American Journal of Plant Sciences
Vol.09 No.11(2018), Article ID:87869,22 pages
10.4236/ajps.2018.911158
A Phenotyping Protocol for Detailed Evaluation of Apple Root Resistance Responses Utilizing Tissue Culture Micropropagated Apple Plants
Yanmin Zhu1*, Melody Saltzgiver1, Jingxian Zhao2
1United States Department of Agriculture, Agricultural Research Service, Tree Fruit Research Laboratory, Wenatchee, WA, USA
2Hebei Academy of Forestry, Shijiazhuang, China
Copyright © 2018 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY 4.0).
http://creativecommons.org/licenses/by/4.0/



Received: September 21, 2018; Accepted: October 16, 2018; Published: October 19, 2018
ABSTRACT
In the post-genomics era, reliable phenotypes are considered the bottleneck for unraveling the genetic control over the biology of interest. Phenotyping resistance response of roots to infection by soilborne pathogen is more challenging compared to that of plant aerial parts. In additional to the hidden nature and small stature of fine roots where infection occurs, extra obstacles exist for rosaceae tree crops such as apple. Due to self-incompatible reproduction and high-level heterozygosity of apple genome, genetically identical apple plants cannot be produced through apple seed germination. Here we report an established phenotyping protocol which includes a streamlined tissue culture procedure for micropropagation of uniform apple plants, standardized inoculation procedure using Pythium ultimum, and multilayered evaluating methods on apple root resistance traits. Because of the implementation of tissue culture based micropropagation procedure, constant availability of the uniform plants with defined genetic background, equivalent age and non-contaminated roots overcame a longstanding barrier of systematic and detailed phenotypic characterization of apple root resistance traits. Repeated infection assays by root-dipping inoculation demonstrated the reproducible and wide-range plant survival rates, from single-digit to over 90% survived plants for a given genotype. Genotype-specific values due to P. ultimum inoculation on shoot and root biomass reduction, maximum root lengths, leaf number and cumulative leaf areas were quantified between mock-inoculated and P. ultimum infected plants. Use of a glass-box container offered enhanced accessibility and minimized invasiveness for continuous and non-disruptive observation on the necrosis progression patterns along inoculated roots. With the assistance of a dissecting microscope, the genotype-specific resistance responses along the infected apple roots were captured and analyzed in detail. This reported phenotyping protocol represents a major development and should be easily adopted for other rosacea tree fruit crops with minor modifications.
Keywords:
Tissue Culture, Micropropagation, Apple Rootstocks, Root Resistance Responses, Phenotyping Methods

1. Introduction
In the post-genomics era, the ability to acquire reliable phenotypes is a well-acknowledged limiting factor as comparison to the relatively easy access to genomic information for most crops [1] . Phenotyping resistance trait in plant roots in response to infection by soilborne pathogen is more difficult than that of aerial parts of a plant. Because of the hidden nature and the small stature of individual young roots where infection often occurs, it is difficult to monitor and assess the pathogenesis process and genotype-specific root resistance responses to infection by soilborne pathogens. For some perennial crops such as apple, additional obstacles exist. Apple reproduction is self-incompatible, and the apple genome has high-level heterozygosity [2] [3] . As a result, germination of apple seeds will not produce genetically identical apple plants. The lack of consistent availability of uniform plants has been along-standing barrier for in-depth and systematic analysis of genotype-specific apple root resistance responses.
Many horticultural traits of apple rootstocks, including disease tolerance, were traditionally evaluated under field conditions using bare root rootstock “sticks” or in the form of commercial trees consisting of specific scions grafted to selected rootstocks [4] . The availability of these commercial nursery-produced one-year old bare root plants is generally restricted to a few elite commercial varieties, and these plants are only available for a limited time during the year. In addition, these rootstock plants have often been exposed to various soil microbes or impacted by inadvertent abiotic conditions during nursery production. While it is a viable approach to assess the overall performance of rootstock selections to disease pressure under field conditions, it is problematic to use these trees as primary plant materials for careful phenotyping root resistance responses aimed for subsequent genetic analysis. The reliable expression of genotype-specific resistance traits in apple root towards infection by soilborne pathogens depends on repeated infection assays with minimized confounding factors. The constant supply of uniform apple plants with defined genetic background, equivalent age, and non-contaminated roots is one of the critical requirements for obtaining the reliable and detailed apple root resistance phenotypes.
Plant tissue culture technique, based on the concept of to tipotency, has existed for more than a century [5] . This practice represents an unparalleled way to propagate and keep plants that are free of disease [6] [7] [8] [9] . Tissue culture technique based micropropagations was originally used in the production of ornamental plants but now is extended to over 1000 species including various vegetable and fruit crops such as apple [9] [10] . Implementing the tissue culture based micropropagation of apple plants can pave the way for consistent supply of genetically uniform plants for any desired genotypes [11] . Specific to the pathosystem of apple root interacting with soilborne pathogens, using tissue culture based in vitro platform to generate uniform and clean plants is a critical requirement for careful evaluation on root resistance traits [7] . Utilizing these uniform apple plants, coupled with a standardized infection protocol, should efficiently remove bias from inconsistency in plant age, unintended microbial influences, most abiotic stresses, and even physiological status due to nutritional discrepancies [12] [13] [14] .
High quality phenotypic data is a prerequisite for establishing the association between genotype and phenotype [15] [16] , which in turn is a foundation for developing and deploying molecular tools for targeted and efficient crop improvement [17] . An elevated level of consistency in the plant materials confers the power to reveal minute or quantitative differences between treatments, infection events or genotypes [18] [19] . Reported here is a protocol which includestissue culture based micropropagation for generating uniform plants for various apple rootstock genotypes, standardized and quantifiable inoculum preparation, and various evaluation methods on apple root resistance responses. Oomycete pathogen Pythium ultimum, which incites apple replant disease [20] , was used as the model soilborne pathogen in this study. Many apple rootstock genotypes including the progenies froma cross between “Ottawa 3” × “Robusta 5” (O3R5), two elite apple rootstock parents, were tested by this phenotyping protocol. This established protocol represents the first systematic and comprehensive method to evaluate the resistance responses in apple root to infection by soilborne pathogens. With minor modifications, it should be readily adapted toother pathosystems related to perennial tree crops.
