American Journal of Plant Sciences
Vol.4 No.6A(2013), Article ID:33297,7 pages DOI:10.4236/ajps.2013.46A004
Arabidopsis Transcription Factor WRKY33 Is Involved in Drought by Directly Regulating the Expression of CesA8
![]()
College of Life Science, Capital Normal University, Beijing, China.
Email: *qixiaoting@cnu.edu.cn
Copyright © 2013 Xinjing Wang et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received April 21st, 2013; revised May 21st, 2013; accepted June 9th, 2013
Keywords: WRKY33; Drought; CesA8; Target Gene; Inverse Yeast One-Hybrid Assay
ABSTRACT
Arabidopsis (Arabidopsis thaliana) WRKY33 is a key transcription factor in pathogen-induced defense signaling, but its function in abiotic stresses remains largely unclear. In this study, we report on the use of a reverse-genetic approach, as well as a yeast (Saccharomyces cerevisiae) expression system, to determine the role of WRKY33 in drought. A T-DNA insertion deletion mutant of WRKY33 is more sensitive to dehydration. Through genome-wide screening the target genes of WRKY33 in yeast, we identified 23 candidate genes including a drought tolerance gene CesA8. Further results revealed that WRKY33 repressed CesA8 expression through binding to the W-box elements of CesA8 distal promoter region and probably interacting with the transcriptional activator of CesA8, MYB46. These findings revealed the primary molecular mechanism underlying the function of WRKY33 in response to drought.
1. Introduction
In plants, WRKY transcription factors are one of the largest families of transcriptional regulators with up to 74 representatives in Arabidopsis [1]. WRKY transcription factors are defined by the presence of the highly conserved WRKY domain containing the sequence WRKYGQK and a distinct zinc finger-type motif [1]. They form integral parts of signaling webs that modulate many plant processes including biotic stresses, abiotic stresses, and developments [2]. WRKY transcription factors bind to a specific cis-acting DNA element, W-box (5’-TTGACC/ T-3’), within the promoter of target gene [3].
Arabidopsis WRKY33 (At2g38470) is strongly activated by pathogen-induced infection [4,5] and serves as a key transcription factor in pathogens defense signaling [6-8]. WRKY33 is regulated by multiple abiotic stresses including high salinity [9], osmotic stress [10], cold [10], heat shock [10,11], heavy metal stress [12], and UV exposure [5], indicating that WRKY33 may also be involved in abiotic stresses. Recently, WRKY33 has been proved to confer tolerance to salt stress [10] and heat stress [11], but its molecular mechanism in response to abiotic stresses is not well understood.
Considering the facts that WRKY33 confers salt stress and drought may result from a physiological water deficit caused by salt stress, we explored whether WRKY33 also conferred drought. In this study, we showed that WRKY33 was involved in drought through a direct transcriptional regulation of CesA8, an Arabidopsis drought tolerance gene. Our findings advanced understanding of the molecular mechanism underlying the role of WRKY- 33 in response to drought stress.
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
The wrky33-1 mutant (SALK_006603), carrying a TDNA insertion in At2g38470 of the Col ecotype, was obtained from the Arabidopsis Biological Resource Center. RT-PCR was used to verify homozygous T-DNA insertion for wrky33-1 using WRKY33-E1 (5’-GTCACAACAATCCGGAAGAAC-3’) and WRKY33-E3 (5’- CAAACTGGCATTGTACACAGC-3’). The seeds of wrky- 33-1 and wild-type were surface-sterilized and sown on 1/2 Murashige and Skoog (MS)-agar plates at 22˚C with 16-h/8-h light/dark cycles.
2.2. Drought Stress Experiments
For dehydration-rehydration assay, rosette leaves were detached from 2-week-old plants, weighed immediately on a plate and then slowly dried at room temperature (22˚C). After the rosette leaves lost 60% of their initial fresh weight, the plants were immersed in water and grown in the greenhouse.
For the phenotype tests under drought stress, seedlings grown on 1/2 MS-agar medium for 7 days were transferred to mixed soil (rich soil:vermiculite, 2:1, v/v) and grown for two weeks with sufficient watering. Then the plants were subjected to drought treatment by withholding irrigation. To ensure reproducibility for the phenotype tests, four pots (four seedlings/pots) of plants were grown for wild-type and wrky33-1, respectively.
2.3. RNA Isolation and cDNA Synthesis
Total RNA was extracted from Arabidopsis seedlings using the RNAprep Pure Plant kit with on-column DNase digestion (Tiangen Biotech, Beijing, China) according to the manufacturer’s protocol. RNA (2 μg) was used to synthesize the first-strand cDNA with an oligo (dT) primer according to the instruction of PrimeScriptTM 1st strand cDNA synthesis kit (Takara, Dalian, China).
