Advances in Microbiology
Vol.4 No.4(2014), Article ID:43993,9 pages DOI:10.4236/aim.2014.44027
Virulence Factors in Methicillin-Resistant Staphylococcus aureus Isolated from ICU Units in Brazil
Simone G. Souza1, Guilherme B. Campos2, Pollianna S. Oliveira1, Daniel S. Sousa1, Danilo C. C. Da Silva1, Verena M. Santos1, Aline T. Amorim1, Angelita M. O. G. Santos2, Jorge Timenetsky2, Mariluze P. Cruz1, Regiane Yatsuda1, Lucas M. Marques1,2*
1Instituto Multidisciplinar em Saúde, Núcleo de Tecnologia em Saúde, Universidade Federal da Bahia, Vitória da Conquista, Brazil
2Instituto de Ciências Biomédicas, Departamento de Microbiologia, Universidade de São Paulo, São Paulo, Brazil
Email: *lucasm@ufba.br
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/


Received 10 February 2014; revised 10 March 2014; accepted 17 March 2014
ABSTRACT
Species of Staphylococcus are common in hospital infection (HI). Methicillin resistant S. aureus (MRSA) has also become a serious problem in Brazilian HI. The aim of this study was to characterize the pathogenicity of methicillin-resistant S. aureus (MRSA) and methicillin-sensitive Staphylococcus aureus (MSSA) isolated in public hospitals. The clinical isolates were obtained from intensive care unit. The MRSA and MSSA strains were genotyped by PCR for detection genes related to virulence factors. Moreover, the strains were tested for biofilm formation and cytokine induction in macrophages. Three strains of MRSA (9.68%) expressed the Sea gene, one (3.23%) Seb, 17 (54.84%) Spa and seven (22.58%) had PVL. Two MSSA strains (2.98%) expressed the Sea gene, three (4.48%) Seb, 18 (26.87%) Spa and 11 (16.42%) showed positive results for the PVL gene. There was no expression of Sec and CflA between MRSA and MSSA strains. Among MRSA and MSSA isolates, none statistical differences were observed in biofilm production. The analysis of cytokine induction in the inflammatory response of J774 macrophages by MRSA and MSSA isolates did not show statistical difference. Understanding the mechanisms of pathogenesis of S. aureus could provide important clues for both preventing and treating infection caused by these organisms.
Keywords:MRSA; MSSA; Virulence Factor; Hospitalar Infection

1. Introduction
The Brazilian Ministry of Health defines Hospital Infection (HI) as that which is acquired during or after hospitalization, being possibly related to hospitalization or hospital procedures. HI has epidemiological significance and important economic costs [1] . HI usually depends on the severity of patient disease at hospitalization, the nutritional status, diagnostic and therapeutic procedures, length of hospital stay, routine care, technical procedures and other features [2] . HI occurs mainly in patients with multiple invasive procedures that are in Intensive Care Unit (ICU), usually with a kind of imunosupression that requires antibiotic therapy [3] .
Species of Staphylococcus are common in HI [4] and S. aureus methicillin resistant (MRSA) has also become a serious problem in Brazilian HI. A total of 28% prevalence of S. aureus resistance to methicillin/oxacillin was found in HI at a Hospital in Salvador/BA. The highest detection was observed at patients in the ICU (59%), hemodialysis (43%), infectious diseases (34%) and neonatal units (18.5%) compared with the other local hospital [5] .
MRSA isolates require phenotypic and genotypic characterization. Staphylococcal Chromosome Cassette mec (SCCmec) is a mobile genetic fragment in the chromosome of methicillin-resistant S. aureus (MRSA), of the mecA gene. Other genetic elements may also be present, such as genes for resistance to ß-lactam antibiotics and heavy metals. Six types of SCCmec have been identified [6] .
Knowledge of S. aureus virulence and pathogenicty aids in better understanding the diversity of infections due to exotoxins and surface virulence factors with adhesive properties for a range of molecules (MSCRAMMs). Among these, exotoxins are superantigens, some named enterotoxins A-E, G-K, M-O and Q, exfoliative toxins A and B, toxic shock syndrome toxin-1 as well as, Panton-Valentine leukocidin [7] . Biofilm production is another mechanism of therapy resistance and pathogenicity. Extracellular polysaccharide substances cause bacterial clusters in multilayer biofilm and inhibit the action of antibiotic and the immune system [8] . Based on these data, the aim of this study was to characterize the pathogenicity of MRSA and methicillin-sensitive Staphylococcus aureus (MSSA) isolated in two public hospitals in Vitória da Conquista—Bahia State (BA).
2. Methods
2.1. Staphylococcus aureus Isolates
The clinical isolates were obtained in a previous study [9] from intensive care unit environments and equipment surfaces in two public hospitals in the city of Vitória da Conquista, Bahia State, Brazil. The sampled sites were: floors, hospital cots, hospital cot control panels, heart monitors, hospital ventilator control panels, infusionpump control panels, blood-gas analyzer control panels, hospital incubators, telephones, scales, doors, tables, hospital beds, cabinets, emergency carts, medication carts, computers, air conditioners, faucets, handles, hospital countertops and prescription documents. S. aureus was isolated from 98 sites and 31 were MRSA. All MRSA isolates showed SCCmec type III genotype characteristics of the Brazilian epidemic clone associated with nosocomial infection [9] . And also, 60 MSSA were isolated from the sites of the hospital. The assays were done with 31 MRSA and 60 MSSA isolates.
