Food and Nutrition Sciences
Vol.4 No.11A(2013), Article ID:38904,6 pages DOI:10.4236/fns.2013.411A004
Autolytic and Proteolytic Properties of Strains of Lactococcus lactis Isolated from Different Vegetables, Raw-Milk Cheeses and Commercial Starter Cultures
![]()
Chemistry Faculty, University Autonomous of Chihuahua, Chihuahua, Mexico.
Email: ngutierrez@uach.mx
Copyright © 2013 Carolina Nájera-Domínguez, Nestor Gutiérrez-Méndez. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received August 14th, 2013; revised September 14th, 2013; accepted September 21st, 2013
Keywords: Com Autolysis; Proteolysis; L. lactis; Wild-Strains
ABSTRACT
The autolysis and proteolysis are important features in the strains of L. lactis used in the manufacture of cheese. The autolytic and proteolytic activity of L. lactis has been linked with the development of flavor and texture in the cheese. On the other hand, there is a growing interest in new strains isolated from raw-milk cheeses and vegetables. These wild-strains have showed different features of industrial importance in comparison with those observed in commercial cultures. However, it still not clear if the autolytic and proteolytic properties of these wild-strains differ from the industrial strains. The objective of this work was to assess the autolytic and proteolytic activities of 21 strains of L. lactis isolated from diverse sources. The rates of autolysis and proteolysis observed in vitro were highly strain-dependent. The pH and the NaCl concentration in the media affected significantly the autolysis of L. lactis. The strains isolated from vegetable showed in general low and medium autolytic activity, whereas the strains isolated from raw-milk cheeses had medium to high autolytic activity. The strain with highest proteolytic activity was a strain isolated from corn leaves. Although still not clear how this strain acquired this pronounced characteristic.
1. Introduction
Lactococcus lactis of the lactic acid bacteria (LAB) are more widely used as starter dairy culture on the manufacture of cheese [1]. The autolysis of Lactococcus lactis is a desirable trait, because most of the enzymes related with the production of flavor compounds are intracellular. In this sense, the early release of intracellular enzymes during the ripening of cheese may accelerate the development of flavor compounds [2-4]. Autolysis is the spontaneous disintegration of cell wall peptidoglycan by endogenous mureinases named autolysins [3,5-7]. Cellular lysis of LAB improves the interaction between bacterial enzymes and cheese substrates [8,9]. The lysis of LAB may occur as the result of age, stress, unfavorable physiological conditions or attack of lytic phages [6]. Environmental factors such as salt concentration, pH and temperature may affect the autolysis of L. lactis [2]; however few studies have assessed the effect of each of these factors and their possible interaction in the autolysis of L. lactis.
Proteolytic activity of L. lactis strains is another feature of industrial relevance. The proteolytic system of L. lactis is composed essentially of proteinases that initially cleave the milk protein to peptides, and then these peptides are cleaved into smaller peptides and amino acids by intracellular peptidases. Some of these amino acids are subsequently catabolized, producing a variety of flavor compounds like aldehydes, alcohols, carboxylic acids, esters and sulfur compounds [8,10,11].
On the other hand, recently there is a growing interest in the identification of new strains isolated from different ecosystems, including vegetables and diverse types of raw-milk cheeses [12]. It has been observed that strains isolated from niches with adverse conditions such as high microbial competition (like raw-milk cheeses) or niches with scare availability of nutrients (like the surface of vegetables) have a more efficient metabolism [13]. Some authors have reported that these wild-strains have different characteristics of industrial interest like lactose and citrate fermentation, or the production of certain flavor compounds from milk components [12,14-16]. However, still not clear if the autolytic and proteolytic properties of these wild-strains differ from the industrial strains. The objective of this work was to assess the autolytic and proteolytic activities of 21 strains of L. lactis isolated from diverse sources including industrial cultures, vegetables and raw-milk cheeses.
2. Materials and Methods
2.1. Microorganisms
In this study were used twenty strains of Lactococcus lactis (Table 1) isolated and identified in a previous work from raw-milk dairy products, vegetables and different commercial starter cultures [12]. Additionally, one reference ATCC strain of L. lactis was included in this work. All the strains were kept at −20˚C in M17 broth (Difco laboratories, Ditroit Mich) with 40% glycerol before their use.
