Natural Science
Vol.5 No.4A(2013), Article ID:30825,8 pages DOI:10.4236/ns.2013.54A004
Reproductive isolation in the Elegans-Group of Caenorhabditis
![]()
Department of Biological Sciences, Wright State University, Dayton, USA; *Corresponding Author: scott.baird@wright.edu
Copyright © 2013 Scott Everet Baird, Sara R. Seibert. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received 21 January 2013; revised 20 February 2013; accepted 7 March 2013
Keywords: Caenorhabditis; Pre-Mating Isolation; Assortative Fertilization; Post-Zygotic Isolation
ABSTRACT
Reproductive isolation is the basis of the Biological Species Definition and can be a driving force of speciation. Theoretical studies have provided models of how reproductive isolation can arise within individual species. Genetic tests of these models are limited to populations in which reproductive isolation is present but not complete. Here, reproductive isolation in the ElgansGroup of the nematode genus Caenorhabditis is reviewed. Pre-mating barriers, assortative fertilization and post-zygotic barriers all have been observed in this clade. In some combinations of species, fertile F1 hybrids can be obtained. Therefore, the Elegans-Group of Caenorhabditis is poised to become an important experimental system for the study of reproductive isolation.
1. INTRODUCTION
Reproductive isolation refers collectively to all genetic mechanisms that prevent or limit gene flow between populations [1]. These mechanisms are thought to result from allele-specific dysgenic interactions between two or more genes [2-5]. Reproductive isolation is the basis of the Biological Species Definition [1]. As reproductive isolation restricts gene flow, it also can facilitate genomic divergence between populations [6-9]. Categories of reproductive isolating mechanisms include pre-mating isolation, assortative fertilization and post-zygotic isolation [1,5,10,11]. Pre-mating isolation refers to mechanisms that prevent matings between individuals from different species. These mechanisms include species-specific differences in reproductive structures, behavior, ecology, and seasonality of reproduction. Assortative fertilization refers to mechanisms that prevent fertilization after mating has occurred. These mechanisms include species-specific differences in sperm chemotaxis and sperm-ova fusion. Post-zygotic isolation refers to mechanisms that decrease the fitness of hybrids relative to that of the parental populations. These mechanisms include hybrid lethality, hybrid sterility and hybrid breakdown.
Early speciation models required the origin reproductive isolation in allopatric populations [1]. In this way, dysgenic combinations of alleles would not be subjected to negative selection before they became fixed. More recently, models of speciation with gene flow have been developed [8,9,12]. In such models, reproductive isolation does not act uniformly across the entire genome [6]. Rather, gene flow will be restricted only in regions linked to loci involved in reproductive isolation [6,9,12].
To gain a broad understanding of speciation, empirical studies in diverse taxa are required. To date, nematodes in general, and Caenorhabditis in particular, have contributed little to these studies [13-20]. However, in recent years, a number of new Caenorhabditis species have been discovered [18,21]. With these new species, four pairwise combinations that yield fertile F1 hybrids have been described [17-19]. With these new resources, Caenorhabditis is poised to become an important speciation genetics model system. This review provides an overview of reproductive isolation in the Elegans-Group of Caenorhabditis.
2. THE ELEGANS-GROUP
Caenorhabditis is a genus of microbiophagus nematodes that reproduce on rotting fruit [21,22]. Caenorhabditis species reproduce rapidly with generation times of three to four days at 20˚C. Like other nematodes, Caenorhabditis species form developmentally arrested, alternative third stage, dauer larvae in conditions of environmental stress [23]. Most if not all species of Caenorhabditis form phoretic associations with other soil invertebrates as dauers [24]. These associations are used for transport to microenvironments conducive to reproduction.
One member of Caenorhabditis, C. elegans is a model organism that has been used extensively in genetic and genomic studies [e.g. 25-27]. More recently, additional members of Caenorhabditis have contributed to a variety of evolutionary studies [28-30]. Mostly, other the species of Caenorhabditis used in these studies have been from the Elegans-Group.
The Elegans-Group is a monophyletic clade within Caenorhabditis (Figure 1). Live cultures of ten ElegansGroup species are available. C. elegans is the basal member or this group [21]. The development of this species has been characterized extensively [31,32]. All species within the Elegans-Group are morphologically similar and follow similar patterns of development [14,33]. The genomes of several Elegans-Group Species have been sequenced or are in the process of being sequenced and assembled [34-36]. Curated lists of orthologous genes can be found at wormbase.org.
Most species in the Elegans-Group of Caenorhabditis

Figure 1. Phylogenetic relationships within the Elegans-Group of Caenorhabditis [18,21]. C.oncomelaniae, C. formosana and C. clavopapillata are included within the ElegansGroup based on morphological characters [37]. live cultures of these three species currently are not available, which precludes their inclusion in phylogenies, such as this one, that are based on DNA sequence comparisions.
are known only from geographically limited collection sites [21]. However, at least four, C. elegans, C. briggsae, C. remanei and C. brenneri have cosmopolitan distributions [21,37]. Of these, populations are most highly structured in C. briggsae [38].
Many pairwise combinations of Elegans-Group species are cross-fertile (Figure 2). In most cross-fertile combinations, F1 hybrids arrest during embryogenesis [13,14,21]. In six combinations, F1 adults are obtained (Table 1) [13,16-18]. In four of these combinations, fertile F1 adults are obtained [17,18]. These results demonstrate that the Elegans-Group of Caenorhabditis affords ample opportunities for genetic and genomic studies of reproductive isolation.