2.4. Semi-Quantitative Duplex RT-PCR Analysis
The semi-quantitative duplex RT-PCR analysis of CesA8 and TUA4 was performed at annealing temperature 56˚C for 30 or 33 cycles using the EX TaqTM polymerase (Takara, Dalian, China). The following primers were used: CesA8, 5’-TGGAGGTGTCTCAGCTCATCTC-3’ and 5’- GAGAAAGGGAAGTCGTATCGG-3’; TUA4, 5’-CTCTACCTCCGTTGTTGAGCCTTAC-3’ and 5’-CACCCACATACCAGTGAACGAAAG-3’.
2.5. Construction of W-Box Sequences Enriched Genomic Library
Arabidopsis genomic DNA fragments enriched for Wbox sequences (5’-TTGAC(T/C)-3’) were obtained by PCR using forward primer (5’-CGGAATTCTT-GAC (C/T)- 3’, EcoRI site is underlined) and reverse primer (5’- GGACTAGT(G/A)GTCAA-3’, SpeI site is underlined). The PCR was carried out in a total volume of 50 µl, containing 50 pmol each primer, 0.5 U EX TaqTM polymerase (Takara, Dalian, China), and 500 ng genomic DNA. The first cycle of amplification is 95˚C 2 min, 30˚C 10 min, 42˚C for 20 min and then another 25 cycles of 94˚C 30 s, 42˚C 30 s, 72˚C 30 s, at last 72˚C for 7 min. PCR products were digested with EcoRI/SpeI and then inserted to the same restriction sites of pHIS 2.1 (Clontech). The recombined DNA was transformed to E. coli Top 10 strain. The transformants grown on LB-ampicillin plates were washed into a flask containing LB medium and then plasmids were isolated using standard procedures. The insert size of library plasmids was determined by PCR with Insert-F (5’-GCCAGGGTTTTCCCAGTCAC-3’) and Insert-R (5’-CTTCGTTTATCTTGCCTGCTCA-3’) using library plasmids and empty pHIS2.1 plasmid (negative control template), respectively.
2.6. Inverse Yeast One-Hybrid Screen
The primer pairs 5’-CGGAATTCGCTGCTTCTTTTCTTACAATGG-3’ (EcoRI site is underlined) and 5’-CGGGATCCTCAGGGCATAAACGAATCGAAA-3’ (BamHI site is underlined) were used to amplify the coding sequence of WRKY33. The resulting PCR products were cloned into the EcoRI/BamHI sites of pGAD424 vector (Clontech) and sequenced to yield bait plasmid ADWRKY33. AD-WRKY33 plasmids were transferred to yeast strain Y187 (Clontech) by the standard LiAc-PEGmediated transformation procedure.
The library plasmids were transferred to Y187 containing AD-WRKY33. Aliquots (100 μl) of each distilled water-diluted transformants were spread onto synthetic defined (SD) medium lacking Trp, Leu, and His and containing 25 mM 3-amino-1,2,4-triazole (3-AT). The library plasmids were rescued from positive yeast clones, amplified in E. coli Top 10, and verified by sequencing using Insert-F and Insert-R.
2.7. Yeast Two-Hybrid Analysis
For the construction of prey, the coding sequence ofMYB46 or VND7 was amplified from Arabidopsis cDNA. The following primers were used: MYB46, 5’- CGGAATTCGCAGCTAGTACTCACCAAGTAAA G-3’ and 5’-AACTGCAGTAAGAATCTTGAGTATAAACCGC-3’, VND7, 5’-CGGAATTCGATAATATAATGCAATCGTCAATG C-3’ and 5’-AACTGCAGTTACGAGTCAGGGAAGCATCC-3’.
The underlined sequences of above primers indicate the EcoRI and PstI sites. The resulting PCR products were then inserted into EcoRI/PstI sites of pGBKT7 (Clontech) to fuse to GAL4 BD, designed BD-MYB46 and BD-VND7, respectively. The different combinations of AD-WRKY33/BD-MYB46, AD-WRKY33/BD-VND7, and pGAD424/pGBKT7 (negative control) were cotransformed into yeast strain PJ69-4A. The interaction was evaluated by spot assay and LacZ-filter lift assay.
The bait and prey plasmids were rescued from yeast clones grown on selective medium, amplified in E. coli Top 10, and verified by sequencing.
2.8. Transcriptional Activation Activity Assay in Yeast
Transcription activation assay of CesA8 promoter and AD-WRKY33 in yeast cells was designed as described previously [13]. A XbaI-SacI fragment containing β- glucuronidase (GUS) coding region from pBI121 (Clontech) was inserted into the XbaI/SacI sites of YEplac181 to construct YEplac181-GUS. The XbaI-SacI fragment containing the GUS coding region from YEplac181-GUS was subcloned into pYES to generate pYES-GUS. Two antiparallel oligonucleotides of an internal deleted CesA8 promoter fragment (ProCesA8) and its mutated version (ProCesA8m) with overhanging HindIII and XbaI sticky ends were synthesized and cloned to the upstream HindIII/XbaI sites of pYES-GUS to yield ProCesA8:GUS and ProCesA8m:GUS, respectively.
