get more accurate value of authentication process. Furthermore, Blosum matrix is reconstructed to increase the similarity degree based of the convergence degree of letters in the keyboard. The experiments proved that Needleman is very promising in extracting user patterns with accuracy rate 80% and precision rate 90.3%. A comparison with other classifiers proved that the proposed approach achieves significantly better results.
References
- Revett, K. (2007) A Bioinformatics Based Approach to Behavioural Biometrics. Proceedings of the 2007 Frontiers in the Convergence of Bioscience and Information Technologies, Jeju City, 11-13 October 2007, 665-670.http://dx.doi.org/10.1109/FBIT.2007.143
- Joyce, R. and Gupta, G.K. (1990) Identity Authentication Based on Keystroke Latencies. Communications of the ACM, 33, 168-176. http://dx.doi.org/10.1145/75577.75582
- Chaudhari, R.D., Pawar, A.A. and Deore, R.S. (2013) The Historical Development of Biometric Authentication Techniques: A Recent Overview. International Journal of Engineering Research & Technology (IJERT), 2, 3921-3928.
- Nanavati, S., Thieme, M. and Nanavati, R. (2002) Biometrics: Identity Verification in a Networked World. John Wiley & Sons.
- Voth, D. (2003) Face Recogniition Technology. IEEE Intelligent Systems, 3, 4-7.
- Shu, W. and Zhang, D. (1998) Automated Personal Identification by Palmprint. Optical Engineering, 37, 2359-2362.http://dx.doi.org/10.1117/1.601756
- Shaughnessy, D.O. (1986) Speaker Recognition. IEEE ASSP Magazine, 3, 4-17. http://dx.doi.org/10.1109/MASSP.1986.1165388
- Tappert, C. (1984) Adaptive On-Line Handwriting Recognition. Proceedings of Seventh International Conference on Pattern Recognition, Montreal, 30 July-2 August 1984, 1004-1007.
- Herbst, N.M. and Liu, C.N. (1977) Automatic Signature Verification Based on Accelerometry. IBM Journal of Research and Development, 21, 245-253. http://dx.doi.org/10.1147/rd.213.0245
- Lin, D.-T. (1997) Computer-Access Authentication with Neural Network Based Keystroke Identity Verification. Proceedings of the International Conference on Neural Networks, Houston, 9-12 June 1997, 174-178.
- Kumar, P. and Sahoo, G. (2013) Survey On Bioinformatics And Computational Biology. International Journal of Engineering Research & Technology (IJERT), 2, 108-114.
- Dayhoff, M.O., Schwartz, R.M. and Orcutt, B.C. (1978) A Model of Evolutionary Change in Proteins. Atlas of Protein Sequence and Structure, 5, 345-351.
- Henikoff, S. and Henikoff, J.G. (1992) Amino Acid Substitution Matrices from Protein Blocks. Proceeding of the National Academy of Sciences of the United States of America, 89, 10915-10919.
- Needleman, S.B. and Wunsch, C.D. (1970) A General Method Applicable to the Search for Similarities in the Amino Acid Sequence of Two Proteins. Journal of Molecular Biology, 48, 443-453. http://dx.doi.org/10.1016/0022-2836(70)90057-4
- Eger, S. (2013) Sequence Alignment with Arbitrary Steps and Further Generalizations, with Applications to Alignments in Linguistics. Information Sciences, 237, 287-304. http://dx.doi.org/10.1016/j.ins.2013.02.031
- Wangsuk, K. and Anusas-amornkul, T. (2013) Trajectory Mining for Keystroke Dynamics Authentication. Procedia Computer Science, 24, 175-183. http://dx.doi.org/10.1016/j.procs.2013.10.041
- Stefan, D., Shu, X. and Yao, D. (2012) Robustness of Keystroke-Dynamics Based Biometrics against Synthetic Forgeries. Computers & Security, 31, 109-121.
- Alpar, O. (2014) Keystroke Recognition in User Authentication Using ANN Based RGB Histogram Technique. Engineering Applications of Artificial Intelligence, 32, 213-217.
- Smith, T.F. and Waterman, M.S. (1981) Identification of Common Molecular Subsequences. Journal of Molecular Biology, 147, 195-197. http://dx.doi.org/10.1016/0022-2836(81)90087-5
- Higgins, D.G. and Sharp, P.M. (1988) CLUSTAL: A Package for Performing Multiple Sequence Alignment on a Microcomputer. Gene, 73, 237-244. http://dx.doi.org/10.1016/0378-1119(88)90330-7
- Dayhoff, M.O. and Foundation, N.B.R. (1979) Atlas of Protein Sequence and Structure. National Biomedical Research Foundation.