3. REPRODUCTIVE ISOLATION IN THE ELEGANS-GROUP
3.1. Pre-Mating Isolation
Within the Elegans-Group of Caenorhabditis, all pairwise combinations of species that have been tested will mate with each other [13,17,18,21]. Therefore, premating mechanisms do not obviously contribute to reproductive isolation. This conclusion has two major caveats.
The first caveat is that all mating tests between Caenorhabditis species were conducted on agar plates with E. coli as a food source. In natural populations, Caenorhabditis species reproduce on rotting fruit while feeding on a diverse microbial community [21,22]. Pre-mating mechanisms not apparent in laboratory conditions may act in natural populations. For example, C. elegans and C. briggsae both reproduce on rotting fruit and have been found to co-occur on the same fruit [22]. These species appear
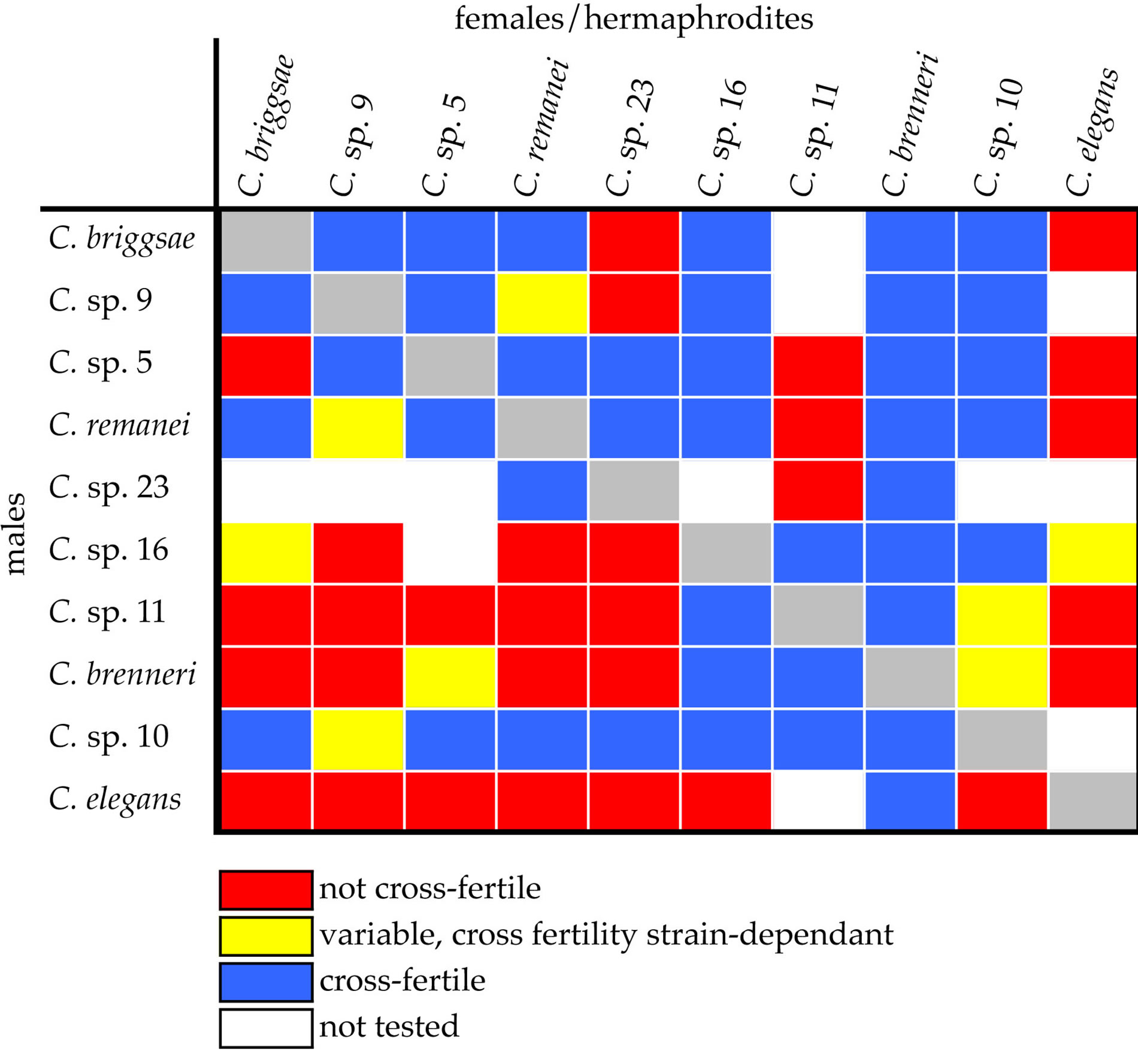
Figure 2. Cross fertility among Elegans-Group species [13-18, 21,57].