2.2. Determination of the Autolytic Capacities of Lactococal Strains
Autolysis was determined measuring changes in optical density (OD). Six solutions with diverse pH (5 and 7)

Table 1. Lactococcus lactis strains used in this study.
and NaCl concentration (1%, 2%, and 3% w/w) were used in the analysis of lytic behavior of L. lactis strains. The pHs of these solutions were adjusted with lactic acid before their sterilization. Before the autolysis assays, each strain was growing (12 - 16 hours) in M17 broth at 37˚C and then bacterial cells were harvested by centrifugation at 2000 × g for 10 min at 4˚C. The pellets obtained were washed twice with phosphate buffer (50 mM) pH 7 and ultimately suspended in the corresponding lysis solution. Immediately, cell suspensions were poured into sterile microplates (96-well U bottom shape) adding 250 μL by well. Changes in OD at 595 nm were monitored throughout 48 hours of incubation at 37˚C with a microplate reader (ELx808, Biotek, USA). The percentage of autolysis was calculated as follows: % Autolysis = (OD0 – OD48) × 100/OD0, where OD0 was the initial optical density, and OD48 the optical density observed after 48 hours of incubation. The strains were assessed by triplicated in each lysis solution.
2.3. Determination of Proteolytic Activities of Lactococal Strains
Each strain was inoculated in 40 mL of sterile M17 broth and incubated overnight at 37˚C. Bacterial cells were harvested by centrifugation at 2000 × g for 10 min at 4˚C and then suspended into 35 mL of phosphate buffer (50 mM) pH 7. Cellular suspensions were subject to ultrasonic lysis applying an acoustic power of 472 mW∙mL−1 (calculated experimentally according to [17] and a constant frequency of 20 kHz, using a sonifier with a 1/2) diameter horn (Branson Sonifier S450, USA). The ultrasound treatment was applied three times to each solution by cycles of five minutes at 10˚C [18].
Cellular extracts (CE) were analyzed for protein content using the Bradford methodology [19]. The proteolytic activities (global activities of proteases and peptidases) of the CEs were measured by the TNBS (2,4,6- trinitrobenzene-sulfonic acid) procedure as described by Boutrou et al. [20] and Ustunol et al. [21]. An aliquot of 30 μL from each CE was added to 1 mL of casein solution (0.01% w/vol) and incubated 20 min at 30˚C. Afterward, 400 μL from the CE-casein solution was mixed with 50 μL of (0.03 M) TNBS (Sigma-Aldrich, St. Louis, MO) and 1.6 mL of sodium tetraborate (0.1 M). The mixture was incubated 30 min at 30˚C and then was measured the absorbance at 420 nm. The reagent blank consisted of 50 μL of (0.03 M) TNBS and 2 mL of (0.1 M) sodium tetraborate. Additionally, the mean absorbance obtained from the TNBS assay of casein solution (0.01% w/v) without CE was used as a correction factor. A standard curve was prepared with glycine, and the specific proteolytic activities were expressed as units of glycine (U-Gly) per milligrams of protein.
2.4. Statistical Analysis
Data collected from autolysis of 21 strains of L. lactis were analyzed as a factorial design using the model yijk = μ + τi + βj + γk + τβ(ij) + eijk. The factors in the model were the concentration of NaCl and the pH of the lysis media. Variations among strains were blocked in the model to reduce the experimental error. The responses from proteolysis were analyzed as one factor completely randomized design. Additionally, a multiple mean comparison analysis (Tukey-Kramer test) was carried out with the software Minitab 16 (Minitab Inc., Pennsylvania, USA) to determine differences among treatments.
3. Results
Each strain of L. lactis had different behavior depending on the lysis solution used. In such a way that some conditions enhanced the autolysis of certain strains while disfavored the autolysis of other strains (Figure 1). Large variation in the percentage of autolysis was observed among the 21 strains of L. lactis (Figure 1). Statistical analysis showed that pH, NaCl concentration and its interaction had a significant influence (P < 0.05) in the autolysis of L. lactis. In general, most strains of L. lactis had higher percentages of autolysis under acidic conditions and low salt concentration (pH 5% - 1% NaCl), but also at neutral pH and high salt content (pH 7% - 5% NaCl). In contrast, the acidic media with high salt content (pH 5% - 5% NaCl) did not favor the autolysis of most strains of L. lactis (Figure 2).