2. Materials and Methods
2.1. Chemicals, Reagent and Plant Genotypes
1) Items used: Laminar flow hood Class II type A/B3 (Nuaire, Plymouth MN), PhytoTechnology Laboratories® PTL-100 Magenta™ boxes, Sunshine® All-Purpose Potting Mix (SUN GRO Horticulture Ltd., Bellevue, WA), forceps, Feather® disposable scalpels (Feather Saftey Razor Company, Osaka, Japan), Wheat Germ Oil (Sigma-Aldrich, St. Louis, MO), PhytoTechnology Laboratories® Murashige & Skoog Basal Salt Mixture, 1 MSodium Hydroxide (NaOH) Sigma-Aldrich, St. Louis, MO), 6 M Hydrochloric Acid (HCl), Clorox Bleach, Tween 20 (Sigma-Aldrich, St. Louis, MO), PhytoTechnology Laboratories® Kao & Michayluk Vitamin Solution (100 x), PhytoTechnology Laboratories® myo-Inositol, bioWORLD® Agar (Phytoagar), 95% (EtOH) Ethanol (Decon Labs Inc, King of Prussia, PA), Petri dishes, 3 M® micropore tape, Plant cell culture grade: 6-Benzylaminopurine, Indole-3-Butyric Acid, Gibberellic Acid, Thiamine Hydrochloride (Sigma-Aldrich, St. Louis, MO), and Sucrose (Fisher Scintific, Waltham, MA).
2) Media composition and preparation of Magenta box: Proliferation, rooting, and elongation media were used in the progression of the procedure. The components of each medium are listed in Table 1. All the components were added to the media, and the pH was established prior to autoclaving. The pH of each media was adjusted with 1 M NaOH and 6 M HCl, respectively. The media was autoclaved for 15 minutes at 121˚C, poured into magenta boxes, and autoclaved for an additional 15 minutes to eliminate any contamination. The boxes were allowed to completely solidify before use and any excess condensation was poured off.
The media was autoclaved for 15 min at 121˚C, poured into containers, and then autoclaved for an additional 15 min. Prior to autoclaving the pH of each media was adjusted with 1 M NaOH and 6 M HCl respectively. The hormones and myo-inositol were made as the stock solutions listed and stored at 4˚C until use.
3) Source of shoots for micropropagation: Many apple rootstock genotypes, including several elite commercial cultivars such as “Bud 9” (B9) and “Malling 26” (M26), as well as more than 90 progenies from a rootstock cross population between “Ottawa 3” and “Robusta 5” (O3R5) were available for testing this inoculation protocol. Both parents, “Ottawa 3” and “Robusta 5”, have a strong genetic background of wild apples. Several agronomical traits such as dwarfness, cold hardness, and disease resistance are known to be segregating among O3R5 progenies [21] .
Table 1. The components of the three medias utilized in this paper.
2.2. A Step-Wise Procedure for Apple Plant Micropropagation
The apple micropropagation procedure was established based on previous publications [22] [23] [24] and practical instructions from Dr. Norelli (USDA, ARS, personal communication), with updates on available reagents as well as an adapted step specifically designed for preparing root before infection assay. Explant sterilization: The apical ends of branches from desired genotypes were gathered from vigorously growing shoots (1.5 - 4.0 mm diameter) from rootstock plants grown in the field or greenhouse. The shoots were cut into 3 - 5 cm lengths and, if possible, each included 4 nodes. These shoot segments were placed in wet paper towels to keep them from drying out. All the leaves were removed prior to sterilization. Explant sterilization was carried outaseptically in a laminar flow hood using a sterilization solution consisting of Clorox Bleach (F.C. 10%), Tween 20 (F.C. 0.01%) and ddH2O. The shoots were placed in a 50 mL falcon tube (Corning Brand, Corning, NY) filled with sterilization solution. The shoots were completely submerged in the solution and soaked for 15 - 20 minutes. After pouring off the sterilization solution, the shoots were washed three times using sterile H2O. The shoots soaked in the sterile H2O for 10 - 15 min between each wash. The shoots were taken from falcon tubes and placed onto a sterile petri dish. Using a sterile scalpel blade, a small section of shoot was removed from either end (~5 mm) to expose tissue uninjured by sterilization. Each shoot was trimmed to 1 to 2 nodes and placed vertically (~5 mm deep) in a magenta box containing proliferation media (Figure 1). All the proliferation boxes were formed by pouring 75 mL of proliferation media per magenta box resulting in roughly a 2-inch-thick media. Each box was then sealed with micropore tape to prevent moisture loss and introduction of foreign matter. The boxes were then kept at 25˚C ± 1˚C, 7000 lux for 12 hr/12 hr light/dark periods. The micropropagation shoots stayed in the proliferation media for 6 weeks allowing them to establish axillary buds that developed into a multiple shoot structures.
Initial establishment of sterile explants can have varying degrees of success depending on the genotype, time of year, and physiological condition of the plant [25] . Shoots are often gathered in the spring after bud break or during summer [23] [25] . There is a fine line to the sterilization of the initial explants: longer soaking time in sterilization solution does lower the instances of contamination, but it leads to more tissue damage at the ends, and depending on the severity, may result in death of the shoot. It is important to add only a few explants to each Magenta box to prevent overcrowding. Beyond overcrowding, an even more important reason is to avoid contamination spreading to all the explants that share that box.
Shoot proliferation: The multiple shoot structures produced from the explants were sub-cultured for auxiliary proliferations aseptically in a laminar flow hood. The new shoots chosen for proliferation were healthy without any evidence of hyperhydricity (glass-like appearance) or fasciation (fused stems). The shoots were cut at the base arisen from the original explant then each shoot was separated. The shoots were trimmed of all leaves including the leaves around the crown which were carefully removed as to leave the meristem tissue intact (Figure 2). This resulted in a bare stem much like the original explant. A fresh box of proliferation media was used. The stems were placed into the media at a shallow horizontal angle with the meristem completely clear of the media. The base of the shoot was required to be in the media for nutrient absorbance. Two to three stems were added to each fresh box with an alternate directional arrangement of the meristems (Figure 3A). The shoot could also be inserted at a completely vertical angle with the base into the media. Each box was then wrapped with 3 M micropore tape. The boxes were then kept at 25˚C ± 1˚C, 7000 lux for 12 h/12 h light/dark for 6 to 8 weeks before being used for rooting (Figures 3B-3D).
Figure 1. Explant sterilization and shoot establishment. (A) Sterilized explants showing new growth two weeks after micropropagation. (B) A close-up of a new shoot emerging from a node.