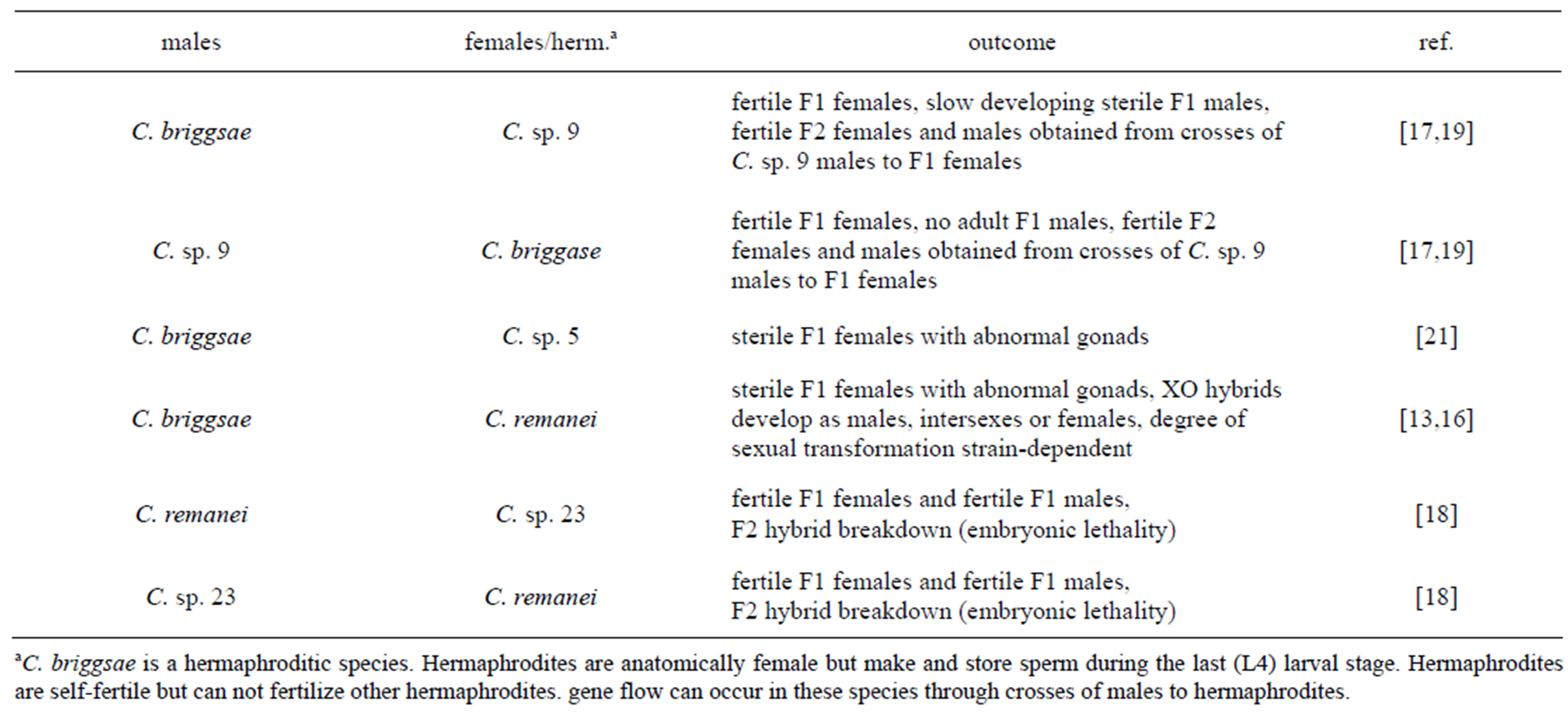
Table 1. Hybrid sterility and hybrid breakdown in the Elegans-Group.
to be temporally isolated with C. briggsae being found primarily at warmer temperatures (e.g. during summer and early fall and C. elegans being found primarily at cooler temperatures (e.g. during late fall) [22]. Caenorhabditis species also may be isolated based on host preferences. Most if not all Caenorhabditis species form phoretic associations with soil invertebrates [24]. These associations are as dauer larvae and are used for transport to suitable reproductive environments. Species-specific host preferences may result in differences in reproductive environments utilized. This does not appear to be true of C. briggsae, C. elegans and C. remanei. All of these species form phoretic associations with a variety of soil invertebrates including terrestrial isopods, snails and beetle larvae [22,39,40]. Host preferences of other members of the Elegans-Group remain to be characterized.
The second caveat is that no quantitative mate choice experiments have been conducted for any combination of Caenorhabditis species. In Drosophila, many species will mate with each other but quantitative differences between conand inter-specific matings in these species can be detected in singleand multiple-choice tests [41, 42]. Similar differences may be present in Caenorhabditis. If present, these differences are likely to be mediated by the male-specific ray sensilla. Caenorhabditis males exhibit an exploratory (wandering) behavior that is suppressed in the presence females or hermaphrodites [43]. The suppression of this behavior requires contact with females or hermaphrodites and is mediated by the neurons of the ray sensilla and the male-specific EF interneurons [44]. Male exploratory behavior and its suppression by contact with conspecific females or hermaphrodites has been described in C. elegans, C. briggsae, and C. remanei [43]. Contact with congeneric females and hermaphrodites also suppresses male-exploratory as males are retained on plates during interspecific crosses [13,21]. The efficacy of intervs con-specific in suppressing male-exploratory behavior has not been determined.
3.2. Assortative Fertilization
Assortative fertilization results from species-specific interactions between sperm and ova. In sea urchins, chemotaxis of sperm toward ova, activation of the acrosome reaction and fusion of the sperm and egg all are species-specific [45-47]. In species with internal fertilization, responses of the reproductive tract to interspecific sperm also may limit or prevent fertilization after mating has occurred. In some combinations of Drosophila species, the insemination reaction may serve this purpose [48- 50].