The strains isolated from vegetables had low and medium autolytic activities at pH 5 and the strains isolated from raw-milk cheeses showed a medium activity, whereas some strains isolated from industrial starter cultures (KK05, PK04, EZ02a) showed high autolytic activeity at pH 5 (Figure 1). In contrast, at pH 7 one strain isolated from an artisanal cheese (strain DE01b) and one strain isolated from an industrial culture (KK05) were those with the highest autolytic activity. The strains isolated from vegetables showed low and medium autolytic activity at pH 7, as well as most of the strains isolated from commercial starter cultures (Figure 1).
On the other hand, cellular extracts (CE) obtained from each strain of L. lactis were analyzed for their proteolytic activities (U-Gly mg of protein−1) on casein solution (Figure 3). CE from the strains MH05, EZ03b, and Li had the higher activities (19.1 to 14.6 U-Gly mg of protein−1), and the strains BB07, FDVSBS10, MM101, C272, Rq07, and PK04 obtained the lower activities (2.4 to 1.6 U-Gly mg of protein−1). It is worth noting that the strain isolated from corn leaves (MH05) was the one with the highest proteolytic activity. However not all the strains isolated from vegetables had a high proteolytic
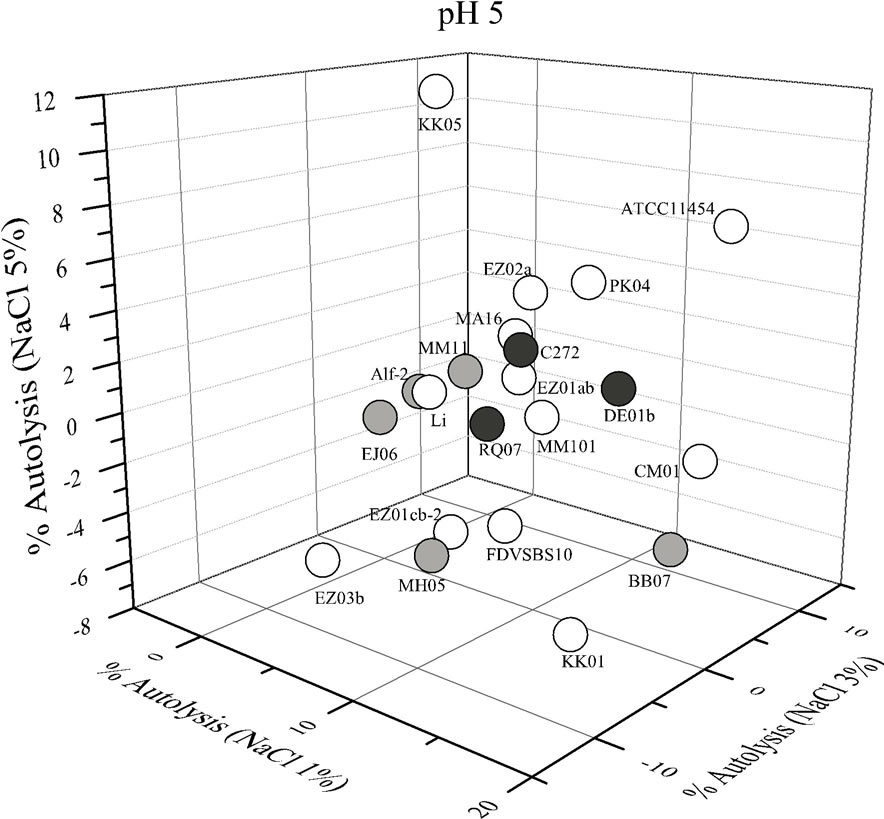
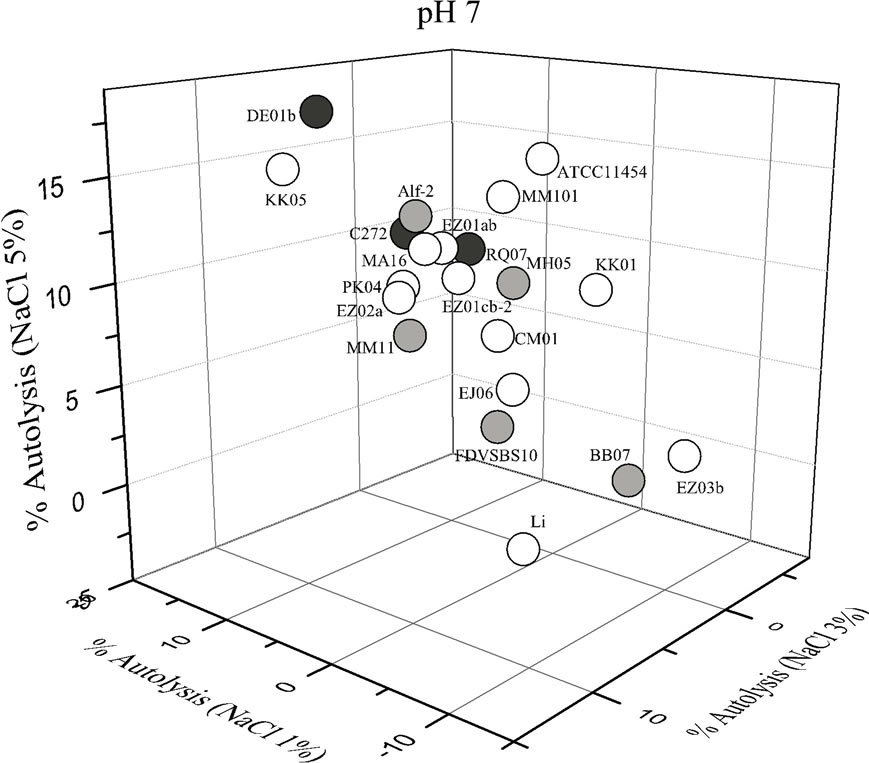
Figure 1. Autolysis of Lactococcus lactis strains in phosphate buffer pH 5 and pH 7 added with different concentrations of NaCl. White circles strains isolated from comercialstarter cultures; gray circles strains isolated from vegetables; black circles strains isolated from raw-milk cheeses.
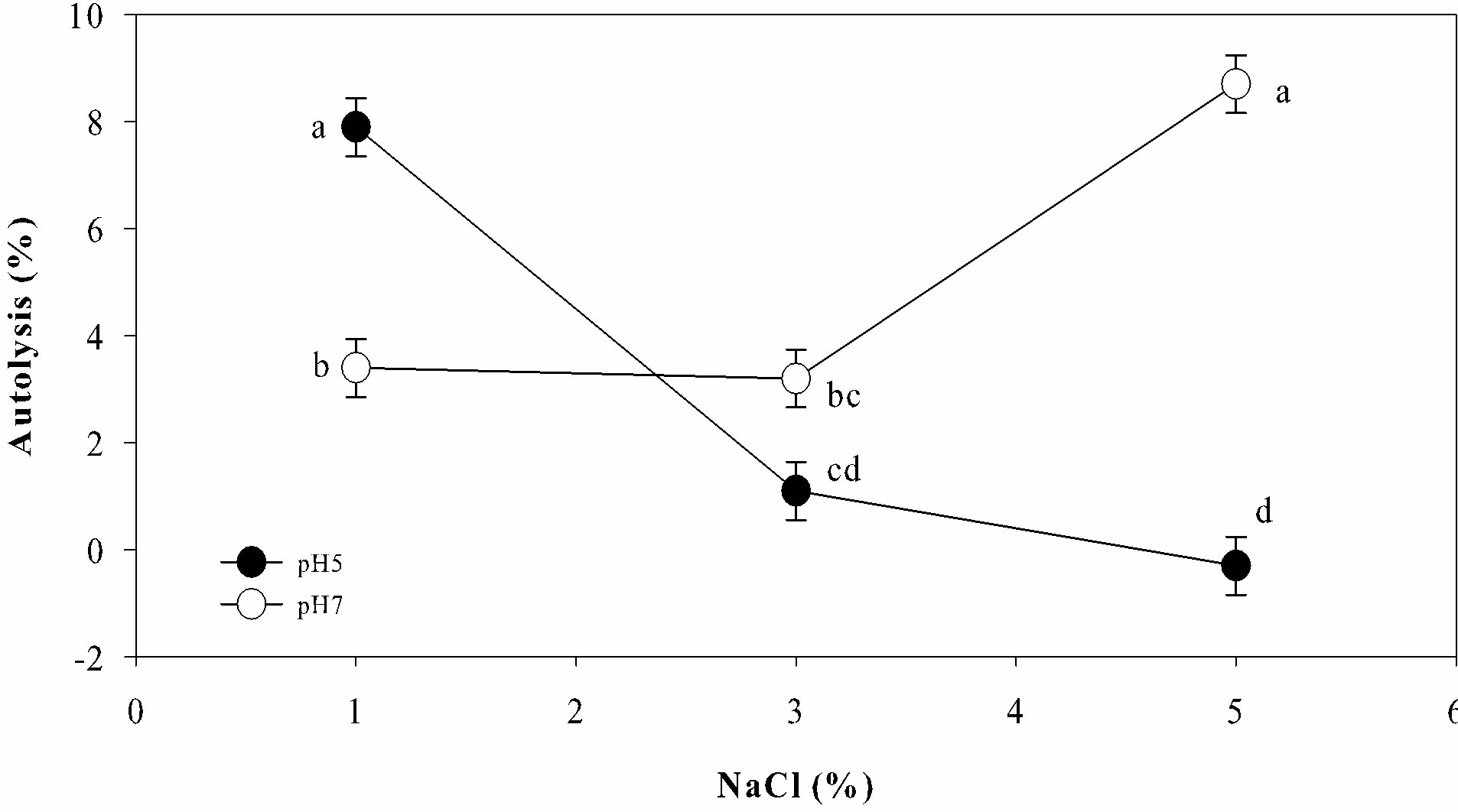