Figure 2. Shoot proliferation. (A) A multiple shoot structure produced from proliferation of explant. (B) Shoots were cut at the base. (C) A single shoot separated from the multiple shoot structure being prepared for proliferation. (D) The shoot trimmed off all leaves, including the leaves around the crown, to leave the meristem tissue intact. This stem was place in a new proliferation box.
Figure 3. Proliferation subcultures time-line. (A) 1-day post proliferation; (B) 2-weeks post proliferation; (C) 1-month post proliferation; (D) 2-months post proliferation; the shoots are ready to be reproliferated or used for rooting to generate individual plants.
Most of the issues encountered during proliferation were linked to the treatment of the meristem. There were observed deleterious side effects to sublimating the meristem portion of the shoot into media, including growth inhibition, hyperhydricity, and death of the shoot. It was also important that the plant material was not allowed set in one proliferation box longer than 2 months due to reduced vigor. The number of shoots and vigor of plantlet growth varied between genotypes. This issue could have mediated to an extent if the cytokinin (6-benzylaminopurine) and auxin (indole-3-butyric acid) were specifically optimized to each genotype, given the time is available. Using this reported media and procedures ample amounts of healthy plant material were produced for almost all the genotypes tested.
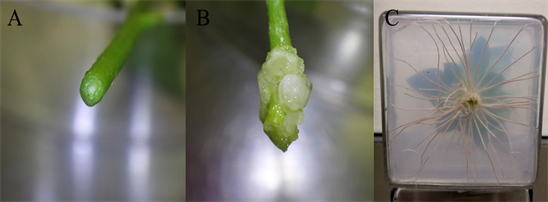
Root induction and elongation: The shoots selected for rooting were treated the same way as proliferations with exception for the removal of the leaves, which were only removed half-way up the stem to create a miniature tree like appearance (Figure 4A and Figure 4B). Five trimmed shoots or trees were placed to each magenta box containing rooting media (Figure 4C). Root at the bottom end was prepared by a fresh cut (Figure 5A). Only enough of the stem (~5 - 8 mm) was inserted into the media to hold the tree upright. The rooting boxes were formed by pouring 25 mL of rooting media per magenta box, resulting in roughly a 1/2-inch-thick media box. The rooting boxes were sealed with 3 M micropore tape and placed in 24 hr darkness at 22˚C for one week. After a week in the rooting boxes, the shoots would form callus (Figure 5B).
After a week in darkness, each shoot was placed in a magenta box, one per box, containing elongation media. The elongation boxes were formed by pouring 50 mL of rooting media per magenta box resulting in roughly a one-inch-thick media box. The shoot was placed directly in the middle of the magenta box. The entire callus formation was then inserted into the media. The boxes were sealed with micropore tape and placed at 25˚C, 7000 lux for 12 hr/12 hr light/dark period for one month. Roots became well-formed (Figure 5C) for most tested genotypes and sufficiently functional under soil medium condition for the in-soil acclimation of the root system for the tissue culture generated plantlets.
For root induction, it was important to only insert a small portion of the prepared shoot into the rooting media because tissue that encountered the media grew a callus. If the shoot was inserted too far into the medium it resulted in the loss of the “trunk” of the plantlet. Low hanging leaves that rested on the rooting media would at times form a callus as well. Interestingly, if a plantlet began growing roots from a leaf callus it would develop those roots rather than developing roots from the call us at the base of the stem. For root elongation, the shoots taken from rooting boxes were placed on the exact middle of the elongation boxes so that growth was not restrained by the Magenta box within four weeks. The time for root elongation was generally not longer than a month for multiple reasons. The first reason is the obvious depletions of nutrients in the magenta box. The second was that prolonged process of root elongation could make it difficult to remove rooted plants from the tissue culture media.
Figure 4. Root induction. (A) A single shoot separated from the multiple shoot structure being prepared for rooting; (B) A single shoot with the leaves removed half-way up the stem to create a miniature tree; (C) Five trees were added to each magenta box containing rooting media, with only a small portion (5 - 8 mm in depth) of the stem inserted into the media.
Figure 5. Root induction and elongation for individual proliferated shoot. (A) The end of a shoot prepared for rooting. (B) The callus formed at the end of a shoot placed in 24 h darkness at 22˚C for one week in rooting media. (C) The roots formed from the callus after 1-month in elongation media at 25˚C, for 12 h/12 h light/dark.
In-soil acclimation of root system: At the end of the four-week period of root elongation and before pathogen infection assay, the root system generated in tissue culture medium was set for a one-week in-soil acclimation for its further differentiation. Several soil media have been used for various experimental objectives including natural orchard soil (natural sandy loam soils from Columbia view experimental orchard), commercial Sunshine® All-Purpose Potting Mix (SUN GRO Horticulture Ltd., Bellevue, WA), construction sands or the mixtures of perlite/vermiculite/sands (with the ratio of 1:1:1 by volume). Soil substrates were autoclaved for 90 minutes before filling the pots or trays. The micropropagated plants (or rooted plantlets) at the end of root elongation were gently removed from the agar using forceps, and then floated in a container with room temperature tap water. Any excessive tissue culture media attached to the roots was carefully removed without damaging root system. To minimize the transplanting effects (from MS medium to soil condition), humidity retainment was applied by placing a transparent 7 Vented Plastic Dome (Greenhouse Megastore, Danville, IL) on top of a 25 × 50 cm tray (Greenhouse Megastore, Danville, IL) holding the pots. In-soil acclimation provides a process for root tissue full differentiation and functional adaption from growing in the artificial semi-solid MS medium to the solid medium. Through observation this step is believed to be a critical requirement for reliable expression of root resistance phenotypes.
2.3. Pathogen Inoculum Preparation, Inoculation and Plant Maintenance
The oomycete pathogen Pythium ultimum, which was used during establishing this protocol, was originally isolated from the roots of “Gala”/M26 apple grown at Moxee, WA, USA [20] . Inoculum solution of P. ultimum was prepared by cultivating in potato carrot broth (20 g of carrots and 20 g of potatoes) with two drops of wheat germ oil added per liter of medium. The P. ultimum cultures were grown in the broth in Petri dishes at 22˚C for 5 - 6 weeks (Figure 6A) until the presence of dark-color oospores (Figure 6B). The resultant hyphae mat consisting of oospores and mycelium was ground in anelectric blender (commercial name: Magic Bullet) for 30 s. The oospores and hyphal fragments were resuspended in 0.5% methyl cellulose solution, and a final oospore concentration was adjusted to approximately 2000 oospores per mL based on counting under microscope (Figure 6C).