2.2. Genotypic Characterization to Pathogenic Genes
Staphylococci cultures in 2 mililiter (mL) of TSB medium were harvested for DNA extraction [10] . The isolates were submitted to PCR for detection of genes; sea (Staphylococcal enterotoxins type A), seb (Staphylococcal enterotoxins type B), sec (Staphylococcal enterotoxins type C), PVL (Panton-Valentine Leucocidin), ClfA (Clumping factor) and spa (IgG-binding and X-region of protein A fragments) [11] . The primer sequences of the sea, seb, sec, PVL, spa and CflA genes are described in Table 1. The reaction volume was 50 mL containing 5 mL PCR buffer (500 mM KCl, 200 mM Tris, pH 8.4), 50 mM of each dNTP (dATP, dCTP, dGTP, and dTTP), 2.0 mM MgCl2, 20 pmol of each primer, 2.5 mL of the chromosomal DNA to be tested, and 1.5 units Ampli Taq DNA polymerase. For detection of sea, seb and sec genes, the amplification reaction consisted of one cycle at 94˚C for 5 minutes, followed by 35 cycles at 94˚C for 1 minute, 58˚C for 30 seconds and 72˚C for 1 minute, and a final cycle at 72˚C for 5 minute. For detection of PVL, ClfA and Spa genes, the amplification reaction consisted of one cycle at 94˚C for 5 minutes, followed by 35 cycles at 94˚C for 1 minute, 58˚C for 30 seconds and 72˚C for 1 minute, and a final cycle at 72˚C for 5 minute. The Polymerase chain reaction (PCR) amplified products were electrophoresed in 1% agarose with 0.5 mg ethidium bromide in 0.5 x Tris-EDTA electrophoresis
Table 1. Sequence of primers used for detection of virulence factors in methicillin-resistant Staphylococcus aureus (MRSA) and methicillin-sensitive Staphylococcus aureus (MSSA) isolated from two public hospitals in Vitória da Conquista, Bahia, Brazil.
buffer at 100 V and photographed in a UV transilluminator.
2.3. Biofilm Production
Biofilm assays were performed in 96-well polystyrene microplates, using trypticase soy broth (TSB) with 1% (w/v) glucose (TSB-1% Glc) [12] . Briefly, cultures of staphylococi in 5 mL were incubated in a shaker with 250 rpm at 37˚C for 18 hours (h). Cultures were diluted 1:100 in TSB-1% Glc and 200 mL were inoculated into each well. The microplate was incubated at 37˚C for 20 h. Supernatants were removed from each well and biofilms were gently washed twice with PBS, then dried and fixed at 65˚C for 1 h. Finally, the plates were stained with crystal violet 1% used in Gram-stain and gently washed twice with PBS. The absorbance at 492 nanometers (nm) was calculated in a spectophometer. The samples were compared with cultures of Streptococcus pyogenes ATCC75194. The S. aureus isolates were classified as non-biofilm producers, weak producers, moderate producers, producers, and strong producers. Because the production of biofilm depends on phase variation, tests were repeated four times. At least two independent experiments were carried out for each test. The cutoff point for the production was taken into account, the absorbance obtained by S. pyogenes (O.D.492 0.07). The mean value was used for the statistical calculation.
In addition, to confirm the differences between biofilm phenotypes, as determined by BU values, confocal laser scanning microscopy (CLSM) was used to obtain the structural images of the biofilms [13] . Here, the biofilm assays were performed at the same way, but after being fixed, the bacterial cells were stained with 25 nanomoles (nM) SYTO9 and propidium iodide (Live/Dead Bacteria—Invitrogen) for 15 minutes (min) in the dark. The stain was gently removed and biofilms were observed with a Confocal Laser Scanning Microscope—CLSM (Carl Zeiss LSM 510, Germany, equipped with Argon laser, 488 nm, and 2 helium/neon 543 nm wavelengths) to visualize the luminescence of fluochromes.
2.4. Cytokine Induction in Murine Macrophages
Staphylococcal cells were homogenized in 0.9% sodium chloride solution and the suspensions were adjusted to 0.5 × 108 CFU/mL by spectrophotometer. Then an aliquot of 100 mL was mixed with 2 ml of Minimum Essential Medium-MEM with 2mM of L-glutamine and Earl´s balanced salts, supplemented with 10% of fetal calf serum (Cult Lab, São Paulo, Brazil), and incubated in a shaker at 250 rpm at 35˚C for 24 hours. Subsequently, the cultures were filtrated through 0.22 micrometer (mm) pores. The filtrates were inoculated into J774 murine macrophages. The sets of inoculated cells were incubated at 37˚C in 5% CO2 atmosphere for 24 h. The supernatants were removed and the cytokines Tumor necrosis factor alpha (TNF-α), Interleukin 1 (IL-1), Interleukin 6 (IL-6) and Interleukin 10 (IL-10) were measured using enzyme-linked immunosorbent assay (ELISA), according to manufacturer instructions (eBioscience, San Diego, CA).