Figure 2. General behavior of autolysis observed in 21 strains of Lactococcus lactis under different conditions of NaCl concentration and pH. a-dTreatments with different letter were significantly different (multiple mean compareson, Tukey-Kramer test, α = 0.05).
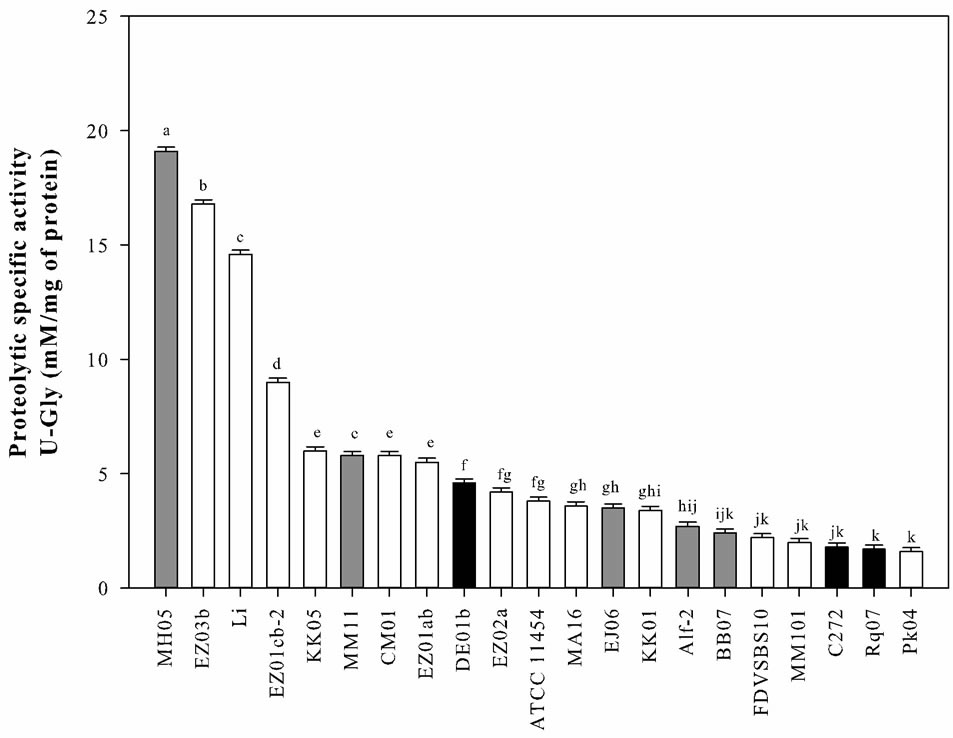
Figure 3. Proteolytic activity observed in casein solutions treated with cell crude extracts (CCE) of Lactococus lactis strains. White bars strains isolated from commercial starter cultures; gray bars strains isolated from vegetables; black bars strains isolated from raw-milk cheeses. a-kBars with different letter were significantly different (multiple mean comparison, Tukey-Kramer test, α = 0.05).
activity. The strains isolated from raw-milk cheeses showed low and medium proteolytic activities, whereas most of the strains isolated from industrial starter cultures had medium and high activities.