Inoculation of apple plant roots was performed by dipping the root system into P. ultimum inoculum solution for 5 s, excessive inoculum solution was then allowed to drip off the roots. Inoculated plants were then immediately planted into the pot filled with autoclaved soil substrates. Control plants were mock-inoculated by dipped the roots into 0.5% methyl cellulose solution and planted into the same soils. All plants were maintained under the identical water and light regimes in growth chambers. A transparent plastic cover with adjustable windows was placed over the tray holding pots for at least 48 h, to minimize
Figure 6. Preparation of Pythium ultimum inoculum. (A) Petri dishes with P. ultimum cultures. (B) Microscopic image of hyphae mat with oospores. The insert small image is the 8x enlarged view of oospores. (C) Dissection microscope used for adjusting oospore concentration, as well as examining and documenting root resistance responses.
the negative effects of transplanting. Gradually decreased humidity was achieved by partially opening adjustable windows on the high-dome plastic cover after 48 h, which allowed plants to adapt to the ambient humidity level in the growth chamber.
2.4. Evaluation of Plant Overall Survival Rates, Biomass Reduction, Root Length and Leaf Area
Depending on the objectives for the specific experiments, apple root resistance traits were evaluated at different time points or with various intervals after P. ultimum inoculation. For preliminary screening of whole plant growth responses, partiallywilt symptomsor plant mortality were visually evaluated daily and recoded at 3, 7, 10, 14, 21 and 28 dpi. Although the overall survival rate was assigned based on the data at 28 dpi, the rapidity of the exhibition of wilting symptoms also provided the valuable information for genotype-specific resistance responses. For biomass reduction assay, mock-inoculated and P. ultimum inoculated and survived plants were excavated at 28 dpi from pots. Roots were gently rinsed under tap water. Care was taken to untangle roots between different plants in the same pot. Individual plants were wrapped into wet paper towels before shoot and root tissues were separated and weighted using an electric balance. After collecting biomass data, maximum root lengths for each plant can also be measured. The shoot lengths were recorded by removing individual leaf, and all leaves from the same plant were assembled and photographed for subsequent image analyzing process. The total root lengths were recorded by dissembling the whole root system and arranged the root branches in the petri dish for image acquisition individually for individual plants. The images were processed by Image J (https://imagej.nih.gov/ij/) for quantifying the root lengths and leaf areas.
2.5. Continuous and Non-Disruptive Observation of Root Resistance Responses and Necrosis Progression
In-house madelarge glass-box containers (30 × 45 × 10 cm) were designed for continuing observation of genotype-specific root growth in response to P. ultimum infection. Evaluation of plant growth rate was carried out by measuring root lengths every other day or on modified schedules as needed. A smaller glass box container (2.5 × 7.5 × 10 cm) was designed for monitoring the symptom development along the infected roots with the assistance of a dissection microscope. Apple roots were aligned against one side of the glass box to maximize the exposure of apple root system for direct observation. A layer of paper towel was used to separate soil from roots. For continuing and non-destructive observation of genotype-specific necrotic progression patterns, the specific sections of root system were continuously observed at the same interval time (6 or 12 h) for comparability between genotypes or infection assays. The glass plate was marked with grid-lines to facilitate the localization of the same segments of the root system. The obtained series of images were analyzed to determine the genotype-specific necrosis development features. Except the time for observation, aluminum foil was used to wrap around the glass box to minimize the unintended effect from excessive light exposure of the root system [26] .
2.6. Microscopic Observation and Documentation of Resistant Responses at Tissue Level
Detailed microscopic observations including various view backgrounds (lighter or darker) were designed to uncover the features of root resistance responses to P. ultimum infection. The root system was carefully excavated from the pot at designated time points. Soils associated with root tissues were gently removed by rinsing under tap water. Root branches were kept in a beaker with autoclaved water until examined under a microscope within two hours. Root branches and shoots were separated to facilitate the easy handle of individual root segments. At the time of observation, a glass slide was used to put on top of the targeted root segments for immobilization in a petri dish with autoclaved water. An Olympus SXZ12 dissecting microscope was used to examine the genotype-specific features of root necrotic symptom from P. ultimum inoculation. Images were obtained by an amounted DP73 digital camera with accompanied software suite of Celsense (Olympus, Center Valley, PA).
White view background with top illumination was designed for observing tissue color changes associated with root necrosis progression. Reflecting light from bottom with dark viewing background allowed for observing feature associated with pathogen hyphae growth along the infected roots. A publicly available software FastStone Image Viewer 5.5 (http://www.faststone.org/), was used for slight image processing including cropping, resizing the images, or adjusting brightness and contrast to enhance the image quality as needed.
2.7. Statistical Analysis
All reported values represent the average for at least three biological replications. Data were analyzed using paired or unpaired Student’s t test in Excel. Difference of means with P < 0.05 was considered as significant.
3. Results and Discussion
3.1. A Streamlined Micropropagation Procedure Provided Uniform Apple Plants with Clean Roots
Because of destructive infection assays, the data reliability and comparability on resistance assessment require repeated experiments using consistent plant materials between infection events. The continuing supply of the uniform young apple plants of desired genotypes, through the implementation of the apple micropropagation procedure, overcame a major barrier in phenotyping apple root resistance traits. The applicability of this micropropagation procedure has been tested among over 100 different apple rootstock genotypes (Table 2). These genotypes included several elite commercial apple rootstock varieties such as B9, M26, M9 (Malling 9), G41 (Geneva 41) and many progenies from “Ottawa 3” × “Robusta 5” (O3R5) apple rootstock cross population. With rare exceptions, plants with functional and uniform root systems have been routinely generatedusing this micropropagation procedure (Figure 7A and Figure 7B). Notably, the step of one-week “in-soil acclimation” facilitated the necessary tissue differentiation and the functional adaptation tissue culture medium to the environment of soil substrate (Figure 7C and Figure 7D). Use of high doom cover for the humidity management has facilitated the acclimation process and reducing the transplanting effect. Preliminary test from this micropropagation protocol also suggested its applicability on different commercial apple scion varieties and even pear genotypes (Table 2).
Table 2. Various germplasm being tested using this micropropagation procedure.