Fertilization in Caenorhabditis is internal. During mating amoeboid sperm are injected into the uterus and then crawl to the spermathecae where fertilization occurs (Figure 3) [15,51,52]. In C. elegans, migration of sperm to the spermathecae is in response to prostaglandins signals that are secreted from oocytes [51,52]. Also in C. elegans, several genes have been identified that are required for fertilization [53]. Two of these, spe-9 and spe-38, are expressed on the plasma membranes of sperm pseudopods and may encode sperm ligands [54,55]. Another two, egg-1 and egg-2 are expressed on the surface of oocytes and may encode receptors for sperm ligands (although not necessarily for SPE-9 and/or SPE-38) [56].
Defects in sperm chemotaxis have been observed in some, but not in all, cross-infertile combinations of species in the Elegans-Group [15,56]. Moreover, a limited

Figure 3. Sperm chemotaxis in Caenorhabditis. (A) An adult C. remanei female. the position of the vulva is indicated by the arrow. The positions of the spermathecae are indicated by the arrowheads; (B) Flourescently labeled C. remanei sperm localized to the spermathecae in a C. remanei female; (C) Flourescently labeled C. remanei sperm randomly distributed throughhout the uterus in a C. sp. 9 female. Scales bars, panel A = 100 µm, panels B and C = 50 µm.
amount of cross fertilization can occur even when defects in sperm chemotaxis are apparent [57]. Therefore, species-specific chemotaxis does contribute to assortative fertilization in the Elegans-Group, but other reproductive barriers, such as species-specific receptor-ligand interactions involved in sperm fusion must act as well.
In the combination of C. briggsae and C. sp. 9, interactions between sperm and the reproductive tract also limit cross fertility [17]. Reciprocal matings between these species are cross-fertile. However, C. briggsae hermaphrodites become sterile two days after mating with C. sp. 9 males despite the continued presence of sperm within their spermathecae. The cause of this mating-induced sterility is not known.
3.3. Post-Zygotic Isolation
Most cross-fertile combinations of Elegans-Group species are reproductively isolated by hybrid lethality [13, 21]. Most lethality occurs during embryogenesis [13,21]. Terminal phenotypes of hybrid embryos in four combinations of Elegans-Group species have been characterized [14]. The embryonic cell lineage of C. elegansis known and is nearly invariant [#]. Embryonic cell lineages of other members of the Elegans-Group are similar, if not identical to that of C. elegans [14]. Because of this, terminal phenotypes can be used to identify which developmental processes are defective in hybrid embryos. Most hybrid embryos either failed to gastrulate or failed to elongate from an ovate ball of cells to a tube-shaped worm.
Transcriptionally repressed C. elegans embryos arrest at the onset of gastrulation indicating that zygotic transcription is first required at this stage [59]. As gastrulation is the earliest arrest stage observed in ElegansGroup hybrids, dysgenic interactions in these embryos likely involve at least one zygotically expressed gene. However, in this context it should be noted that in C. elegans, the paternally delivered PEEL-1 protein causes lethality well after the completion of gastrulation [60]. Embryonic elongation occurs during the later half of embryogenesis. During elongation, the embryo changes from an ovoid ball of cells into a tube-shaped worm. Many hybrid embryos either failed to initiate elongation or arrested at the two-fold stage. The initial stages of elongation are driven by circumferential actin microfilaments in the epidermis [61]. Continued elongation beyond the two-fold stage requires proper development of the body wall muscles that underlie and are connected to the basement membrane and epidermis [62,63]. ElegansGroup hybrids that arrest at the onset of elongation likely have defects in epidermal development and those that arrest at the two-fold stage likely have defects in the development or function of the body wall musculature. PEEL-1-induced lethality in C. elegans intraspecific hybrids occurs at the two-fold stage and results from defects in muscular and epidermal tissues [60].
In a few combinations of Elegans-Group species, F1 hybrids arrest during larval development. The arrest stages of these hybrid larvae have not been well characterized. Characterizations of terminal phenotypes in these larvae may provide insights into the development processes that are disrupted by dysgenic interactions.
In six pairwise combinations of Elegans-Group species, adult F1s are obtained (Table 1) [16-18,21]. Sterile female F1 hybrids were obtained from crosses of C. briggsae males to C. remanei females and from crosses of C. briggsae males to C. sp. 5 females [16,21]. Gonad development in F1 females derived from both of these crosses was abnormal.
Fertile F1 female hybrids were obtained reciprocal crosses of C. briggsae to C. sp. 9 [17,19]. Fertile female and male F2 hybrids were obtained when F1 females were mated to C. sp. 9 males. The frequencies of viable and fertile F2 males were consistent with the segregation of dysgenic C. briggsae alleles at a small number of autosomal loci. When C. briggsae males were mated to F1s, F2 hybrids arrested during embryogenesis. Sterile F1 adult males were obtained from crosses of C. briggsae males to C. sp. 9 [17,19]. These males were small and sickly and took twice as long to reach adulthood as did F1 females. F1 adult males were absent or present at much lower frequencies from crosses of C. sp. 9 males to C. briggsae hermaphrodites [17,19]. This difference likely resulted from differences in X-chromosomes, mitochondrial cytotype and/or maternal inheritance in reciprocal F1 males [19].
From reciprocal crosses of C. remanei to C. sp. 23 fertile F1 females and males were obtained [18]. Most F2 progeny from these crosses died during embryogenesis. Results from crosses of F1 progeny back to parental species have not been reported. However, this combination of Elegans-Group species presents an opportunity to characterize the genetic architecture of hybrid breakdown.