2.5. Statistical Analysis
Data were analyzed using GraphPad software. A nonparametric test, the Mann-Whitney U test was used to compare continuous variables between MRSA and MSSA data. Data was considered statistically significant at the p < 0.05 level.
3. Results
3.1. Genotypic Characterization to Pathogenic Genes
The MRSA and MSSA samples were genotyped by PCR for detection of genes sea (enterotoxin A), seb (B), sec (C), PVL (Panton-Valentine Leukocidin), ClfA (Clumping Factor A) and spa (protein A). Three strains of MRSA (9.68%) expressed the sea gene, one (3.23%) Seb, 17 (54.84%) spa and seven (22.58%) had PVL. Two MSSA strains (2.98%) expressed the sea gene, three (4.48%) Seb, 18 (26.87%) spa and 11 (16.42%) showed positive results to PVL gene. There was no expression of sec and CflA between MRSA and MSSA strains (Table 2).
3.2. Biofilm Production
The biofilm production of S. aureus isolates were observed among MRSA and MSSA strains of both hospitals. Among the 31 MRSA, six (19.35%) were not biofilm producers, six (19.35%) were low producers, five (16.13%) moderate producers, six (19.35%) were producers and eight (25.81%) were high producers. Among the 60 MSSA samples, four (6.67%) were low producers, 14 (23.33%) were low producers, 14 (23.33%) moderate producers, 17 (28.33%) were producers and 11 (18.33%) were high producers (Table 3). Biofilm formation with a thickness of about 19 µm is shown in Figure 1. There was no statistical difference in biofilm production between MRSA and MSSA isolates (p > 0.05), Mann Whitney test, GraphPad Prism® (Figure 2).
3.3. Cytokine Induction in Murine Macrophages Assay
The analysis of cytokine induction in the inflammatory response of J774 macrophages by MRSA and MSSA isolates did not show statistical difference in the levels of IL-6 (Figure 3), TNF-a (3B), IL-1 (3C) and IL-10 (3D)
Table 2. Determination of the sea, seb, sec, PVL, spa and CflA genes in methicillin-resistant Staphylococcus aureus (MRSA) and methicillin-sensitive Staphylococcus aureus (MSSA) isolated from two public hospitals in Vitória da Conquista, Bahia, Brazil.
Table 3. Biofilm production of MRSA and MSSA isolates obtained in two public hospitals in Vitória da Conquista, Bahia, Brazil.
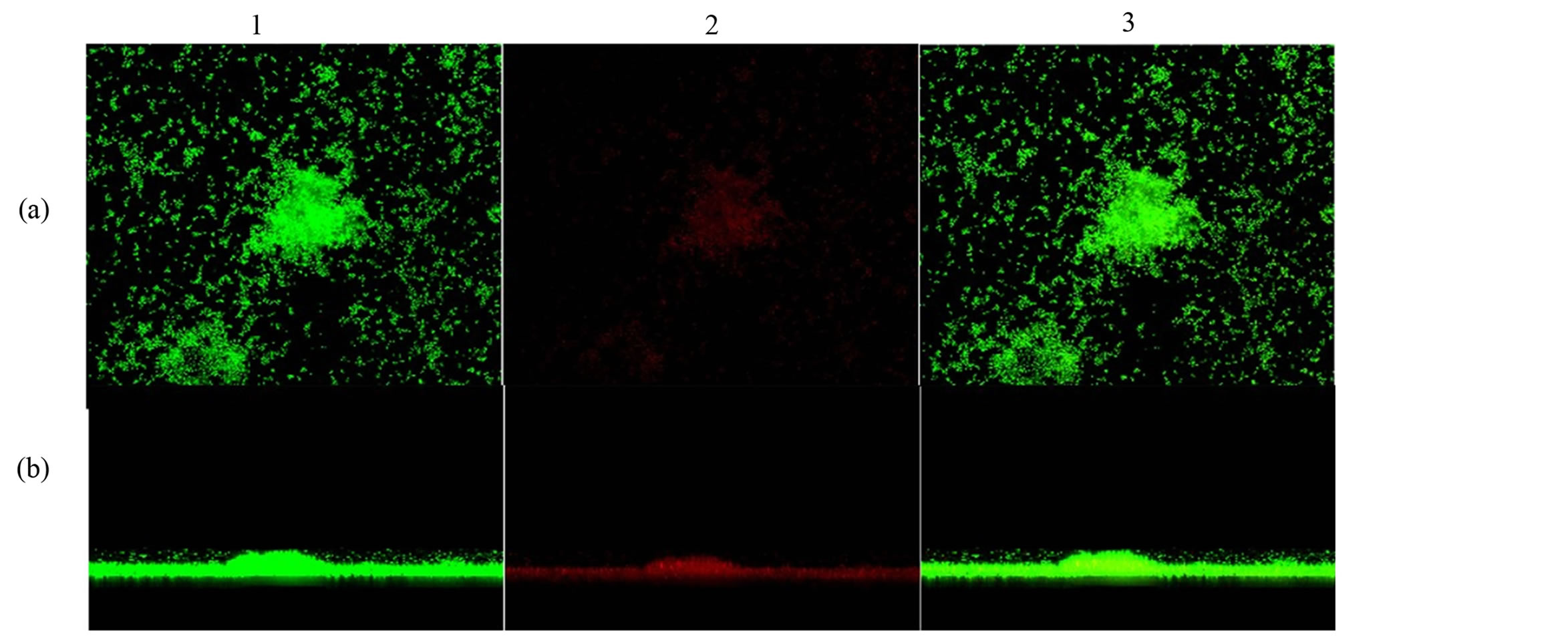
Figure 1. Confocal microscopy showing biofilm formation of Staphylococcus aureus samples isolated from hospital environments, and (a) shows the top view of the biofilm and (b) the side view of the biofilm. The microorganisms were marked with SYTO9 (green, 1) and unviable with propidium iodide (red, 2). Image 3 is an overlay of images 1 and 2. Magnification X2.
