Advances in Microbiology
Vol. 3 No. 2 (2013) , Article ID: 32341 , 6 pages DOI:10.4236/aim.2013.32024
Molecular Evaluation of the Enterotoxigenicity of Clostridium difficile and Clostridium perfringens Swine Isolates by PCR Assays
Department of Veterinary Medical Science—Unit of Infectious Diseases and Microbiology, University of Parma, Parma, Italy
Email: *mariacristina.ossiprandi@unipr.it
Copyright © 2013 Maria Cristina Ossiprandi, Laura Zerbini. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received March 20, 2013; revised April 20, 2013; accepted May 20, 2013
Keywords: Clostridium difficile; Clostridium perfringens; Toxinotyping; Swine; PCR Assays
ABSTRACT
Clostridium difficile and C. perfringens are enteric pathogens affecting a variety of mammals. This study evaluated the molecular enterotoxigenicity of Clostridium swine isolates by PCRs. One hundred and ten swine faeces were analyzed by culture assay. The faecal samples were from sixty-seven healthy animals and 43 with gastrointestinal tract disease. C. difficile strains were PCR-screened for the presence of tcdA/tcdB and cdtA/cdtB genes. All C. perfringens isolates were tested for the characterization of the toxinotype. Overall, sixty-five swine resulted positive: 38 for C. difficile and 17 for C. perfringens. One sample tested C. perfringens and C. difficile-positive, at the same time: on the whole, 39 C. difficile strains were isolated. Thirty-eight C. difficile isolates (all from healthy animals) resulted tcdA/tcdB and cdtA/cdtBnegative by PCRs and toxins A/B-negative by immunological tests. All C. perfringens strains were type A; eight were also cpb2-positive. In the sample (diarrhoeic), with double infection, C. difficile tested tcdA/tcdB and cdtA/cdtB-positive by PCRs and toxins A/B-positive by immunoassays; C. perfringens resulted cpb2-positive. The molecular genotypeing/toxinotyping should be applied to establish a final diagnosis and to assess properly the full implications and the epidemiological impact of these findings in particular in samples of healthy animals and aid in the development of effective intervention methods for controlling clostridial disease outbreaks.
1. Introduction
Over the past decade Clostridium difficile has emerged as an important enteric pathogen in human [1] and veterinary medicine [2]. Clostridium perfringens has been associated with enterocolitis in animals, including horses and humans [3,4].
Clostridium difficile is a Gram-positive, anaerobic, bacterium forming environmentally hardy spores. Enteric infection caused by C. difficile has emerged as a common diagnosis in neonatal pigs in recent years. This pathogen is known to cause disease in a variety of other animals, including calves, lambs, dogs, and horses. C. difficile has also been associated with hospitalization and antibiotic use in humans, and recently there have been epidemic outbreaks of C. difficile-infection (CDI) due to the emergence of a hypervirulent strain in hospitals worldwide.
This strain is a toxinotype III (ribotype 027) strain, contains the binary toxin (CDT, adenosine diphosphate-ribosyltransferase), and has an 18-bp deletion in the tcdC regulatory gene [5].
Lesions in non-human mammals are similar to those in humans, but vary widely in severity and distribution within the gastrointestinal tract. This variation is evident for different species and different age groups within a species [2].
Clinical signs and lesions may be mild, as in porcine neonatal colitis, but range to elevated leukocyte count, abdominal pain, profuse watery diarrhoea, anorexia, lethargy, and death in humans. Collective pathology is comprised of pseudomembrane formation, inflammation, necrosis, and an intercryptal exudate of neutrophils and fibrin (“volcano lesions”) [6,7]. Diarrhoea is variably present and some pigs with mild disease are apparently obstipated. Other clinical signs of disease include dyspnea, mild abdominal distension, and scrotal edema [2]. More than 50% of preweaning deaths in intensively raised calves may be due to diarrhoeal disease [7].
The pathophysiology of CDI involves colonization of the intestinal tract with C. difficile and a production of specific toxins [8].
Virulent strains of C. difficile are associated with two toxins: the enterotoxin TcdA (toxin A) and the cytotoxin TcdB (toxin B) [5].