The different combinations of AD-WRKY33/ProCesA8:GUS and AD-WRKY33/ProCesA8m: GUS were cotransformed into PJ69-4A. The positive transforms were grown in SD/-Leu-Ura liquid medium. The method for measuring GUS activity was adapted from [13]. Briefly, 7.5 × 107 cells of each transformated yeast line were collected by centrifugation at 8000 rpm for 5 min. 100 μl GUS staining buffer were added to the yeast pellet, followed by vortexing and three freeze-thaws in liquid nitrogen. The reaction was incubated at 37˚C for 12 h. Three independent clones for each transformant were analyzed.
3. Results and Discussion
3.1. Mutation in WRKY33 Causes a Moderate Decrease in Drought Tolerance
To explore whether WRKY33 was involved in drought, we analyzed the drought tolerance in a homozygous T-DNA insertion mutant line (wrky33-1) from the Arabidopsis Biological Resource Center (Figure 1(a)). The primers around the T-DNA insertion site produced a single larger RT-PCR product (783 bp) in wrky33-1 compared with the wild type (724 bp) as previously described [10], demonstrating that wrky33-1 is a homozygous mutant line (Figure 1(b)). By cloning and sequencing of this larger RT-PCR product, we revealed that an extra 59-bp sequence is inserted between the exon2 and the exon 3. This extra sequence T-DNA insertion leads to a frame-shift and premature stop codon within the exon 3. This could result in the production of small truncated WRKY33 protein which has 216 amino acid in length (referred to as S-WRKY33). S-WRKY33 contains a Nterminal region of WRKY33 (residues 1 to 150) and an additional 66 amino acids (residues 151 to 216). Thus, wrky33-1 is a deletion mutation line. Because SWRKY33 lacks two WRKY domains, it cannot act as a WRKY33.
With this wrky33-1 deletion mutant line, we performed dehydration-rehydration assay and soil drought experiment. Rehydration for 4 days recovered the dehydration-treated detached rosette leaves of wild-type and wrky33-1. After another 6 days of rehydration, most of wrky33-1 rosette leaves became bleached and were
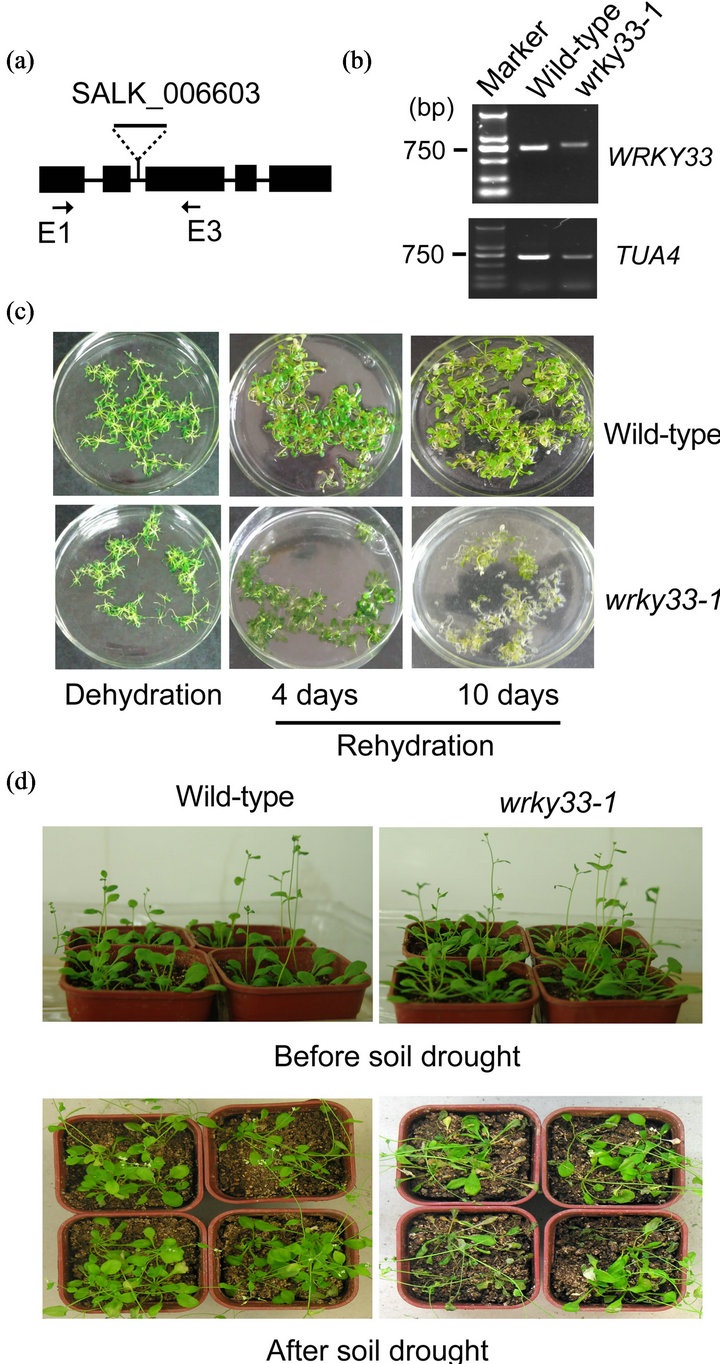
Figure 1. Response of wrky33-1 seedlings to drought stress (a) The T-DNA was inserted in the second intron of the WRKY33 genomic DNA. Black boxes and lines denote exons and introns, respectively. Arrows indicated the primers that were used to RT-PCR confirmation of wrky33-1 mutant. (b) RT-PCR analysis of wild-type and wrky33-1 plants using the exon 1-specific forward primer (E1) and the exon 3-specific reverse primer (E3). TUA4 (At1g04820), encoding an α-tubulin isoform, served as a loading control. (c) Dehydration-rehydration assay of detached rosette leaves of wild-type and wrky33-1 was performed as described in Materials and methods. The photographs were taken after dehydration and at 4 days or 10 days of rehydration, respectively. (d) Phenotype analysis of three-week-old wrky33-1 or wild-type plants under soil drought (without watering). The photographs were taken before and two weeks after water stress, respectively.