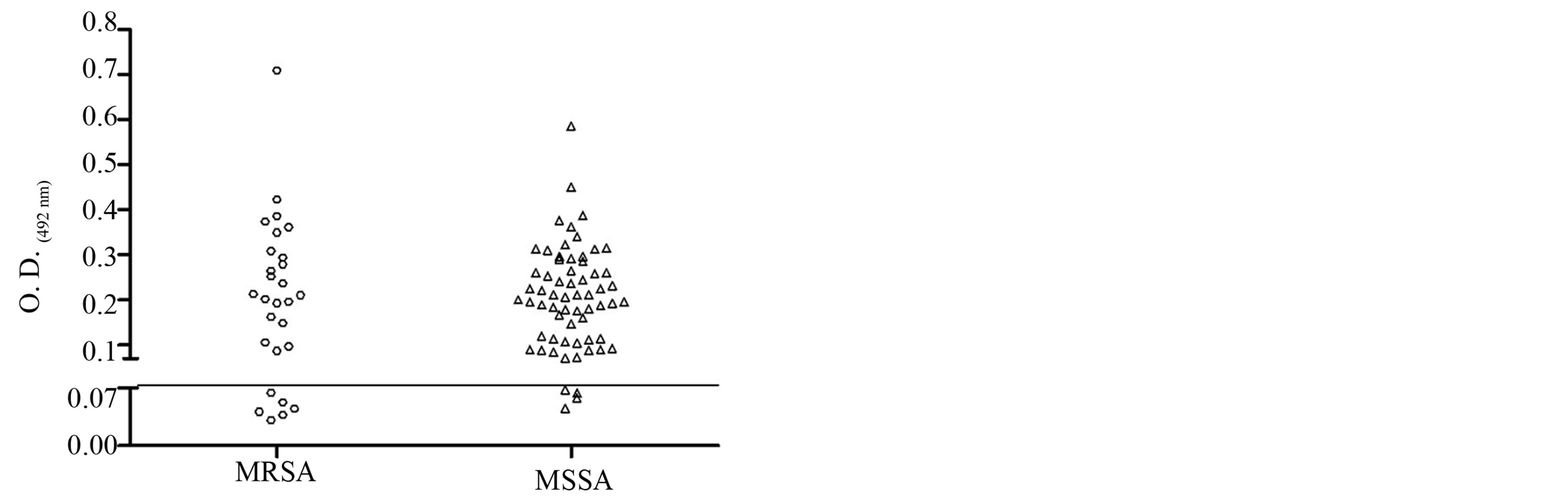
Figure 2. Dispersion analysis of the samples in relation to production of biofilm of MRSA and MSSA strains isolated in two hospitals in Vitória da Conquista, Bahia, Brazil. As the cutoff point for the production was taken into account, the absorbance obtained by S. pyogenes (O.D.492 0.07). There was no statistical difference in biofilm formation between MRSA and MSSA isolates obtained (p > 0.05, Mann Whitney test, GraphPad Prism®).
production (p > 0.05, Mann Whitney test, GraphPad Prism®).
4. Discussion
Critically, ill patients in ICU are especially vulnerable to HI compared to patients in other hospital units. Some studies have found that ICU patients are 5 to 10 times more likely to acquire an HI, and that this sector can accommodate approximately 20% of all hospital infections [14] [15] . S. aureus is part of the human microbiota; however, it is an important causative agent of infections related to health care service, including bacteremia, pneumonia, osteomyelitis, endocarditis and toxic shock syndrome [16] [17] . This bacterium can also cause disease in both healthy individuals as well as in immunocompromised individuals [18] .