3.4. Haldane’s Rule in the Elegans-Group
In many crosses between closely related species a gender bias is observed in the fitnesses of F1 hybrids [5,64, 65]. In almost all instances, it is the heterogametic gender that is less fit. This association between sexual karyotype and hybrid fitness is referred to as Haldane’s Rule. Haldane’s Rule is of interest because it is thought to result from a common genetic mechanism of speciation in diverse taxa [5].
Caenorhabditis species are diploid with five pairs of autosomes [66]. Females and hermaphrodites are diplo-X and males are haplo-X [30]. Therefore, hermaphrodites and females are homogametic, producing only X-bearing gametes, and males are heterogametic, producing both haplo-X and nullo-X sperm. Haldane’s rule is observed in Caenorhabditis. In four combinations of species a gender bias is observed in the viabilities and/or fertilities of F1 hybrids [13,16,17,19,21]. In all cases, F1 hybrid males are less fit than F1 hybrid females.
From crosses of C. briggsae males to C. remanei females, sterile F1 adult hybrids were obtained [13]. The genders of adult hybrids obtained in this combination varied depending upon the strains of C. briggsae and C. remanei used [16]. In most combinations of strains, haplo-X hybrids that should have developed as males were partially or completely transformed into females. Variation in the degree of sexual transformation of XO hybrids was linked to allelic variation in the C. briggsaeCbr-tra-2 sex determination gene [67]. To our knowledge, this is the only instance in which Haldane’s Rule results from sexual transformation [65].
From cross of C. briggsae males to C. sp. 5 females, sterile F1 females with gonadal abnormalities were obtained [21]. F1 males were not observed. It was not determined whether the absence of F1 males was due to male-specific hybrid lethality or sexual transformation of XO hybrids.
From reciprocal crosses of C. briggsae to C. sp. 9, fertile F1 female hybrids were obtained [17,19]. F1 males were obtained from crosses of C. briggsae males to C. sp. 9 females. As noted above, these males were sickly, sterile and took twice as long to reach adulthood than did their sibling females. F1 adult males were absent or extremely rare from crosses of C. sp. 9 males to C. briggsae hermaphrodites [17,19]. These results indicate that the viability of F1 hybrid males was impacted by the source of the X chromosome, the mitochondrial cytotype and/or the maternal genotype [19].
3.5. Genic Isolation
Nascent reproductive isolation is not expected to restrict gene flow throughout the entire genome [6]. Rather, restrictions to gene flow are expected only in regions linked to loci involved in dysgenic interactions. In the Elegans-Group of Caenorhabditis, two examples of localized restriction of gene flow are known [20,68].
In C. elegans, gene flow is restricted between “Bristolcompatible” and “Hawaii-compatible” strains in a 33 kb region of chromosome I [68]. In Bristol-compatible strains, this region contains peel-1 and zeel-1. These genes are absent from Hawaii-compatible strains. peel-1 is a paternal-effect gene that encodes a sperm-delivered toxin that causes lethality during embryonic elongation [60]. zeel-1 is a zygotic-effect gene that suppresses PEEL-1-induced lethality. Lethality is observed in F2 hybrids that lack zeel-1.
In C. briggsae, gene flow is restricted on chromosome III in crosses between temperate strains and the AF16 tropical strain [20,69]. Approximately 20% of the F2 progeny in these crosses exhibit a delayed development phenotype (DDP) in which affected animals take four days to reach sexual maturity in contrast to their non-delayed siblings the reach sexual maturity in three days. This delay in maturation causes a 25% decrease in the intrinsic growth rate. The DDP results from a dysgenic interaction between recessive AF16 alleles at a zygotic acting gene on chromosome III and temperate alleles at an unmapped maternal-effect gene [20]. Because of the decreased intrinsic growth rate in delayed animals AF16 alleles on chromosome III were under-represented in recombinant inbred lines constructed from crosses of AF16 to the HK104 temperate strain [69,70].
4. CONCLUSION
C. elegans has long been an important model system for developmental genetics. With the maturation of this system, increasing attention has been given to other species within Caenorhabditis. This has resulted the recent discovery of multiple new species and in a rapid increase in evolutionary studies in this genus. Within the ElegansGroup of Caenorhabditis, many cross-fertile combinations of species have been identified. These include four combinations that yield fertile F1 hybrids. Also, intraspecific reproductive isolation has been described C. elegans and in C. briggsae. With these resources, genetic and genomic studies of reproductive isolation and speciation are possible in the Elegans-Group Caenorhabditis.
5. ACKNOWLEDGEMENTS
We thank E. E. King for helpful comments on this manuscript.
REFERENCES
- Mayr, E. (1963) Animal species and evolution. Belknap Press, Harvard.
- Bateson, W. (1909) Heredity and variation in modern lights. In: Seward, A.C. Ed., Darwin and Modern Science, Cambridge University Press, Cambridge, 85-101.
- Dobzhansky, T. (1936) Studies on hybrid sterility. II. Localization of sterility factors in Drosophila pseudoobscura hybrids. American Naturalist, 71, 404-420.
- Muller, H.J. (1942) Isolating mechanisms, evolution, and temperature. Biology Symposium, 6, 71-125.
- Coyne, J.A. and Orr, H.A. (2004) Speciation. Sinauer, Sunderland.
- Wu, C.-I. (2001) The genic view of the process of speciation. Journal of Evolutionary Biology, 14, 851-865. doi:10.1046/j.1420-9101.2001.00335.x
- Payseur, B.A. (2010) Using differential introgression in hybrid zones to identify genomic regions involved in speciation. Molecular Ecology, 10, 806-820.