dead; whereas, wild-type still survived (Figure 1(c)). After two-week soil drought treatment, wrky33-1 plants showed more severe leaf-wilting phenotype than wildtype plants (Figure 1(d)). These results confirmed that wrky33-1 is more sensitive to drought than wild-type. The decrease in drought tolerance caused by WRKY33 mutation suggested the involvement of WRKY33 in drought. Moreover, the moderate drought-sensitive phenotype of wrky33-1 may reflect WRKY33 functional redundancy with other transcription factors in drought tolerance. Similarly, WRKY33 is involved in thermotolerance interacting with WRKY25 and WRKY26 in Arabidopsis [11].
3.2. Inverse Yeast One-Hybrid Assay Identifies 23 Candidate WRKY33 Target Genes
Identifying the downstream target genes of WRKY33 is crucial in understanding its molecular mechanism in response to drought stress. Zeng et al. [14] develop an inverse yeast one-hybrid (Y1H) system which allows rapid and genome-wide identification of transcriptional binding targets. We modified this inverse Y1H system to screen the candidate target genes of WRKY33 (Figure 2(a)). In this system, yeast Gal4 activation domain (AD)- WRKY33 fusion protein was used as bait to screen a genomic library enriched with W-box sequences.
W-box enriched genomic fragments were obtained by PCR using primers containing W-box sequences (Figure 2(b)) and then cloned upstream of the HIS3 reporter gene to construct W-box enriched genomic library. The recombinant ratio and insert size of genomic library plasmids were estimated by PCR using the primers around the insertion sites (Figure 2(a)). Most of PCR products amplified from the library plasmids were larger than that from empty plasmid control (Figure 2(c)), suggesting that most of library plasmids contained insert DNA. The size of insert DNA varied from approximately 50 bp to 200 bp by subtracting 197 bp representing the empty vector control (Figure 2(c)).
After transforming the library plasmids into the yeast containing AD-WRKY33, we got sixty positive clones by using HIS-free selective medium. Using the sequenced insert DNA as queries, 23 candidate target genes were identified by screening 3000 bp of upstream sequence of all known genes in the Arabidopsis genome. These putative target genes of WRKY33 encode proteins related with various biological functions (Table 1). Three salt stress-related genes (CesA8, HOS3-1, and EDGP) and two defense-related genes (MED8 and At4g13580) were included. This result was consistent with the roles of WRKY33 in salt stress [10] and pathogens-induced defense [4,5].