According to Souza and Figueredo [16] , about 70% of the isolates of S. aureus causing infections related to primary health care in Brazilian hospitals are MRSA. The range of prevalence of HI by S. aureus in Brazil is from 17% to 26%, and about 70% to 100% are caused by multidrug-resistant strains [19] . In the United States
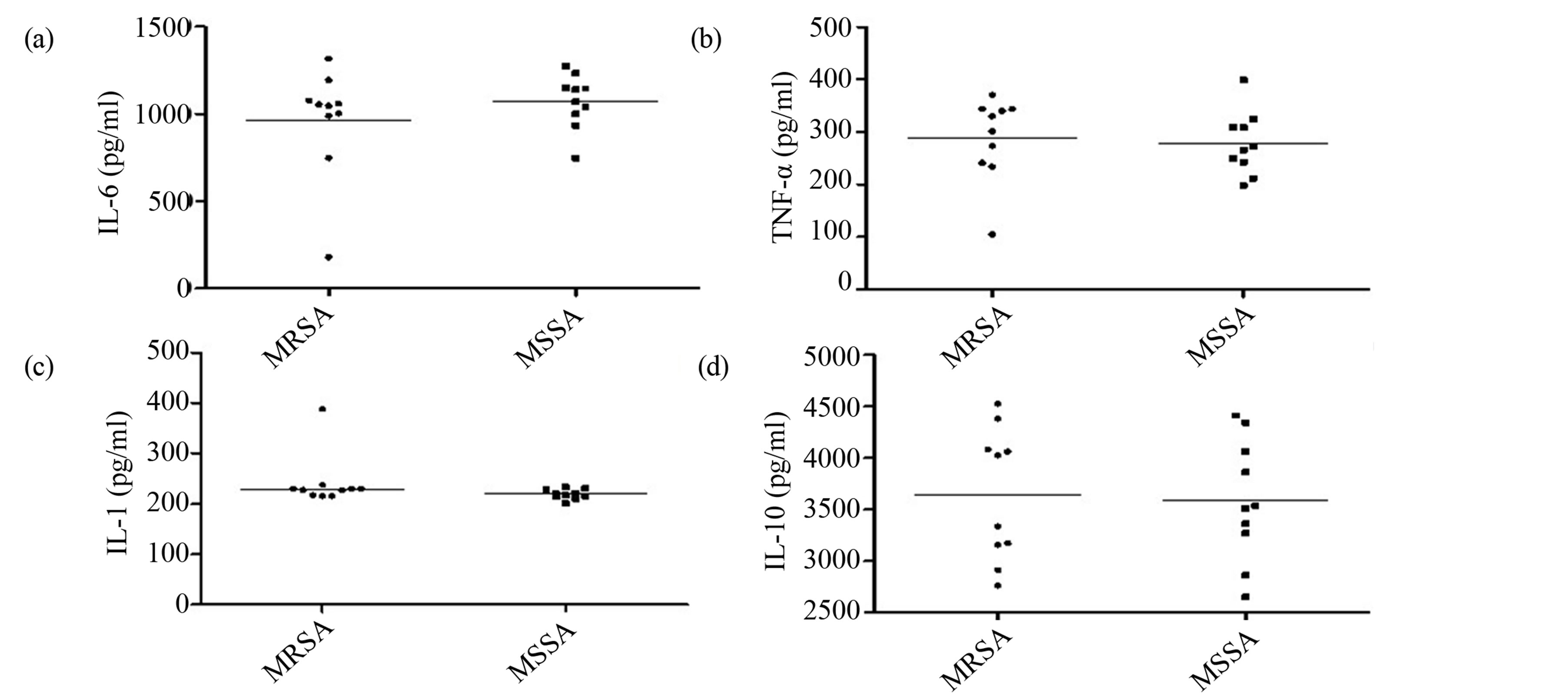
Figure 3. Production of cytokines involved in the inflammatory response by MRSA and MSSA strains isolated in two hospitals in Vitória da Conquista, Bahia, Brazil. There was no statistical difference in IL-6 (a), TNF-a (b), IL-1 (c) and IL-10 (d) induction between MRSA and MSSA isolated (p > 0.05, Mann Whitney test, GraphPad Prism®).
there is about 94,000 individuals infected with MRSA and about 18% result in death. Most invasive MRSA infections (86%) occurred in individuals with previous exposure to a hospital environment or the health care service [20] .
S. aureus has different mechanisms of virulence, pathogenicity and favors the development of antibiotic resistance and increases vulnerability to infection [19] . Infections associated with biofilm production are generally recurring, since the conventional antimicrobial therapy predominantly eliminates planktonic forms, leaving the sessile cells free to reproduce and propagate the biofilm after treatment. In even worse situations, the bacteria in biofilms are more protected against the host immune system. Typical examples of biofilm-associated diseases include infections caused by heart valve implants, catheters, and contact lenses, among others [21] .
In the present study, the biofilm production of MRSA and MSSA isolates was detected, however, there was no statistical difference among both types of S. aureus and sampled hospitals. There are few studies in the literature about the biofilm production of staphylococci isolates recovered from hospital environments. Smith et al. [22] studied MRSA isolates from a hospital and found that 25.7% were low biofilm producers, 53.8% were moderate and 20.5% were biofilm producers. In the same study, compared to MSSA isolates, 28.5% were non-biofilm producers, 43.5% were moderate and 28.0% were biofilm producers. The data indicate that there is no correlation of methicillin resistance and the ability to produce biofilm. Similarly, other study [23] found that 45% of MRSA and 66% of MSSA were biofilm producers, and there was also no relationship with antibiotic resistance. The differences of the biofilm producers and antibiotic resistant isolates may be related to places of isolation and geographical variation in the different genotypes of MRSA. The non relationship of the MRSA and biofilm-producing isolates may be explained by the adhesion phase of bacterial cells, which is an approximation of the surface of the biofilm microorganisms making the bacteria susceptible to the antimicrobial action [22] [24] .