- Smadja, C.M. and Butlin, R.K. (2011) A framework for comparing processes of speciation in the presence of gene flow. Molecular Ecology, 20, 5123-5140. doi:10.1111/j.1365-294X.2011.05350.x
- Feder, J.L., Egan, S.P. and Nosil, P. (2012) The genomics of speciation-with-gene-flow. Trends in Genetics, 28, 342-350. doi:10.1016/j.tig.2012.03.009
- Markow, T.A. (1997) Assortative fertilization in Drosophila. Proceedings of the National Academy of Sciences of USA, 94, 7756-7760. doi:10.1073/pnas.94.15.7756
- Turelli, M., Barton, N.H. and Coyne, J.A. (2001) Theory and speciation. Trends in Ecology & Evolution, 16, 330- 343.
- Servidio, M.R. (2009) The role of linkage disequilibrium in the evolution of premating isolation. Heredity, 102, 51- 56. doi:10.1038/hdy.2008.98
- Baird, S.E., Sutherlin, M.E. and Emmons, S.W. (1992) Reproductive isolation in Rhabditidae (Nematoda: Secernentea); mechanisms that isolate six species of three genera. Evolution, 46, 585-594. doi:10.2307/2409629
- Baird, S.E. and Yen, W.-C. (2000) Reproductive isolation in Caenorhabditis: Terminal phenotypes of hybrid embryos. Evolution & Development, 2, 9-15. doi:10.1046/j.1525-142x.2000.00031.x
- Hill, K.L. and L’Hernault, S.W. (2001) Analyses of reproductive interactions that occur after heterospecificmatings within the genus Caenorhabditis. Evolution & Development, 232, 104-114. doi:10.1006/dbio.2000.0136
- Baird, S.E. (2002) Haldane’s rule by sexual transformation in Caenorhabditis. Genetics, 161, 1349-1353.
- Woodruff, G.C., Eke, O., Baird, S.E., Félix, M.-A. and Haag, E.S. (2010) Insights into species divergence and the evolution of hermaphroditism from fertile interspecies hybrids of Caenorhabditis nematodes. Genetics, 186, 997-1012. doi:10.1534/genetics.110.120550
- Dey, A., Jeon, Y., Wang, G.-X. and Cutter, A.D. (2012) Global population genetic structure of Caenorhabditisremanei reveals incipient speciation. Genetics, 191, 1257- 1269. doi:10.1534/genetics.112.140418
- Kozlowska, J.L., Ahmad, A.R., Jahesh, E. and Cutter, A.D. (2011) Genetic variation for postzygotic reproductive isolation between Caenorhabditisbriggsae and Caenorhabditis sp. 9. Evolution, 66, 1180-1195. doi:10.1111/j.1558-5646.2011.01514.x
- Baird, S.E. and Stonesifer, R. (2012) Reproductive isolation in Caenorhabditisbriggsae: Dysgenic interactions between maternaland zygotic-effect loci result in a delayed development phenotype. Worm, 1, in press. doi:10.4161/worm.23535
- Kiontke, K.C., Félix, M.-A., Ailion, M., Rockman, M.V., Braendle, C., Pénigault, J.-B. and Fitch, D.H.A. (2011) A phylogeny and molecular barcodes for Caenorhabditis, with numerous new species from rotting fruits. BMC Evolutionary Biology, 11, 339. doi:10.1186/1471-2148-11-339
- Félix, M.-A. and Duveau, F. (2012) Population dynamics and habitat sharing of natural populations of Caenorhabditiselegans and C. briggsae. BMC Biology, 10, 59. doi:10.1186/1741-7007-10-59
- Hu, P.J. (2007) Dauer. In: Worm Book, The C. elegans Research Community.
- Kiontke, K.C. and Sudhaus, W. (2006) Ecology of Caenorhabditis species. In: The C. elegans Research Community, Ed., WormBook.
- Avila, D., Helmcke, K. and Aschner, M. (2012) The Caenorhabditiselegans model as a reliable tool in neurotoxicology. Human & Experimental Toxicology, 31, 236- 243. doi:10.1177/0960327110392084
- Pukkila-Worley, R. and Ausubel, F.M. (2012) Immune defense mechanisms in the Caenorhabditis intestinal epithelium. Current Opinion in Immunology, 24, 3-9. doi:10.1016/j.coi.2011.10.004
- Furuhashi, H. and Kelly, W.G. (2010) The epigenetics of germ-line immortality: Lessons from an elegant model system. Development, Growth & Differentiation, 52, 527- 532. doi:10.1111/j.1440-169X.2010.01179.x
- Fitch, D.H.A. and Thomas, W.K. (1997) Evolution. In: Riddle, D.L., Blumenthal, T., Meyer, J. and Priess, J.R., Eds. C. elegans II, CSHL Press, Cold Spring Harbor.
- Cutter, A.D., Dey, A. and Murray, R.L. (2009) Evolution of the Caenorhabditiselegans genome. Molecular Biology and Evolution, 26, 1199-1234. doi:10.1093/molbev/msp048
- Haag, E.S. (2005) The evolution of nematode sex determination: C. elegans as a reference point for comparative biology. In: The C. elegans Research Community, Ed., WormBook.
- Wood, W.B. (1988) The Nematode, Caenorhabditiselegans. Cold Spring Harbor Press, New York.