Figure 7. Uniform apple plants within or between genotypes from the synchronized micropropagation processes. (A) and (B) Comparable root systems during root elongation process for two different O3R5 genotypes at 14 days of root elongation. (C) and (D) Similar plant sizes during in-soil acclimation for two O3R5 rootstock genotypes.
Apple reproduction is self-incompatible, and its genome has high-level heterozygosity and numerous chromosome-level duplications [2] [27] . Therefore, genetically identical apple plants cannot be produced by seed germination, as each seed represents a unique genetic identity. This biological feature creates an undeniable burden in studying the traits of apple roots, particularly for pathogen resistance, which requires the repeated assays as infection can be destructive for some genotypes. Continuing supply of genetically uniform apple plants with functional and non-contaminated root systems overcame this long-standing barrier. This micropropagation procedure avoided several limitations associated with stool-bed produced one-year old bare root trees, or “rootstock sticks” [28] . Restricted availability on genotypes and time window, roots with unintended exposure to biotic and abiotic stresses, and uneven root system between individual trees are a few known factors which can severely impact the expression of genotype-specific root resistance phenotype. Consistent availability of these uniform apple plants with defined genetic background and non-contaminated root system should facilitate the identification of quantitative variations at the resistance responses. The primary plant materials were the progenies from “Ottawa 3” × “Robusta 5” (O3R5) cross, which represents an elite set of apple rootstock germplasm. These genotypes were known to contain high-level wild apple genetic compositions and segregate several agronomically important traits [21] .
3.2. Contrasting Resistance Responses among O3R5 Progenies from Repeated Infection Assays
Distinctive resistance responses were identified based on standardized inoculation procedure and the use of age-equivalent plants among apple rootstock genotypes. As shown in Figure 8A, three tested O3R5 genotypes displayed the contrasting plant survival rates in response to inoculation of P. ultimum. A resistance genotype O3R5-58 at the top row showed 100% survival rate without noticeable growth inhibition at 10 dpi (days post inoculation). In a sharp contrast, two susceptible genotypes, O3R5-106 and O3R5-47, in the right sections of the middle and bottomrows, showed less than 30% survival. All six mock-inoculated plants as experimental control for the corresponding genotypes remained healthy under the identical growth conditions (Figure 8A, left column). In addition to the distinguishable plant survival rates, the more susceptible genotypes demonstrate the stunted growth for those survived plants as indicated by the reduced size of shoot and root system (Figure 8B), compared to the those of mock-inoculation control plants. The darker yellow to brownish colorationofnecrotic roots for P. ultimum inoculated roots was easily discernable from those of mock-inoculated roots (Figure 8C). Among the tested O3R5 genotypes, a wide-spectrum of plant survival rate was observed (Figure 8D), suggesting that the appropriate inoculum level was used to distinguish the resistance levels among these genotypes.
A synchronized micropropagation process for different rootstock genotypes and the simultaneous inoculation using the identical inoculum preparation effectively minimized the potential bias associated with entire phenotyping procedure. Therefore, the data reliability was greatly enhanced by removing the variations in the plant ages (between genotypes and/or inoculation events), pathogen inoculum preparations, root system size and reduced exposure to unintended biotic and abiotic factors. During the establishment of this phenotyping protocol, various inoculation methods were tested including by soil-drenching or targeted deliver of inoculum solution to the vicinity of plant root system at the end of in-soil acclimation (without extra step of transplanting), orpre-mixing inoculum solution intosoils before re-planting. Dipping root system into inoculum solution appeared to be a more feasible and effective method for evenly distributing inoculum along individual root systems. Minimizing the transplanting effects after inoculation by pacing the humidity-conserving plastic covers over the trays have also significantly reduce the unnecessary abiotic stresses, as evidenced by the 100% survivability of mock-inoculated control plants through the infection events. Consistent and repeatable plant survival rates for a given O3R5 genotype between infection assays were routinely observed indicating the applicability of this phenotyping protocol.
3.3. Quantifiable Evaluations on Genotype-Specific Resistance Responses
Overall plant survival rates have been shown to be distributed as the continuum among tested O3R5 progenies ranging from single digit for those more susceptible genotypes to close to 100% of the more resistant ones. The ability to discern
Figure 8. Distinguishable resistance responses to Pythium ultimum infection among tested genotypes. (A) Top row represents a resistant genotype (O3R5-58); bottom two rows represent two susceptible lines (O3R5-106 and O3R5-47). Plants in pots at the left column are the mock-inoculated plants for corresponding genotypes. The rest of plants were simultaneously inoculated with P. ultimum using the same inoculum preparation, adjusted to the concentration of 2 × 103 oospores per mL. Each pot contained six plants. (B) Reduced shoot and root size due to P. ultimum inoculation (right panel), compared with those for the mock-inoculated control plants (left panel). (C) The darker brown color of necrotic root tissue from P. ultimum infection (left panel) as compared to the light-yellow color between the mock inoculated roots (left panel). All plants were grown under the identical conditions after inoculation. Images were taken at 10 dpi.
the subtle variations is critical for defining the inherent resistance responses with accuracy and precision. Quantitative values for several physiological parameters were designed to distinguish the quantitative variations between O3R5 genotypes. These parameters included root and shoot biomasses, shoot length, maximum root length, leaf number, accumulative leaf areas and the ratio between necrotic and healthy root segments (Figures 9A-9C). A few example son the measurements for root biomasses, shoot biomasses, leaf number and shoot length were shown as an example between mock-inoculated control plants and P. ultimum inoculated plants for a moderate resistant O3R5-135 (Figures 9D-9G).
Quantifiable parameters are needed for assigning the appropriate resistance levels for tested genotypes, which will be crucial for subsequent genetic studies to identify the minor-effect genes or mapping of QTLs (quantitative trait loci). It is conceivable that different resistance mechanisms function in various aspects of interaction between apple roots and P. ultimum. Values of overall plant mortality reflect only one aspect of such genotype-specific resistance responses. The detailed phenotypic traits or wide-range biological and physiological parameters, at both macro- or micro-level and for both belowground and aboveground organs, should facilitate the proper and reliable assessment on the genotype-specific reactions to the same target pathogen. Use of uniform plants with non-contaminated root systems is certainly advantageous for such detailed and consistent phenotype data.