Biofilm producing bacteria are also important to human health mainly in hospital transmission [25] . This feature is related to the high incidence of local and systemic infections, the restriction of the antimicrobial treatment, increase of health care costs and mortality [21] . Another study [26] reported that 65% of bacterial human infections may be associated with biofilm production. This may be caused by devices implanted in patients, such as vascular and urinary catheters, which may increase morbidity and mortality.
In the present study, some virulence genes were analyzed in MRSA and MSSA isolates. The sea, seb, spa and PVL genes were detected in both staphylococci biotypes, but not the sec and CflA genes. The virulence genes of S. aureus described in the literature show variations. In Brazil, a study [26] detected that PVL was rarely present in MRSA and MSSA hospital isolates. In the present study, the PVL gene was detected in seven MRSA isolates and eleven isolates MSSA, a total of eighteen (18/50). Souza et al. [26] , for the gene seb, found that it was detected in three MSSA isolates (3/50) and in four isolates MRSA (4/50), collectively accounting for 3.3% of the total isolates analyzed (7/214). Kim et al. [27] observed that none of the MRSA isolates of the SCCmecIII type carried the seb and sec genes. These results are consistent with the present study in regard to sec gene detection. Another authors studied MRSA and MSSA isolates obtained from a University Hospital and more frequently detected the genes related to toxins (sea, seb, sed, seg, sei, sej, and eta), and, the pvl, tst and sec genes were more frequent in MSSA [28] . Aung et al. [7] verified that the MRSA clinical strains had only a few or no staphylococcal enterotoxin (SE) genes, whereas the PVL gene was detected in MSSA and MRSA isolates recovered from a healthy adult possessing an enterotoxin gene cluster (seg, sei, sem, sen, seo, and selu).
In another study, approximately 50% of all isolates produced at least one enterotoxin and 21.5% of the S. aureus isolates from produced PVL. Genes encoding clumping factor B, and elastin and laminin binding proteins were detected in almost all isolates (80%), irrespective of the geographical origin [29] . Despite the fact that these genes are carried by mobile genetic elements and, thus, could theoretically be present or absent in different isolates of a specific lineage, the existence of a correlation of a specific clone type and superantigen profiles, in a hospital or in a geographical area, should be investigated in order to trace potential staphylococcal virulence syndrome-associated isolates.
No statistical difference was obtained among the studied staphylococcal isolates for the production of inflammatory cytokines. In fact, these compounds are induced mainly by the exocellular lipoteichoic acid of S. aureus [30] . In animal models, lipoteichoic acid can induce features of sepsis such as delayed circulatory failure with hypotension and multiple organ failure [31] . Jones et al. [32] demonstrated that the staphylococcal exocellular lipoteichoic acid is a potent activator of pro-inflammatory cytokines (TNF-a, IL-6 and IL-1) and nitric oxide in a murine macrophage cell line. The exocellular lipoteichoic acid is significantly more active than that of lipoteichoic acid, peptidoglycan or wall teichoic acid, especially for TNF-a and nitric oxide production. Other virulence factors could be associated with the intensive inflammatory response, such as PVL [33] or entetotoxin [34] but in the present study, the relationship between the presence of these genes and increased production of cytokines was not observed.
The incidence of infections by staphylococci, mainly S. aureus, has risen significantly over the past two decades. This follows the increased use of implanted medical devices such as central venous catheters, continuous ambulatory peritoneal dialysis catheters, prosthetic hip joints, and cardiac and vascular prostheses. Understanding the mechanisms of pathogenesis of S. aureus could provide important clues to both preventing and treating infection caused by these organisms. Although products such as extracellular slime, lipase, haemolysins and receptors for collagen, lami laminin, vitronectin and fibronectin enhance pathogenicity of S. aureus, no single determinant has proven to be essential for virulence. The importance of identify early S. aureus in hospitals is becoming increasingly urgent because of its high prevalence and their resistance mechanisms, which are crucial to its considerable virulence. Consequently, patients submitted to antimicrobial therapy, prolonged hospitalization and the use of invasive devices are more inclined to acquire a HI. The findings of this study should assist in reducing the occurrence of nosocomial infections and, therefore, the morbidity, mortality and socio-economic burden caused by prolonged hospitalization.
Acknowledgments
We thank the hospitals for valuable assistance and AcademicEnglishSolutions.com for proofreading.
Funding
This study was supported by FAPESB (N.SUS0012/2009) and PIBIC/CNPq.
Transparency Declaration
The authors declare that they have no conflicts of interest.
References
- Pereira, M.S., Souza, A.C.S., Tipple, A.F.V. and Prad, M.A. (2005) Infecção hospitalar e suas Implementações para o cuidado da enfermagem. Texto & Contexto Enfermagem, 14, 250-257. http://dx.doi.org/10.1590/S0104-07072005000200013
- Pilonetto, M., Rosa, E.A.R., Brofman, P.R.S., et al. (2004) Hospital Gowns as a Vehicle for Bacterial Dissemination in an Intensive Care Unit. Brazilian Journal of Infectious Diseases, 8, 206-210. http://dx.doi.org/10.1590/S1413-86702004000300003
- Martins, S.T., Moreira, M. and Furtado, G.H. (2004) Application of Control Measures for Infections Caused by Multi-Resistant Gram-Negative Bacteria in Intensive Care Unit Patients. The Memórias do Instituto Oswaldo Cruz, 99, 331-334. http://dx.doi.org/10.1590/S0074-02762004000300017
- Dos Santos, A.L., Santos, D.O., Freitas, C.C., et al. (2007) Staphylococcus aureus: visitando uma cepa de importância hospitalar. Jornal Brasileiro de Patologia e Medicina Laboratorial, 43, 414.