- Riddle, D.L., Blumenthal, T., Meyer, B.J. and Priess, J.R. (1997) C. elegans II. Cold Spring Harbor Press, New York.
- Sudhaus, W. and Kiontke, K. (1996) Phylogeny of rhabditis subgenus Caenorhabditis (Rhaditidae, Nematoda). Journal of Zoological Systematics and Evolutionary Research, 34, 217-233. doi:10.1111/j.1439-0469.1996.tb00827.x
- C. elegans (1998) Genome sequence of the nematode C. elegans: A platform for investigating biology. Science, 282, 2012-2017.
- Stein, L.D., Bao, Z., Blasiar, D., Blumenthal, T., Brent, M. R., Chen, N., Chinwalla, A., Clarke, L., Clee, C., Coghlan, A., Coulson, A., D'Eustachio, P., Fitch, D.H.A., Fulton, L.A., Fulton, R.E., Griffiths-Jones, S., Harris, T.W., Hillier, L.W., Kamath, R., Kuwabara, P.E., Mardis, E.R., Marra, M.A., Miner, T.L., Minx, P., Mullikin, J.C., Plumb, R.W., Rogers, J., Schein, J.E., Sohrmann, M., Spieth, J., Stajich, J.E., Wei, C., Willey, D., Wilson, R.K., Durbin R., and Waterston, R.H. (2003) The genome sequence of Caenorhabditisbriggsae: A platform for comparative genomics. PLoS Biology, 1, e45. doi:10.1371/journal.pbio.0000045
- Barrière, A., Yang, S.-P., Pekarek, E. Thomas, C.G., Haag, E.S. and Ruvinsky, I. (2009) Detecting heterozygosity in shotgun genome assemblies: lessons from obligately outcrossing nematodes. Gen. Res., 19, 470-480.
- Sudhaus, W. and Kiontke, K. (2007) Comparison of the cryptic nematode species Caenorhabditisbrenneri sp. n. and C. remanei (Nematoda: Rhabditidae) with the stem species pattern of the Caenorhabditiselegans group. Zootaxa, 1456, 45-62.
- Cutter, A.D., Yan, W., Tsvetkov, N. Sunil, S. and Félix, M.-A. (2010) Molecular population genetics and phenotypic sensitivity to ethanol for a globally diverse sample of the nematode Caenrhabditisbriggsae. Molecular Ecology, 19, 798-809. doi:10.1111/j.1365-294X.2009.04491.x
- Baird, S.E., Fitch, D.H.A. and Emmons, S.W. (1994) Caenorhabditis vulgaris sp. n. (Nematoda: Rhabditidae): A necromenic associate of pillbugs and snails. Nematologica, 40, 1-11. doi:10.1163/003525994X00012
- Baird, S.E. (1999) Natural and experimental associations of Caenorhabditisremanei with Trachelipusrathkii and other terrestrial isopods. Nematology, 1, 471-475. doi:10.1163/156854199508478
- Banerjee, P. and Singh, B.N. (2012) Interspecific sexual isolation and phylogeny among different members of the Drosophila bipectinata species complex. Genetica, 140, 75-81. doi:10.1007/s10709-012-9659-4
- Laturney, M. and Moehring, A.J. (2012) Fine-scale genetic analysis of species-specific female preference in Drosophila simulans. Journal of Evolutionary Biology, 25, 1718-1731. doi:10.1111/j.1420-9101.2012.02550.x
- Lipton, J., Kleemann, G. Ghosh, R., Lints, R. and Emmons, S.W. (2004) Mate searching in Caenorhabditiselegans: a genetic model for sex drive in a simple invertebrate. The Journal of Neuroscience, 24, 7427-7434. doi:10.1523/JNEUROSCI.1746-04.2004
- Barrios, A., Nurrish, S. and Emmons, S.W. (2008) Sensory regulation of C. elegans male mate-searching behavior. Current Biology, 18, 1865-1871. doi:10.1016/j.cub.2008.10.050
- Suzuki, N. and Yamaguchi, M. (1986) Species specific respiratory stimulation of sea urchin (Clypeaster japonicas) spermatozoa by an egg-associated factor. Zoological Science, 3, 801-806.