4. Discussion
From results obtained in this study, it may be affirmed that autolysis of L. lactis is strongly strain depended. Different authors have reported that autolysis rate of lactic acid bacteria varied markedly among strains belonging to the same species [3,4,16,20]. It has been suggested that variation of autolysis among strains is related with differences in cell wall composition and autolysins [2,7].
Additionally, in this work it was confirmed that pH and salt concentration had a significant effect on the level of autolysis in L. lactis. In a similar work, MartínezCuesta et al. [22] assessed the effect of different lysis solutions (10 m mM sodium phosphate, pH 6.0 and 6.5; 0.1 M sodium phosphate pH 6.0 and 6.5; 0.1 M citratesodium phosphate pH 5.5 and 6.0) in the level of autolysis of L. lactis, finding maximal response at pH 6.5. Piraino et al. [23] evaluated the autolysis of L. lactis strains using three different lysis solutions (88.5 mM Na-lactate, 0.7 M NaCl, pH 5.1; 0.2 M NaCl, pH 5.5; 50 mM Na-phosphate, pH 7) and found the highest values of autolysis at pH 7. According to our results, the maximum autolysis of L. lactis can be achieved in both acidic and neutral pH, depending of the salt concentration in the media. In this sense, Kang et al. [6] observed that the rate of autolysis of LAB was influenced by the concentration and type of salt used (NaCl, KCl, K2PO4, Na2PO4) in autolysis assays. These authors reported that autolysis of Lb. bulgaricus was favored at low concentration of NaCl (and pH 5.5); nevertheless an increase of salt concentration negatively affected the autolysis. The effect of pH and salt on the rate of L. lactis autolysis remains unclear but may be related with changes in the cell wall (expansion and contraction) as a response to environmental conditions like pH and ionic strength, as well as changes in the expression, activity and attachment to the cell wall of autolysins (AcmA).
The proteolytic activities of Lactococcus lactis strains were strongly strain dependent. This observation has been previously reported by other authors like Boutrou et al. [20]. The proteinases of diverse strains of L. lactis have shown a high degree of genetic, biochemical and immunological differences. Furthermore, the amount of proteinase produced per cell or culture may vary depending of the strain [24]. This may explain the large variations in proteolytic activity among the 28 strains of L. lactis.
On the other hand, it is believed that industrial strains of L. lactis have acquired features linked to the adaptation to milk, such as to degrade casein by a proteinase [15]. In this sense, the gene (prtP) that encodes the cell envelope-associated proteinase is required for the efficient growth of L. lactis in milk. The prtP is commonly encoded by plasmid DNA and this plasmid frequently encodes determinants for other essential industrial traits such as lactose fermentation [25]. For this reason it was expected that the strains isolated from vegetables showed a limited proteolytic activity. However, one of the most interesting observations was that the strain with highest proteolytic activity was a strain isolated from a vegetable source.
5. Conclusion
The results obtained confirmed that autolysis of L. lactis is strain dependent. The pH and salt content significantly modified the rate of autolysis. The maximal responses of autolysis in most of the strains were obtained in vitro at pH 5% - 1% NaCl and pH 7% - 5% NaCl. The strains isolated from raw-milk cheeses and vegetable showed a low and medium autolytic activity. In reference to the proteolytic activity the strain with the highest proteolytic activity was a strain isolated from corn leaves. However, further research is required to understand the highly proteolytic activity of this strain isolated from vegetables.
6. Acknowledgements
The Mexican National Council of Science and Technology (CONACYT) supported this research through the research project No. CB-2008-01/0106209.