- Brites, C., Silva, N. and Sampaio-Sá, M. (2006) Temporal Evolution of the Prevalence of Methicillin-Resistant Staphylococcus aureus in a Tertiary Hospital in Bahia, Brazil. A Nine-Year Evaluation Study. Brazilian Journal of Infectious Diseases, 10, 235-238. http://dx.doi.org/10.1590/S1413-86702006000400003
- Appelbaum, P.C. (2007) Microbiology of Antibiotic Resistance in Staphylococcus aureus. Clinical Infectious Diseases, 45, 165-170. http://dx.doi.org/10.1086/519474
- Aung, M.S., Urushibara, N., Kawaguchiya, M., et al. (2011) Virulence Factors and Genetic Characteristics of Methicillin-Resistant and Methicillin-Susceptible Staphylococcus aureus Isolates in Myanmar. Microbial Drug Resistance, 17, 525-535. http://dx.doi.org/10.1089/mdr.2011.0061
- Antunes, A.L.S., Perez, L.R.R., Reiter, K.C., et al. (2007) Detecção da Produção de Biofilmes em Staphylococcus ssp. por Agar Congo Red. Revista de Saúde da Universidade Católica de Pelotas, 1, 1.
- Campos, G.B., Souza, S.G., Lobão, T.N., et al. (2012) Isolation, Molecular Characteristics and Disinfection of Methicillin-Resistant Staphylococcus aureus from ICU Units in Brazil. New Microbiologica, in Press.
- Fan, H.H., Kleven, S.H. and Jackwood, M.W. (1995) Application of Polymerase Chain Reaction with Arbitrary Primers to Strain Identification of Mycoplasma gallisepticum. Avian Diseases, 39, 729-735. http://dx.doi.org/10.2307/1592409
- Proietti, P.C., Coppola, G., Bietta, A., et al. (2010) Characterization of Genes Encoding Virulence Determinants and Toxins in Staphylococcus aureus from Bovine Milk in Central Italy. The Journal of Veterinary Medical Science, 72, 1443-1448. http://dx.doi.org/10.1292/jvms.10-0158
- Cassat, J.E., Lee, C.Y. and Smeltzer, M.S. (2007) Investigation of Biofilm Formation in Clinical Isolates of Staphylococcus aureus. Methods in Molecular Biology, 391, 127-144. http://dx.doi.org/10.1007/978-1-59745-468-1_10
- Coelho, L.R., Souza, R.R., Ferreira, F.A., et al. (2008) agr RNAIII Divergently Regulates Glucose-Induced Biofilm Formation in Clinical Isolates of Staphylococcus aureus. Microbiology, 154, 3480-3490. http://dx.doi.org/10.1099/mic.0.2007/016014-0
- Vincent, J.L. (2003) Nosocomial Infections in Adult Intensive-Care Units. Lancet, 361, 2068-2077. http://dx.doi.org/10.1016/S0140-6736(03)13644-6
- Lima, M.E., Andrade, D. and Haas, V.J. (2008) Avaliação Prospectiva da Ocorrência de Infecção em Pacientes Críticos de Unidade de Terapia Intensiva. Revista Brasileira de Terapia Intensiva, 19, 134-137.
- Souza, L.B. and Figueiredo, B.B. (2008) Prevalência de Infecções Nosocomiais Provocadas por Staphylococcus aureus Resistente à Meticilina (M.R.S.A.), no Hospital Universitário Regional de Maringá. RBAC, 40, 31-34.
- Calderón, E.J., Monteros, L.E.E. and Beltran, R.A. (2002) Epidemiology of drug resistance: The case of Staphylococcus aureus and coagulase-negative staphylococci infections. Salud Pública de México, 44, 108-112. http://dx.doi.org/10.1590/S0036-36342002000200004
- Menegotto, F.R. and Picoli, S.U. (2007) Staphylococcus aureus oxacilina resistente (MRSA): incidência de cepas adquiridas na comunidade (CA-MRSA) e importância da pesquisa e descolonização em hospital. RBAC, 39, 147-150.
- Almeida, M.I., Bedendo, J., Cavasin, E.D. and Tognim, M.C.B. (2007) Prevalência e perfil de sensibilidade de amostras de Staphylococcus aureus isoladas de casos clínicos de infecções hospitalares. Revista Eletrônica de Enfermagem, 9, 489-495.