- Alves, A.-P., Mulloy, B., Diniz, J.A. and Mourão, A.S. (1997) Sulfated polysaccharides from the egg jelly layer are species-specific inducers of acrosomal reactions in sperms of sea urchins. The Journal of Biological Chemistry, 272, 6965-6971. doi:10.1074/jbc.272.11.6965
- Kamei, N. and Glabe, C.G. (2003) The species-specific egg receptor for sea urchin sperm adhesion is ERB1, a novel ADAMTS protein. Genes & Development, 17, 2502-2507. doi:10.1101/gad.1133003
- Asada, N. and Kitagawa, O. (1988) Insemination reaction in the Drosophila nasuta subgroup. Japanese Journal of Genetics, 63, 137-148. doi:10.1266/jjg.63.137
- Pitnick, S., Miller, G.T., Schneider, K. and Markow, T.A. (2003) Ejaculate-female coevolution in Drosophila mohavensis. Proceedings of the Royal Society B, 270, 1507- 1512. doi:10.1098/rspb.2003.2382
- Bono, J.M., Matzkin, L.M., Kelleher, E.S. and Markow, T.A. (2011) Postmating transcriptional changes in reproductive tracts of conand heterospecifically mated Drosophila mojavenis females. Proceedings of the National Academy of Sciences of the United States of America, 108, 7878-7883. doi:10.1073/pnas.1100388108
- Kubagawa, H.M., Watts, J.L., Corrigan, C., Edmonds, J.W., Sztul, E., Browse, J. and Miller, M.A. (2006) Oocyte signals derived from polyunsaturated fatty acids control sperm recruitment in vivo. Nature Cell Biology, 8, 1143-1148. doi:10.1038/ncb1476
- Edmonds, J.W., Prasain, J.K., Dorand, D., Yang, Y. Hoang, H.D., Vibbert, H., Kubagawa, H.M. and Miller, M.A. (2010) Insulin/FOXO signaling regulates ovarian prostaglandins critical for reproduction. Developmental Cell, 19, 858-871. doi:10.1016/j.devcel.2010.11.005
- Marcello, M.R. and Singson, A. (2010) Fetilization and the oocyte-to-embryo transition in C. elegans. BMB Reports, 43, 389-399. doi:10.5483/BMBRep.2010.43.6.389
- Singson, A., Mercer, K.B and L'Hernault, S.W. (1998) The C. elegansspe-9 gene encodes a sperm transmembrane protein that contains EGF-like repeats and is required for fertilization. Cell, 93, 71-79. doi:10.1016/S0092-8674(00)81147-2
- Chatterjee, I., Richmond, A. Putiri, E. Shakes, D.C. and Singson, A. (2005) The Caenorhabditiselegansspe-38 gene encodes a novel four-pass integral membrane protein required for sperm function at fertilization. Development, 132, 2795-2808. doi:10.1242/dev.01868
- Kadandale, P., Stewart-Michaelis, A., Gordon, S. Rubin, J., Klancer, R. Schweinsberg, P. Grant, B.D., and Singson, A. (2005) The egg surface LDL receptor repeat-containing proteins EGG-1 and EGG-2 are required for fertilization in Caenorhabditiselegans. Current Biology, 15, 2222-2229. doi:10.1016/j.cub.2005.10.043
- Baird, S.E. and Seibert, S.R. (2012) Assortative fertilization in the Elegans-Group of Caenorhabditis. Evolution of Caenorhabditis and other nematodes. Cold Spring, Harbor, 2012.
- Sulston, J.E., Schierenberg, E., White, J.G. and Thomson, J.N. (1983) The embryonic cell lineage of Caenorhabditiselegans. Developmental Biology, 100, 64-119. doi:10.1016/0012-1606(83)90201-4
- Powell-Coffman, J.A., Knight, J. and Wood, W.B. (1994) Onset of C. elegans gastrulation is blocked by inhibition of embryonic transcription with an RNA polymerase antisense RNA. Developmental Biology, 178, 472-483. doi:10.1006/dbio.1996.0232
- Seidel, H.S., Ailion, M., Li, J., van Oudenaarden, A., Rockman, M.V. and Kruglyak, L. (2011) A novel spermdelivered toxin causes late-stage embryo lethality and transmission ratio distortion in C. elegans. PLoS Biology, 9, e1001115. doi:10.1371/journal.pbio.1001115
- Priess, J.R. and Hirsh, D.I. (1986) Caenorhabditiselegans morphogenesis: the role of the cytoskeleton in elongation of the embryo. Developmental Biology, 117, 156-173. doi:10.1016/0012-1606(86)90358-1
- Barstead, R.J. and Waterston, R.H. (1991) Vinculin is essential for muscle function in the nematode. The Journal of Cell Biology, 114, 715-724. doi:10.1083/jcb.114.4.715
- Topf, U. and Chiquet-Ehrismann, R. (2011) Genetic interaction between Caenorhabditiselegansteneurinten-1 and prolyl 4-hydroxylase phy-1 and their function in collagen IV-mediated basement membrane intergrity during late elongation of the embryo. Molecular Biology of the Celll, 22, 3331-3343. doi:10.1091/mbc.E10-10-0853
- Haldane, J.B.S. (1922) Sex ratio and unisexual sterility in hybrid animals. Journal of Genetics, 12, 101-109. doi:10.1007/BF02983075
- Laurie, C.C. (1997) The weaker sex in heterogametic: 75 years of Haldane’s Rule. Genetics, 147, 937-951.
- Hodgkin. J. (2005) Karyotype, ploidy, and gene dosage. In: The C. elegans Research Community, Ed., WormBook.
- Romer, S.H. (2005) The genetics of sexual transformation in Caenorhabditisbriggsae-Caenorhabditisremanei hybrids. M.Sc. Dissertation, Wright State University, Dayton.
- Seidel, H.S., Rockman, M.V. and Kruglyak, L. (2008) Widespread genetic incompatibility in C. elegans maintained by balancing selection. Science, 319, 589-594. doi:10.1126/science.1151107
- Ross, J.A., Koboldt, D.C., Staisch, J.E., Chamberlin, J.E., Gupta, B.P., Miller, R.D., Baird, S.E. and Haag, E.S. (2011) Caenorhabditisbriggsae recombinant inbred lines genotypes reveal inter-strain incompatibility and the evolution of recombination. PLoS Genetics, 7, e1002174. doi:10.1371/journal.pgen.1002174
- Hillier, L.W., Miller, R.D., Baird, S.E., Chinwalla, A., Fulton, L.A., Koboldt, D.C. and Waterston, R.H. (2007) Comparison of C. elegans and C. briggsae genome sequences reveals extensive conservation of chromosome organization and synteny. PLoS Biology, 5, e167. doi:10.1371/journal.pbio.0050167