Figure 9. Quantifiable parameters of genotype-specific responses to Pythium ultimum inoculation. (A) Shoot lengths of mock inoculation control plants and P. ultimum infected plants; (B) The collected leaves from a single plant for cumulative leaf area measurement by a publicly available ImageJ software; and (C) Total root length versus necrotic sections within a root system of an individual infected plants. (D)-(G) Measurements of root biomass, shoot biomass, leaf number and shoot lengths from the mock and P. ultimum infected plants for a resistant line O3R5-172. Values were measured at 28 dpi. Bars with different number of star(s) indicated the means are significantly different (P ≤ 0.05); n = 6 for mock-inoculated control treatment, n = 16 for P. ultimum infected treatment.
3.4. An Adaptable Method for Continuous and Non-Disruptive Observation for Genotype-Specific Root Resistance Responses
The availability of these young apple plants presented feasibility indesigning experiments for non-disruptive, non-destructive and less-invasive evaluationon root resistance response to P. ultimum infection. As shown in Figure 10A, root growth dynamicsin response to P. ultimum inoculation can be tracked for as long as four weeks using the larger glass-box growth container (30 × 45 × 10 cm). The dynamics of root growth rates can be quantified by continuous measurement of image analysis and compared between mock-inoculated and P. ultimum inoculated plants. About 35% reduction at the maximum length of root was observed for O3R5-135 (Figure 10A, inset figure) at 28 dpi. Using a miniatureglass box (3.5 × 7.5 × 10 cm) (Figure 10B), the continuous observation and documentation on symptom development was achieved with the assistance of a dissecting microscope equipped with image-capturing system. The time-lapsed image analysis captured the features of necrosis progression on the specific sections of root system (Figure 10C and Figure 10D). While the mock-inoculated root remained healthy throughout the observation period, the yellow-brownish coloration, due to P. ultimum inoculation, was observed at 120 hpi. This observation provided the critical insight on the real-time necrosis patterns along infected apple roots for the first time.
Continuous observation on the dynamics of root resistance responses towards pathogen inoculation is more challenging compared to those of aerial organs because of the hidden nature of roots in soil substrates. Additionally, young apple feeder roots where infection occurs are miniature in size which often defies the direct visual assessment on symptom development without assistance from a dissecting microscope. Utilizing a small-glass box as growth container makes it feasible to expose infected roots under dissecting microscope. The continuous evaluation and documentation of apple root necrosis progression patterns has never reported. Minirhizotron and MNR technology-based imaging technologies are being actively developed with the aim of uncovering agronomical or horticultural traits of roots such as root growth and root system architecture under natural soil condition [29] [30] . However, these technologies may not immediately applicable to define the detailed root resistance traits; for example, the subtle change of tissue necrosis such as coloration is difficult to detector define by the current form of these technologies. The combination of small glass-box and microscope-assisted observation can be considered as a simple but innovative method for documenting early pathogenesis of apple root under the pathogenic pressure. It opens a new venue for phenotyping apple root resistance traits in a non-disruptive, non-destructive manner with minimal invasiveness. Use of the large glass-box pot offered the chance for uncovering the traits related to root regeneration and architectural alterations beneficial to plant survival by a longer period observation. These micro-propagated young apple plants with non-contaminated root systems enabled the experimental design for such detailed observations of root responses in real-time and with minimized invasiveness.
3.5. Microscopic Features of Genotype-Specific Necrosis Patterns along Infected Roots
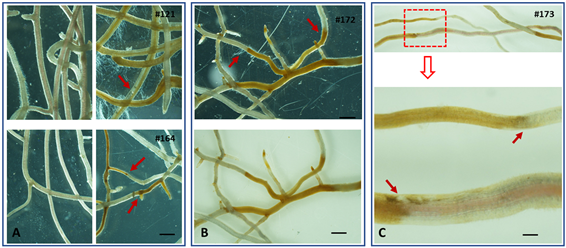
In addition to the use of glass-box container for continuous observation of root necrosis progression, careful and systematic microscopic observation allowed to capture the more detailed feature of interactions between apple root and P. ultimum. Distinguishable patterns of P. ultimum incited necrosis were demonstrated for four O3R5 apple rootstock genotypes. Roots of mock-inoculated control plants remained healthy as expected, demonstrating white-color root branches and unscathed root tissue (without appearance of transparency in root cortex tissue) (Figure 11A, left panels). In response to P. ultimum inoculation, contrasting responses were readily identified between a more susceptible O3R5-121 (Figure 11A, top right image) and a more resistant O3R5-164 (Figure 11A, bottom right image). Almost all inoculated root branches of O3R5-121 exhibited necrotic symptom, with yellow to brownish tissue coloration, and disintegrated tissues showing discernable semi-transparent appearance (arrows); probably more noticeable is the profuse hyphae growth across the observed section of this root system. In contrast, the roots of a more resistant O3R5-164 showed that majority of root section remained healthy with white color and intact appearance of root cortex tissue without sign of transparency. The more noticeable feature is the presence of a defined “line” or “zone” (arrows) which clearly separated necrotic sections from the healthy ones. Expanded examination
Figure 10. Plants growing in glass-box container for continuous observation of resistance responses and microscope-assisteddocumentation of symptom development. (A) Continuing assessment of apple root growth behaviors with or without pathogenic pressure using a large glass-box pot. Left panel showed mock-inoculated plants; and right panel showed Pythium ultimum inoculated plants at 14 dpi. Inset figure: the average values for maximus of root length at 28 dpi. (B) Smaller glass-box growth containers enabled the enhanced accessibility for microscopic observation on specific sections within an inoculated root system. (C) The image series for a section within a root system for a mock-inoculated plant under dissecting microscope. (D) The image series for a section of root system of a P. ultimum inoculated plant. The number on top of each image represents the time points expressed as hour post inoculation (hpi). The bar represents the length of 500 µm.
Figure 11. Microscopic features of root tissue necrosis of four apple rootstock genotypes at 48 hpi. (A) Representing images of infected roots two genotypes. Top left: mock-inoculated O3R5-121; Top right: Pythium ultimum-inoculated O3R5-121; bottom left: mock-inoculated O3R5-164; bottom right: Pythium ultimum-inoculated O3R5-164. (B) Representing images of roots inoculated with P. ultimum; demonstrating coloration and defined zone separating healthy and necrotic sections of root for O3R5-172; top image with black background and reflecting light, bottom image with white background for the same section of roots. (C) Azoom-out view and a close-up image on the defined zone for infected roots from O3R5-173. The bars in (A) and (B) represent 500 µm; and the bar in (C) bottom panel represents 200 µm.
to other genotypes suggested that such defined zone between healthy and necrotic sections appeared to be common among the more resistant genotypes such as O3R5-172 and O3R5-173 (Figure 11B and Figure 11C, arrows). In contrast, the features of profuse growth of P. ultimum hyphae from the infected root sections were more frequently associated with other susceptible O3R5 genotypes.