- Moura, J.P., Gir, E., Rosa, J.O., et al. (2010) Resistência à mupirocina entre isolados de Staphylococcus aureus de profissionais de enfermagem. Acta Paul Enferm, 23, 399-403. http://dx.doi.org/10.1590/S0103-21002010000300014
- Gotz, F. (2002) Staphylococcus and Biofilms. Molecular Microbiology, 43, 1367-1378. http://dx.doi.org/10.1046/j.1365-2958.2002.02827.x
- Smith, K., Perez, A., Ramage, G., Lappin, D., Gemmell, C.G. and Lang, S. (2008) Biofilm Formation by Scottish Clinical Isolates of Staphylococcus aureus. Journal of Medical Microbiology, 57, 1018-1023. http://dx.doi.org/10.1099/jmm.0.2008/000968-0
- Grinholc, M., Wegrzyn, G. and Kurlenda, J. (2007) Evaluation of Biofilm Production and Prevalence of the icaD Gene in Methicillin-Resistant and Methicillin-Susceptible Staphylococcus aureus Strains Isolated from Patients with Nosocomial Infections and Carriers. FEMS Immunology & Medical Microbiology, 50, 375-379. http://dx.doi.org/10.1111/j.1574-695X.2007.00262.x
- Nunes, M.C.P., Cassati, M.Z., Villalpando, K.T. and Cirano, F.R. (2007) Contribution of Dental Biofilm Study to the Treatment of Periodontal Diseases. Revista do Instituto de Ciências da Saúde, 25, 55-61.
- Thomson, C.H. (2011) Biofilms: Do They Affect Wound Healing? International Wound Journal, 8, 63-67. http://dx.doi.org/10.1111/j.1742-481X.2010.00749.x
- Souza, R.R., Coelho, L.R., Botelho, A.M., Ribeiro, A., Rito, P.N., Vieira, V.V., Teixeira, L.A., Ferreira-Carvalho, B.T. and Figueiredo, A.M.S. (2009) Biofilm Formation and Prevalence of lukf-pv, seb, sec and tst Genes among Hospitaland Community-Acquired Isolates of Some International Methicillin-Resistant Staphylococcus aureus Lineages. Clinical Microbiology and Infection, 15, 203-207. http://dx.doi.org/10.1111/j.1469-0691.2008.02118.x
- Kim, J.S., Song, W., Kim, H.S., Chan Cho, H., Lee, K.M., Choi, M.S. and Kim, E.C. (2006) Association between the Methicillin Resistance of Clinical Isolates of Staphylococcus aureus, Their Staphylococcal Cassette Chromosome mec (SCCmec) Subtype Classification, and Their Toxin Gene Profiles. Diagnostic Microbiology & Infectious Disease, 56, 289-295. http://dx.doi.org/10.1016/j.diagmicrobio.2006.05.003
- Sila, J., Sauer, P. and Kolar, M. (2009) Comparison of the Prevalence of Genes Coding for Enterotoxins, Exfoliatins, PantonValentine Leukocidin and Tsst-1 between Methicillin-Resistant and Methicillin-Susceptible Isolates of Staphylococcus aureus at the University Hospital in Olomouc. Biomedical papers of the Medical Faculty of the University Palacky, 153, 215-218. http://dx.doi.org/10.5507/bp.2009.036
- Baba-Moussa, L., Anani, L., Scheftel, J.M., Couturier, M., Riegel, P., Haïkou, N., Hounsou, F., Monteil, H., Sanni, A. and Prévost, G. (2008) Virulence Factors Produced by Strains of Staphylococcus aureus Isolated from Urinary Tract Infections. Journal of Hospital Infection, 68, 32-38. http://dx.doi.org/10.1016/j.jhin.2007.10.010
- Bhakdi, S., Klonisch, T., Nuber, P. and Fischer, W. (1991) Stimulation of Monokine Production by Lipoteichoic Acids. Infection and Immunity, 59, 4614-4620.
- De Kimpe, S.J., Kengatharan, M., Thiemermann, C. and Vane, J.R. (1995) The Cell Wall Components Peptidoglycan and Lipoteichoic Acid from Staphylococcus aureus Act in Synergy to Cause Shock and Multiple Organ Failure. Proceedings of the National Academy of Sciences of the United States of America, 92, 10359-10363. http://dx.doi.org/10.1073/pnas.92.22.10359
- Jones, K.J., Perris, A.D., Vernallis, A.B., Worthington, T., Lambert, P.A. and Elliott, T.S.J. (2005) Induction of Inflammatory Cytokines and Nitric Oxide in J774.2 Cells and Murine Macrophages by Lipoteichoic Acid and Related Cell Wall Antigens from Staphylococcus epidermidis. Journal of Medical Microbiology, 54, 315-321. http://dx.doi.org/10.1099/jmm.0.45872-0
- Lo, W.T., Tang, C.S., Chen, S.J., Huang, C.F., Tseng, M.H. and Wang, C.C. (2009) Panton-Valentine Leukocidin Is Associated with Exacerbated Skin Manifestations and Inflammatory Response in Children with Community-Associated Staphylococcal Scarlet Fever. Clinical Infectious Diseases, 49, e69-e75. http://dx.doi.org/10.1086/605580
- Dauwalder, O., Thomas, D., Ferry, T., Debard, A.L., Badiou, C., Vandenesch, F., Etienne, J., Lina, G. and Monneret, G. (2006) Comparative Inflammatory Properties of Staphylococcal Superantigenic Enterotoxins SEA and SEG: Implications for Septic Shock. Journal of Leukocyte Biology, 80, 753-758. http://dx.doi.org/10.1189/jlb.0306232
NOTES
*Corresponding author.