3.3. WRKY33 Binds to the CesA8 Promoter in Yeast
Among the 23 candidate genes, CesA8 has been reported
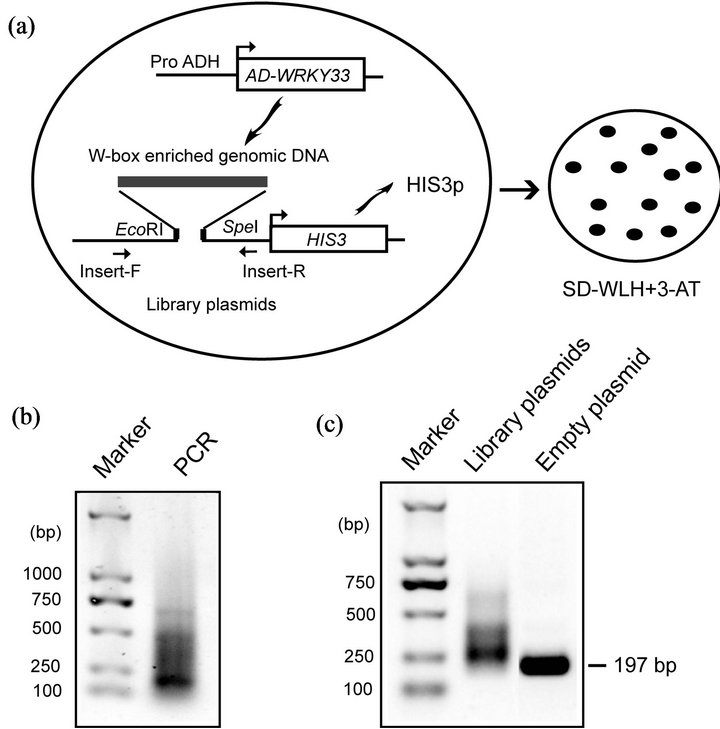
Figure 2. Genome-wide screening target genes of WRKY33 in yeast. (a) Strategy of the modified inverse Y1H screening to identify the WRKY33 target genes. The W-box enriched genomic library plasmids were transformed into the yeast strain Y187 containing AD-WRKY33 bait. AD-WRKY33 binds to the target DNA fragment enriched for W-box sequences to result in generation of HIS3 protein (HIS3p) and permit yeast growth on SD medium lacking Trp, Leu, and His (SD-WLH) and containing 3-amino-1,2,4-triazole (3-AT, a competitive inhibitor of the HIS3p). (b) Agarose gel analysis of PCR products enriched for W-box sequences. (c) Agarose gel analysis of amplicons from library plasmids and empty plasmid using Insert-F/Insert-R (as shown in Figure 2(a)).
to confer drought tolerance. CesA8 encodes a member of the cellulose synthase family required for secondary cell wall biosynthesis [15]. Disruption of this gene enhances tolerance to drought stress [16]. Therefore, we asked whether CesA8 was involved in WRKY33-mediated drought tolerance.
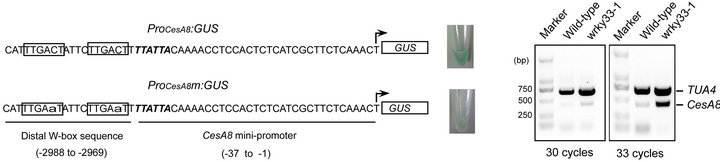
Results from the inverse Y1H experiment (Table 1) suggest that WRKY33 binds to the distal CesA8 promoter region (-2988 to -2969) containing two repeat W-box elements (5’-TTGACT-3’). We used the yeast transcriptional activation assay developed by Yuan et al. [13] to verify this binding. The WRKY33-binding distal CesA8 promoter region or its W-box mutated version fused to the minimal CesA8 promoter (designated ProCesA8 and ProCesA8m, respectively) were cloned into the upstream of the GUS reporter gene to construct two reporter plasmids (ProCesA8:GUS and ProCesA8m: GUS), respectively (Figure 3(a)). As expected, co-expression of AD-WRKY33 and ProCesA8:GUS in yeast cells resulted in strong blue color, whereas, co-expression of AD-WRKY33 and ProCesA8m:GUS only caused
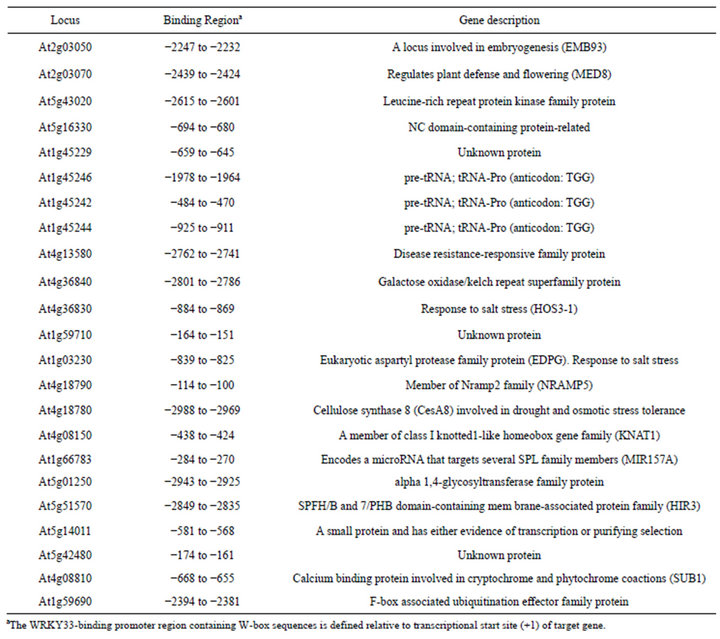
Table 1. Candidate target genes of WRKY33.