Previous phenotyping survey and transcriptome analysis on this pathosystem of apple root-P. ultimum interaction indicated that the time point at 48 hpi appeared to be a critical stage in differentiating the outcome of their interactions [31] [32] . The contrasting resistance responses at this time point may be indicative between resistance and susceptibility for a given genotype. Side by side comparison between genotypes on root necrosis development allowed phenotypic evaluation at root tissue levels. For example, the profuse hyphae growth from infected roots of the susceptible genotypes indicated the lack of ability to deter the pathogen’s aggressiveness, which most likely contributed to its low survival rates. On the other hand, the presence of so-called defined zone or line seemed to function as a physical or biochemical barrier separating the healthy and necrotic sections. The arrest, or the compartmentation, of necrosis advancement could represent a major resistance mechanism in apple roots to this fast-acting pathogen. Consequently, such deterrence of necrosis along the infected roots lead to the high survival rate in response to infection by P. ultimum. The detailed microscopic features on necrosis progression exhibited the necessity of in-depth phenotyping. The combined evaluation methods are crucial in extracting valuable genotype-specific resistance responses which will provide critical insight in elucidating the underlying resistance mechanisms in apple roots. The availability of uniform plants for desired genotypes from the synchronized micropropagation process, as well as simultaneous inoculation using the identical inoculum preparations, contributes enormously to the data reliability and repeatability onappleresistance phenotypes.
4. Summary
The hidden nature of roots limits their accessibility for a non-disruptive, non-destructive phenotypic evaluation. For this reason, it is well acknowledged that phenotyping resistance response of plant root is more challenging compared to those of plant aerial parts. This reported phenotyping protocol offers the capability for capturing the reliable and detailed resistance responses in apple root. The uniform apple plants from a streamlined tissue culture procedure, the standardized inoculation procedure and various evaluating methods are the integral components of this established protocol for obtaining high-quality apple root resistance phenotypes. In particular, the tissue culture based micropropagation overcomes a long-standing barrier of lacking the constant supply of uniform apple plants for repeated infection assays. The availability of these genetically-defined and age-equivalent apple plants with non-contaminated root system enhanced the accurate and reproducible assessment of apple root resistance responses for any desired apple rootstock genotypes. An innovative observation method, which combines the use of glass-box container and a dissecting microscope, provides the enhanced accessibility and minimized invasiveness for continuous and close-up observation. These observations allow, for the first time, the characterization of genotype-specific necrosis progression patterns along inoculated apple roots. The reported protocol represents a significant advancement in methodology of phenotyping apple root resistance traits. With minor modification, this protocol should be applicable to other rosacea tree fruit crops or other perennial species. Therefore, this phenotyping protocol contributes significantly to the subsequent genetic analysis and targeted exploitation of innate resistance in apple root to infection by soilborne pathogens.
Authors’ Contributions
YZ provided guidance for the experimental design and obtained research funding. YZ and MS planned and conducted the experiments, collected and analyzed the data, and wrote the manuscript. JZ collaborated in set-up the experiments using glass boxes for microscopic observation and collection of results. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflicts of interest regarding the publication of this paper.
Cite this paper
Zhu, Y.M., Saltzgiver, M. and Zhao, J.X. (2018) A Phenotyping Protocol for Detailed Evaluation of Apple Root Resistance Responses Utilizing Tissue Culture Micropropagated Apple Plants. American Journal of Plant Sciences, 9, 2183-2204. https://doi.org/10.4236/ajps.2018.911158
References
- 1. Fiorani, F. and Schurr, U. (2013) Future Scenarios for Plant Phenotyping. Annual Review of Plant Biology, 64, 267-291. https://doi.org/10.1146/annurev-arplant-050312-120137
- 2. Velasco, R., Zharkikh, A., Affourtit, J., Dhingra, A., Cestaro, A. and Kalyanaraman, A. (2010) The Genome of the Domesticated Apple (Malus × domestica Borkh.). Nature Genetics, 42, 833-839. https://doi.org/10.1038/ng.654
- 3. Janick, J., Cummins, J., Brown, S. and Hemmat, M. (1996) Apples. Fruit Breeding, 1, 1-77.
- 4. Russo, N.L., Aldwinckle, H., Fazio, G. and Wan, Y. (2007) Investigation on the Inheritance of Strain Specific Resistance to Erwinia amylovora in an Apple Rootstock Segregating Population. XI International Workshop on Fire Blight, Portland, 12-17 August 2007, 331-335.
- 5. Thorpe, T.A. (2007) History of Plant Tissue Culture. Molecular Biotechnology, 37, 169-180. https://doi.org/10.1007/s12033-007-0031-3
- 6. Abdollahi, H., Rugini, E., Ruzzi, M. and Muleo, R. (2004) In Vitro System for Studying the Interaction between Erwinia amylovora and Genotypes of Pear. Plant Cell, Tissue and Organ Culture, 79, 203-212. https://doi.org/10.1007/s11240-004-0661-0
- 7. Armas, I., Pogrebnyak, N. and Raskin, I. (2017) A Rapid and Efficient in Vitro Regeneration System for Lettuce (Lactuca sativa L.). Plant Methods, 13, 58. https://doi.org/10.1186/s13007-017-0208-0
- 8. Kornova, K. and Popov, S. (2011) Obtaining Virus-Free Planting Material of Plum Cultivars by the in Vitro Method. II Balkan Symposium on Fruit Growing, Pitesti, 5-7 September 2011, 507-512.
- 9. Datta, S., Chakraborty, D. and Janakiram, T. (2017) Low Cost Tissue Culture: An Overview. The Journal of Plant Science Research, 33, 181-199.
- 10. Bhatti, S. and Jha, G. (2010) Current Trends and Future Prospects of Biotechnological Interventions through Tissue Culture in Apple. Plant Cell Reports, 29, 1215-1225. https://doi.org/10.1007/s00299-010-0907-8
- 11. Kaushal, N., Modgil, M., Thakur, M. and Sharma, D. (2005) In Vitro Clonal Multiplication of an Apple Rootstock by Culture of Shoot Apices and Axillary Buds. Indian Journal of Experimental Biology, 43, 561-565.