REFERENCES
- E. H. E. Ayad, A. Verheul, J. T. M. Wouters and G. Smit, “Antimicrobial-Producing Wild Lactococci Isolated from Artisanal and Non-Dairy Origins,” International Dairy Journal, Vol. 12, No. 2-3, 2002, pp. 145-150. http://dx.doi.org/10.1016/S0958-6946(01)00133-9
- C. J. Pillidge, P. S. Rallabhandi, X.-Z. Tong, P. K. Gopal, P. C. Farley and P. A. Sullivan, “Autolysis of Lactococcus lactis,” International Dairy Journal, Vol. 12, No. 2-3, 2002, pp. 133-140. http://dx.doi.org/10.1016/S0958-6946(01)00135-2
- O. Kenny, R. J. FitzGerald, G. O Cuinn, T. P. Beresford and K. Jordan, “Autolysis of selected Lactobacillus helveticus Adjunct Strains during Cheddar Cheese Ripening,” International Dairy Journal, Vol. 16, No. 7, 2006, pp. 797-804. http://dx.doi.org/10.1016/j.idairyj.2005.07.008
- C. L. Ma, L. W. Zhang, H. X. Yi, M. Du, X. Han, L. L. Zhang, Z. Feng, Y. C. Zhang and Q. Li, “Technological Characterization of Lactococci Isolated from Traditional Chinese Fermented Milks,” Journal of Dairy Science, Vol. 94, No. 4, 2011, pp. 1691-1696. http://dx.doi.org/10.3168/jds.2010-3738
- V. L. Crow, T. Coolbear, P. K. Gopal, F. G. Martley, L. L. McKay and H. Riepe, “The Role of Autolysis of Lactic Acid Bacteria in the Ripening of Cheese,” International Dairy Journal, Vol. 5, No. 8, 1995, pp. 855-875. http://dx.doi.org/10.1016/0958-6946(95)00036-4
- O. J. Kang, L. P. Vézinz, S. Laberge and R. E. Simard, “Some Factors Influencing the Autolysis of Lactobacillus bulgaricus and Lactobacillus casei,” Journal of Dairy Science, Vol. 81, No. 3, 1998, pp. 639-646. http://dx.doi.org/10.3168/jds.S0022-0302(98)75618-8
- S. Lortal and M.-P. Chapot-Chartier, “Role, Mechanisms and Control of Lactic Acid Bacteria Lysis in Cheese,” International Dairy Journal, Vol. 15, No. 6-9, 2005, pp. 857-871. http://dx.doi.org/10.1016/j.idairyj.2004.08.024
- K. Savijoki, H. Ingmer and P. Varmanen, “Proteolytic Systems of Lactic Acid Bacteria,” Applied Microbiology and Biotechnology, Vol. 71, No. 4, 2006, pp. 394-406. http://dx.doi.org/10.1007/s00253-006-0427-1
- J. Ramírez-Nuñez, R. Romero-Medrano, G. V. NevárezMoorillon and N. Gutiérrez-Méndez, “Effect of pH and Salt Gradient on the Autolysis of Lactococcus lactis Strains,” Brazilian Journal of Microbiology, Vol. 42, No. 4, 2011, pp. 1495-1499. http://dx.doi.org/10.1590/S1517-83822011000400036
- M. V. Santos, Y. Ma, Z. Caplan and D. M. Barbano, “Sensory Threshold of Off-Flavors Caused by Proteolysis and Lipolysis in Milk,” Journal of Dairy Science, Vol. 86, No. 5, pp. 2003, 1601-1607.
- M. Yvon and L. Rijnen, “Cheese Flavour Formation by Amino Acid Catabolism,” International Dairy Journal, Vol. 11, No. 4-7, 2001, pp. 185-201. http://dx.doi.org/10.1016/S0958-6946(01)00049-8
- N. Gutiérrez-Méndez, B. Vallejo-Cordoba, A. F. Gonzá- lez-Córdova, G. V. Nevárez-Moorillon and B. RiveraChavira, “Evaluation of Aroma Generation of Lactococcus lactis with an Electronic Nose and Sensory Analysis,” Journal of Dairy Science, Vol. 91, No. 1, 2008, pp. 49-57. http://dx.doi.org/10.3168/jds.2007-0193
- N. Gutiérrez-Méndez, J. C. Rodríguez, A. F. González, G. V. Nevárez, B. Rivera-Chavira and B. Vallejo-Cordoba, “Phenotypic and Genotypic Characteristics of Lactococcus lactis Strains Isolated from Different Ecosystems,” Canadian Journal of Microbiology, Vol. 56, No. 5, 2010, pp. 432-439. http://dx.doi.org/10.1139/W10-026
- N. Gutiérrez-Méndez, E. Valenzuela Soto, A. F. González-Córdova and B. Vallejo-Cordoba, “α-Ketoglutarate Biosynthesis in Wild and Industrial Strains of Lactococcus lactis,” Letters in Applied Microbiology, Vol. 47, No. 3, 2008, pp. 202-207. http://dx.doi.org/10.1111/j.1472-765X.2008.02405.x
- E. H. E. Ayad, A. Verheul, C. de Jong, J. T. M. Wouters and G. Smit, “Flavour Forming Abilities and Amino Acid Requirements of Lactococcus lactis Strains Isolated from Artisanal and Non-Dairy Origin,” International Dairy Journal, Vol. 9, 1999, pp. 725-735.