 (a) (b)
(a) (b)
Figure 3. Characterization of WRKY33-mediated CesA8 expression. (a) The yeast transcriptional activation assay of WRKY33-mediated CesA8 promoter expression. The sequences of the internal deleted CesA8 promoter and its W-box mutated (lowercase) version were shown in left panel. The arrow indicated transcriptional start site. The W-box sequence was boxed and TATA-box was shown in italics. The corresponding GUS staining was shown in right panel. Three independent positive clones for each transformant were analyzed and similar results were obtained. (b) Validation of differential expression for CesA8 in wild-type and wrky33-1 plants by semi-quantitative duplex RT-PCR. TUA4 was used as a loading control.
very weak blue color. This may result from the failure of WRKY33 in binding to the mutated W-boxes in ProCesA8m. Thus, WRKY33-mediated CesA8 mini-promoter activity required the distal W-boxes in yeast. This result confirmed that WRKY33 bound to the W-box sequences localized in the distal CesA8 promoter region in yeast.
3.4 CesA8 Is Up-Regulated in wrky33-1
To further confirm whether CeA8 is a direct target gene of WRKY33 in Arabidopsis, we therefore analyzed the expression of CesA8 in wrky33-1 plants. Transcript abundance of CesA8 was determined using semi-quantitative duplex RT-PCR assay which allows amplification of CesA8 and an internal control TUA4 gene in a single reaction. Result showed that CesA8 mRNA accumulation was greatly increased in wrky33-1 compared with the wildtype line (Figure 3(b)). The down-regulation of CesA8 in wrky33-1 confirmed that CesA8 was a direct target gene of WRKY33. And WRKY33 may serve as a negative transcription regulator of CesA8.
3.5. WRKY33 Interacts with MYB46 in Yeast
As a transcriptional repressor of CesA8, WRKY33 may interact with other transcriptional factors of CesA8 to regulate CesA8 expression. Recently, VND7 (At1g71930) and MYB46 (At5g12870) are identified to be the transcriptional activators of CesA8 expression [17,18]. We performed yeast two-hybrid (Y2H) assay and showed that WRKY33 could interact with MYB46 but not VND7 to activate the expression of both HIS3 and LacZ reporter genes (Figure 4). The interaction between WRKY33 and MYB46 indicates that WRKY33 may regulate the down target genes of MYB46 including CesA8.
3.6. CesA8 Connects WRKY33 with Its Role in Drought Stress
Collectively, the results from altering expression of CesA8 in wrky33-1 and the CesA8 promoter-binding ability of WRKY33 suggested that CesA8 was a direct target gene of WRKY33. WRKY33 negatively regulated CesA8 expression through binding to the W-box elements of CesA8 distal promoter region and probably through forming a heterodimer with MYB46. Above conclusion is mainly based on a yeast heterologous system and thus poses a limitation in our study. Therefore, we need to be further verified these in Arabidopsis plants.
Because mutation in CesA8 leads to enhanced tolerance to drought stress, salt stress, and pathogen resistance [16,19], the increased accumulation level of CesA8 mRNA in wrky33-1 plants can partly explain its droughtor salt stress-sensitive phenotype [10] and enhanced pathogen susceptibility [4,5]. Therefore CesA8 links
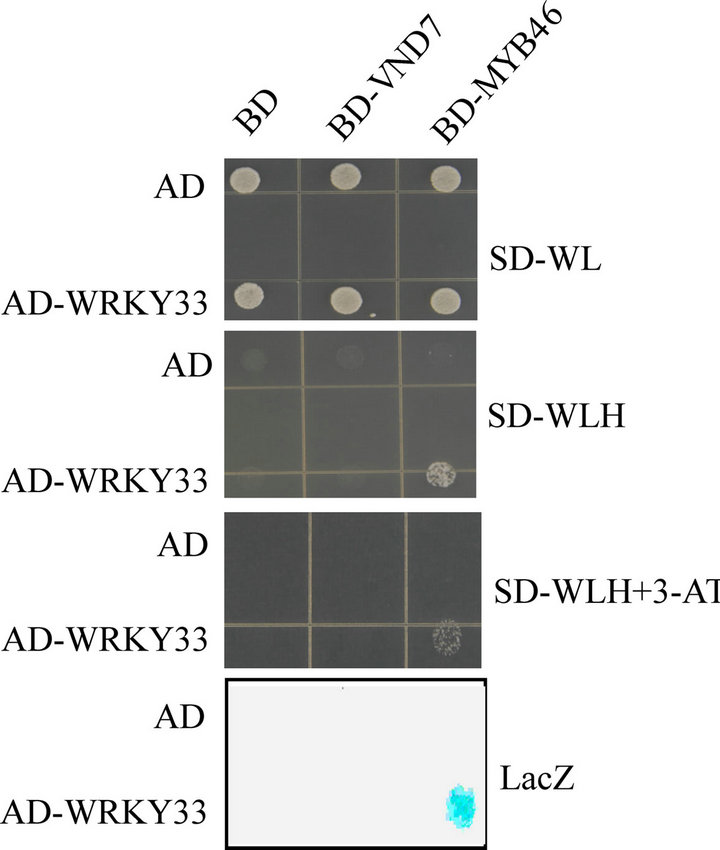
Figure 4. Y2H interaction assay of WRKY33 with MYB46 or VND7. The yeast strain PJ69-4A was co-transformed with bait (AD-WRKY33) and prey (BD-MYB46 or BDVND7) construct. The empty bait (AD) or prey (BD) vector was used as a negative control. Protein-protein interaction was examined by colony filter-lift assay (LacZ) and by cell growth on plates with SD-WLH medium or with same medium containing 5 mM 3-AT. The experiments were performed three times with consistent results.