- 12. Diler, E., Unver, T. and Karakülah, G. (2016) Differential Expression of Hyperhydricity Responsive Peach MicroRNAs. Journal of Integrative Bioinformatics, 13, 57-69. https://doi.org/10.1515/jib-2016-308
- 13. Mu, H., Lin, L., Liu, G. and Jiang, J. (2013) Transcriptomic Analysis of Incised Leaf-Shape Determination in Birch. Gene, 531, 263-269. https://doi.org/10.1016/j.gene.2013.08.091
- 14. Wang, W., Li, H., Lin, X., Yang, S., Wang, Z. and Fang, B. (2015) Transcriptome Analysis Identifies Genes Involved in Adventitious Branches Formation of Gracilaria lichenoides in Vitro. Scientific Reports, 5, Article No. 17099. https://doi.org/10.1038/srep17099
- 15. Ingvarsson, P.K. and Street, N.R. (2011) Association Genetics of Complex Traits in Plants. New Phytologist, 189, 909-922. https://doi.org/10.1111/j.1469-8137.2010.03593.x
- 16. Ogura, T. and Busch, W. (2016) Genotypes, Networks, Phenotypes: Moving toward Plant Systems Genetics. Annual Review of Cell and Developmental Biology, 32, 103-126. https://doi.org/10.1146/annurev-cellbio-111315-124922
- 17. Chiong, K.T., Damaj, M.B., Padilla, C.S., Avila, C.A., Pant, S.R. and Mandadi, K.K. (2017) Reproducible Genomic DNA Preparation from Diverse Crop Species for Molecular Genetic Applications. Plant Methods, 13, 106. https://doi.org/10.1186/s13007-017-0255-6
- 18. Arismendi, M.J., Almada, R., Pimentel, P., Bastias, A., Salvatierra, A. and Rojas, P. (2015) Transcriptome Sequencing of Prunus sp. Rootstocks Roots to Identify Candidate Genes Involved in the Response to Root Hypoxia. Tree Genetics & Genomes, 11, 11. https://doi.org/10.1007/s11295-015-0838-1
- 19. Lee, S. and Choi, D. (2013) Comparative Transcriptome Analysis of Pepper (Capsicum annuum) Revealed Common Regulons in Multiple Stress Conditions and Hormone Treatments. Plant Cell Reports, 32, 1351-1359. https://doi.org/10.1007/s00299-013-1447-9
- 20. Mazzola, M. (1998) Elucidation of the Microbial Complex Having a Causal Role in the Development of Apple Replant Disease in Washington. Phytopathology, 88, 930-938. https://doi.org/10.1094/PHYTO.1998.88.9.930
- 21. Gardiner, S.E., Norelli, J.L., De Silva, N., Fazio, G., Peil, A. and Malnoy, M. (2012) Putative Resistance Gene Markers Associated with Quantitative Trait Loci for Fire Blight Resistance in Malus “Robusta 5” Accessions. BMC Genetics, 13, 25. https://doi.org/10.1186/1471-2156-13-25
- 22. Norelli, J., Aldwinckle, H. and Beer, S. (1988) Virulence of Erwinia amylovora Strains to Malus sp. Novole Plants Grown in Vitro and in the Greenhouse. Phytopathology, 78, 1292-1297. https://doi.org/10.1094/Phyto-78-1292
- 23. Yepes, L.M. and Aldwinckle, H.S. (1994) Micropropagation of Thirteen Malus cultivars and Rootstocks, and Effect of Antibiotics on Proliferation. Plant Growth Regulation, 15, 55-67. https://doi.org/10.1007/BF00024677
- 24. Dobránszki, J. and Teixeira da Silva, J.A. (2010) Micropropagation of Apple—A Review. Biotechnology Advances28, 462-488. https://doi.org/10.1016/j.biotechadv.2010.02.008
- 25. Modgil, M., Handa, R., Sharma, D. and Thakur, M. (2003) High Efficiency Shoot Regeneration from Leaf Explants of in Vitro Grown Shoots of Apple. 7th International Symposium on Temperate Zone Fruits in the Tropics and Subtropics Part Two, Solan, 25 November 2005, 696.
- 26. Lee, H.-J., Park, Y.-J., Ha, J.-H., Baldwin, I.T. and Park, C.-M. (2017) Multiple Routes of Light Signaling during Root Photomorphogenesis. Trends in Plant Science, 22, 803-812. https://doi.org/10.1016/j.tplants.2017.06.009
- 27. Daccord, N., Celton, J.-M., Linsmith, G., Becker, C., Choisne, N. and Schijlen, E. (2017) High-Quality de Novo Assembly of the Apple Genome and Methylome Dynamics of Early Fruit Development. Nature Genetics, 49, 1099-1106. https://doi.org/10.1038/ng.3886
- 28. Russo, N.L., Robinson, T.L., Fazio, G. and Aldwinckle, H.S. (2007) Field Evaluation of 64 Apple Rootstocks for Orchard Performance and Fire Blight Resistance. HortScience, 42, 1517-1525.
- 29. Shi, R., Junker, A., Seiler, C. and Altmann, T. (2018) Phenotyping Roots in Darkness: Disturbance-Free Root Imaging with Near Infrared Illumination. Functional Plant Biology, 45, 400-411. https://doi.org/10.1071/FP17262
- 30. Guo, L., Chen, J., Cui, X., Fan, B. and Lin, H. (2013) Application of Ground Penetrating Radar for Coarse Root Detection and Quantification: A Review. Plant and Soil, 362, 1-23. https://doi.org/10.1007/s11104-012-1455-5
- 31. Zhu, Y., Shin, S. and Mazzola, M. (2016) Genotype Responses of Two Apple Rootstocks to Infection by Pythium ultimum Causing Apple Replant Disease. Canadian Journal of Plant Pathology, 38, 483-491. https://doi.org/10.1080/07060661.2016.1260640
- 32. Shin, S., Zheng, P., Fazio, G., Mazzola, M., Main, D. and Zhu, Y. (2016) Transcriptome Changes Specifically Associated with Apple (Malus domestica) Root Defense Response during Pythium ultimum Infection. Physiological and Molecular Plant Pathology, 94, 16-26. https://doi.org/10.1016/j.pmpp.2016.03.003
Abbreviations
BA: benzylaminopurine; IBA: indolebutyric acid; MS: Murashige and Skoog basal salt mixture; NAA: naphthaleneacetic acid; SD: standard deviation.