- E. Franciosi, L. Settanni, A. Cavazza and E. Poznanski, “Biodiversity and Technological Potential of Wild Lactic Acid Bacteria,” International Dairy Journal, Vol. 19, No. 1, 2009, pp. 3-11. http://dx.doi.org/10.1016/j.idairyj.2008.07.008
- H. Kiani, Z. Zhang, A. Delgado and D.-W. Sun, “Ultrasound Assisted Nucleation of Some Liquid and Solid Model Foods during Freezing,” Food Research International, Vol. 44, No. 9, 2011, pp. 2915-2921. http://dx.doi.org/10.1016/j.foodres.2011.06.051
- H. S. Choonia and S. S. Lele, “Beta-Galactosidase Release Kinetics during Ultrasonic Disruption of Lactobacillus acidophilus Isolated from Fermented Eleusine Coracana,” Food and Bioproducts Processing, Vol. 89, No. 4, 2011, pp. 288-293. http://dx.doi.org/10.1016/j.fbp.2010.08.009
- M. M. Bradford, “A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding,” Analytical Biochemistry, Vol. 72, No. 1-2, 1976, pp. 248-254. http://dx.doi.org/10.1016/0003-2697(76)90527-3
- R. Boutrou, A. Sepulchre, J. C. Gripon and V. Monnet, “Simple Test for Predicting the Lytic Behavior and Proteolytic Activity of Lactococcal Strains in Cheese,” Journal of Dairy Science, Vol. 81, No. 9, 1998, pp. 2321-2328. http://dx.doi.org/10.3168/jds.S0022-0302(98)70121-3
- Z. Ustunol and T. Zecker, “Relative Proteolytic Action of Milk-Clotting Enzyme Preparations on Bovine Alpha, Beta and Kappa-Casein,” Journal of Food Science, Vol. 61, No. 6, 1996, pp. 1136-1138. http://dx.doi.org/10.1111/j.1365-2621.1996.tb10947.x
- M. C. Martínez-Cuesta, C. Peláez, M. Juárez and T. Requena, “Autolysis of Lactococcus lactis ssp. and Lactobacillus casei ssp. casei. Cell Lysis Induced by a Crude Bacteriocin,” International Journal of Food Microbiology, Vol. 38, No. 2-3, 1997, pp. 125-131. http://dx.doi.org/10.1016/S0168-1605(97)00099-8
- P. Piraino, T. Zotta, A. Ricciardi, P. L. McSweeney and E. Parente, “Acid Production, Proteolysis, Autolytic and Inhibitory Properties of Lactic Acid Bacteria Isolated from Pasta Filata Cheeses: A Multivariate Screening Study,” International Dairy Journal, Vol. 18, No. 1, 2008, pp. 81- 92. http://dx.doi.org/10.1016/j.idairyj.2007.06.002
- P. G. Bruinenberg, P. Vos and W. M. de Vos, “Proteinase Overproduction in Lactococcus lactis Strains: Regulation and Effect on Growth and Acidification in Milk,” Applied and Environmental Microbiology, Vol. 58, No. 1, 1992, pp. 78-84.
- J. R. Broadbent, B. T. Rodríguez, P. Joseph, E. A. Smith and J. L. Steel, “Conversion of Lactococcus lactis Cell Envelope Proteinase Specificity by Partial Allele Exchange,” Journal of Applied Microbiology, Vol. 100, No. 6, 2006, pp. 1307-1317. http://dx.doi.org/10.1111/j.1365-2672.2006.02860.x