WRKY33 with its roles in response to drought, salt stress [10] and pathogens deference [4,5].
Besides CesA8, five genes (At3g60120, At4g16260, At5g24540, At5g24550, and At5g24090) encoding glycosyl hydrolase, which play a role in cell wall remodeling, are identified as the target genes of WRKY33 [10]. These data as well as our results in this study indicate that WRKY33 may be involved in transcriptional regulation of cell wall synthesisand remodeling-related genes.
In conclusion, our results revealed that WRKY33 was involved in drought partly through a direct transcriptional repression of CesA8. These findings enriched our understanding of the function and molecular mechanism of WRKY33 in drought stress. Further examination of drought-related genes resulting from high CesA8 mRNA accumulation will be an interesting line to provide more insights into the molecular mechanism of WRKY33 in drought stress signaling.
4. Acknowledgements
We are grateful to Prof. Weiwei Zhang (College of life science, Capital Normal University, China) for critical reading and valuable suggestions. This work was supported by grants from the Beijing Natural Science Foundation (Grant no. 5112005) and the National Natural Science Foundation of China (Grant nos. 31271293 and 30770181) to X. Q.
REFERENCES
- T. Eulgem, P. J. Rushton, S. Robatzek and I. E. Somssich, “The WRKY Super Family of Plant Transcription Factors,” Trends in Plant Science, Vol. 5, No. 5, 2000, pp. 199-206. doi:10.1016/S1360-1385(00)01600-9
- P. J. Rushton, I. E. Somssich, P. Ringler and Q. J. Shen, “WRKY Transcription Factors,” Trends in Plant Science, Vol. 15, No. 5, 2010, pp. 247-258. doi:10.1016/j.tplants.2010.02.006
- B. Ülker and I. E. Somssich, “WRKY Transcription Factors: From DNA Binding towards Biological Function,” Current Opinion in Plant Biology, Vol. 7, No. 5, 2004, pp. 491-498. doi:10.1016/j.pbi.2004.07.012
- Z. Zheng, S. A. Qamar, Z. Chen and T. Mengiste, “Arabidopsis WRKY33 Transcription Factor Is Required for Resistance to Necrotrophic Fungal Pathogens,” The Plant Journal, Vol. 48, No. 4, 2006, pp. 592-605. doi:10.1111/j.1365-313X.2006.02901.x
- B. Lippok, R. P. Birkenbihl, G. Rivory, J. Brümmer, E. Schmelzer, E. Logemann and I. E. Somssich, “Expression of AtWRKY33 Encoding a Pathogenor PAMP-Responsive WRKY Transcription Factor Is Regulated by a Composite DNA Motif Containing W Box Elements,” Molecular Plant-Microbe Interaction, Vol. 20, No. 4, 2007, pp. 420-429. doi:10.1094/MPMI-20-4-0420
- J. L. Qiu, B. K. Fiil, K. Petersen, H. B. Nielsen, C. J. Botanga, S. horgrimsen, K. Palma, M. C. Suarez-Rodriguez, S. Sandbech-Clausen, J. Lichota, P. Brodersen, K. D. Grasser, O. Mattsson, J. Glazebrook, J. Mundy and M. Petersen, “Arabidopsis MAP Kinase 4 Regulates Gene Expression through Transcription Factor Release in the Nucleus,” EMBO Journal, Vol. 27, No. 16, 2008, pp. 2214- 2221. doi:10.1038/emboj.2008.147
- G. Mao, X. Meng, Y. Liu, Z. Zheng, Z. Chen and S. Zhang, “Phosphorylation of a WRKY Transcription Factor by Two Pathogen Responsive MAPKs Drives Phytoalex in Biosynthesis in Arabidopsis,” The Plant Cell, Vol. 23, No. 4, 2011, pp. 1639-1653. doi:10.1105/tpc.111.084996