- Karnan, M. and Krishnaraj, N. (2010) BioPassword—Keystroke Dynamic Approach to Secure Mobile Devices. IEEE International Conference on Computational Intelligence and Computing Research (ICCIC), Coimbatore, 28-29 December 2010, 1-4.
- Gaines, R.S. (1980) Authentication by Keystroke Timing: Some Preliminary Results. Rand, Santa Monica.
- Young, J.R. and Hammon, R.W. (1989) Method and Apparatus for Verifying an Individual’s Identity. United States Patent.
- Hu, J., Gingrich, D. and Sentosa, A. (2008) A k-Nearest Neighbor Approach for User Authentication through Biometric Keystroke Dynamics. Proceedings of IEEE International Conference on Communications, Beijing, 19-23 May 2008, 1556-1560.
- Rodrigues, R.N., Yared, G.F.G., do N. Costa, C.R., Yabu-Uti, J.B.T., Violaro, F. and Ling, L.L. (2006) Biometric Access Control through Numerical Keyboards Based on Keystroke Dynamics. Proceedings of the 2006 International Conference on Advances in Biometrics, Hong Kong, 5-7 January 2006, 640-646.
- Bleha, S.A., Knopp, J. and Obaidat, M.S. (1992) Performance of the Perceptron Algorithm for the Classification of Computer Users. Presented at the Proceedings of the 1992 ACM/SIGAPP Symposium on Applied Computing: Technological Challenges of the 1990’s, Kansas City, 1992.
- Bleha, S.A. and Obaidat, M.S. (1993) Computer Users Verification Using the Perceptron Algorithm. IEEE Transactions on Systems, Man, and Cybernetics, 23, 900-902. http://dx.doi.org/10.1109/21.256563
- Bleha, S., Slivinsky, C. and Hussien, B. (1990) Computer-Access Security Systems Using Keystroke Dynamics. IEEE Transactions on Pattern Analysis and Machine Intelligence, 12, 1217-1222. http://dx.doi.org/10.1109/34.62613
- Bleha, S.A. and Obaidat, M.S. (1991) Dimensionality Reduction and Feature Extraction Applications in Identifying Computer Users. IEEE Transactions on Systems, Man and Cybernetics, 21, 452-456. http://dx.doi.org/10.1109/21.87093
- Obaidat, M.S. (1995) A Verification Methodology for Computer Systems Users. Proceedings of the 1995 ACM Symposium on Applied Computing, Nashville, 26-28 February 1995, 258-262. http://dx.doi.org/10.1145/315891.315976
- Haider, S., Abbas, A. and Zaidi, A.K. (2000) A Multi-Technique Approach for User Identification through Keystroke Dynamics. IEEE International Conference on Systems, Man, and Cybernetics, 2, 1336-1341.
- Gutiérrez, F.J., Lerma-Rascón, M.M., Salgado-Garza, L.R. and Cantú, F.J. (2002) Biometrics and Data Mining: Comparison of Data Mining-Based Keystroke Dynamics Methods for Identity Verification. Proceedings of the 2nd Mexican International Conference on Artificial Intelligence: Advances in Artificial Intelligence, Yucatán, 22-26 April 2002, 460-469.
- Krause, M. and Tipton, H.F. (2011) Handbook of Information Security Management. Vol. 5, CRC Press LLC, Boca Raton.
- Eisner, R., Poulin, B., Szafron, D., Lu, P. and Greiner, R. (2005) Improving Protein Function Prediction Using the Hierarchical Structure of the Gene Ontology. IEEE Symposium on Computational Intelligence in Bioinformatics and Computational Biology, San Diego, 14-15 November 2005, 1-10.
- Lu, Z., Szafron, D., Greiner, R., Lu, P., Wishart, D.S., Poulin, B., Anvik, J., Macdonell, C. and Eisner, R. (2004) Predicting Subcellular Localization of Proteins Using Machine-Learned Classifiers. Bioinformatics, 20, 547-556. http://dx.doi.org/10.1093/bioinformatics/btg447
- Killourhy, K. and Maxion, R. (2009) Keystroke Dynamics-Benchmark Data Set. www.cs.cmu.edu/~keystroke
- Holmes, G., Donkin, A. and Witten, I.H. (1994) WEKA: A Machine Learning Workbench. Proceedings of the 1994 2nd Australian and New Zealand Conference on Intelligent Information Systems, Brisbane, 29 November-2 December 1994.