- R. P. Birkenbihl, C. Diezel and I. E. Somssich, “Arabidopsis WRKY33 Is A Key Transcriptional Regulator of Hormonal and Metabolic Responses toward Botrytis Cinerea Infection,” Plant Physiology, Vol. 159, No. 1, 2012, pp. 266-285. doi:10.1104/pp.111.192641
- Y. Jiang and M. K. Deyholos, “Comprehensive Transcriptional Profiling of NaCl-Stressed Arabidopsis Roots Reveals Novel Classes of Responsive Genes,” BMC Plant Biology, Vol. 6, 2006, p. 25. doi:10.1186/1471-2229-6-25
- Y. Jiang and M. K. Deyholos, “Functional Characterization of Arabidopsis NaCl-Inducible WRKY25 and WRKY33 Transcription Factors in Abiotic Stresses,” Plant Molecular Biology, Vol. 69, No. 1-2, 2009, pp. 91-105. doi:10.1007/s11103-008-9408-3
- S. Li, Q. Fu, L. Chen, W. Huang and D. Yu, “Arabidopsis Thaliana WRKY25, WRKY26, and WRKY33 Coordinate Induction of Plant Thermotolerance,” Planta, Vol. 233, No. 6, 2011, pp. 1237-1252. doi:10.1007/s00425-011-1375-2
- M. Weber, A. Trampczynska and S. Clemens, “Comparative Transcriptome Analysis of Toxic Metal Responses in Arabidopsis Thaliana and the Cd2+-Hypertolerant Facultative Metallophyte Arabidopsis Halleri,” Plant Cell and Environment, Vol. 29, No. 5, 2006, pp. 950-963. doi:10.1111/j.1365-3040.2005.01479.x
- Y. Yuan, H. Wu, N. Wang, J. Li, W. Zhao, J. Du, D. Wang and H. Q. Ling, “FIT Interacts with AtbHLH38 and AtbHLH39 in Regulating Iron Uptake Gene Expression for Iron Homeostasis in Arabidopsis,” Cell Research, Vol. 18, No. 3, 2008, pp. 385-397. doi:10.1038/cr.2008.26
- J. Zeng, J. Yan, T. Wang, D. Mosbrook-Davis, K. T. Dolan, R. Christensen, G. D. Stormo, D. Haussler, R. H. Lathrop, R. K. Brachmann and S. M. Burgess, “Genome Wide Screens in Yeast to Identify Candidate Binding Sites and Target Genes of DNA-Binding Proteins,” Nucleic Acids Research, Vol. 36, No. 1, 2008, p. e8. doi:10.1093/nar/gkm1117
- N. G. Taylor, S. Laurie and S. R. Turner, “Multiple Cellulose Synthase Catalytic Subunits Are Required for Cellulose Synthesis in Arabidopsis,” The Plant Cell, Vol. 12, No. 12, 2000, pp. 2529-2539. doi:10.1105/tpc.12.12.2529
- Z. Chen, X. Hong, H. Zhang, Y. Wang, X. Li, J. K. Zhu and Z. Gong, “Disruption of the Cellulose Synthase Gene, AtCesA8/IRX1, Enhances Drought and Osmotic Stress Tolerance in Arabidopsis,” The Plant Journal, Vol. 43, No. 2, 2005, pp. 273-283. doi:10.1111/j.1365-313X.2005.02452.x
- W. C. Kim, J. H. Ko, J. Y. Kim, J. M. Kim, H. J. Bae and K. H. Han, “MYB46 Directly Regulates the Gene Expression of Secondary Wall-Associated Cellulose Synthases in Arabidopsis,” The Plant Journal, 2012. doi:10.1111/j.1365-313X.2012.05124.x
- M. Yamaguchi, N. Mitsuda, M. Ohtani, M. Ohme-Takagi, K. Kato and T. Demura, “Vascular-Related NAC-Domain7 Directly Regulates the Expression of a Broad Range of Genes for Xylem Vessel Formation,” The Plant Journal, Vol. 66, No. 4, 2011, pp. 579-590. doi:10.1111/j.1365-313X.2011.04514.x
- C. Hernández-Blanco, D. X. Feng, J. Hu, A. SánchezVallet, L. Deslandes, F. Llorente, M. Berrocal-Lobo, H. Keller, X. Barlet, C. Sánchez-Rodríguez, L. K. Anderson, S. Somerville, Y. Marco and A. Molina, “Impairment of Cellulose Synthases Required for Arabidopsis Secondary Cell Wall Formation Enhances Disease Resistance,” The Plant Cell, Vol. 19, No. 3, 2007, pp. 890-903. doi:10.1105/tpc.106.048058
NOTES
*Corresponding author.