TcdA and TcdB are encoded by two separate genes, tcdA and tcdB, located in a 19.6-kb pathogenicity locus (PaLoc). Some strains also produce binary toxin, as above mentioned, which is encoded by the genes cdtA and cdtB, located outside PaLoc. The real role of binary toxin in disease is currently under investigation [8].
Clostridium perfringens is commonly found in the environment and in the gastrointestinal tract of a variety of mammals and birds where it is considered a part of the normal bacterial flora [3,9]. It is also recognized as an important pathogen in domestic animals, wildlife, and humans. C. perfringens can cause gas gangrene and food poisoning in humans; necrotic enteritis in poultry; enterotoxaemia in lambs and calves; and enteritis in pigs, cattle, dogs, and horses [3,10,11].
Clostridium perfringens is a Gram-positive, anaerobic, oxygen-tolerant, rod-shaped bacterium. Like all bacterial species, C. perfringens can be subdivided into strains according to the results of different typing methods. Although subdivision by serotyping was proposed in the past, division into strains according to the combinations of toxins produced (or toxinotypes) is still the most widespread and routinely useful method today. Genotyping is generally used only in PCR analysis of toxin genotype [12]. C. perfringens can produce up to 30 potential toxins, and strains are traditionally classified into five categories (A, B, C, D and E) according to the combination of the four major toxins (α, β, ι ed ε) they produce (Table 1) [12]. These five types can be further subdivided according to the production of two additional toxins: the enterotoxin (CPE) (encoded by the cpe gene)
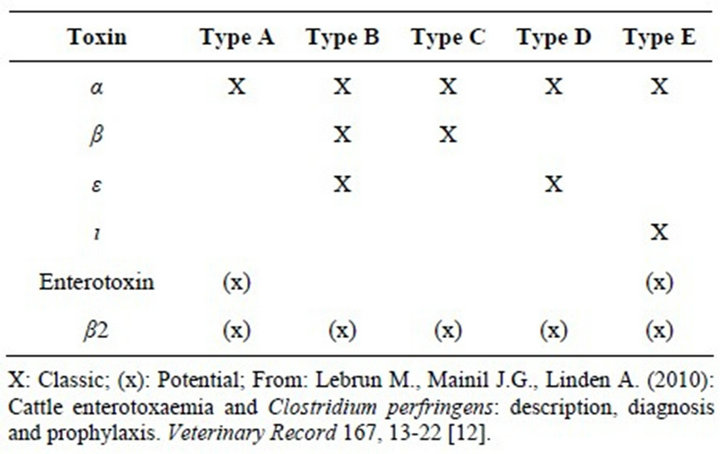
Table 1. Clostridium perfringens conventional toxinotypes.
and the β2 toxin (encoded by the cpb2 gene) and numerous so-called minor toxins (Table 1) [12].
Type A strains cause most pathologies associated with C. perfringens in human beings: gas gangrene (type A, non-enterotoxigenic), sporadic or antibiotic-associated diarrhoea (type A, ± enterotoxigenic, ± cpb2) and food poisoning (type A or D, enterotoxigenic) [12]. Necrotising enteritis (type C) is also seen in human beings [13]. In animals, the five toxinotypes cause numerous forms of enteritis and enterotoxaemia [12].
Regardless of the type, C. perfringens isolates can also produce β2-toxin and CPE. The β2-toxin has been associated with the onset of enteritis in pigs, horses, and cattle, and appears to have similar, but weaker, biological activity as the β-toxin [14]. Enterotoxin has been associated with diarrhoeal disease in some animal species pigs, cats and dogs and, more importantly, with food poisoning in humans [3,11,15].
Several techniques have been used to type C. difficile and C. perfringens in both humans and animal species [5].
The common typing methods include multilocus sequence typing (MLST), pulsed-field gel electrophoresis (PFGE), random amplified polymorphic DNA (RAPD) typing, PCR ribotyping, and toxinotyping [5]. Generally, these methods have been used to type C. perfringens in attempts to differentiate pathogenic strains from commensals and to type C. difficile as an epidemiology tool to identify clusters or strains that are associated with disease outbreaks. Understanding the diversity of toxigenic strains in commercial swine herds may lead to a greater understanding of the pathogenesis of Clostridium in neonatal pigs and aid in the development of effective intervention methods for controlling clostridial disease outbreaks [5].
Therefore, the objective of the current study was to investigate the molecular characteristics of various strains of C. difficile and C. perfringens isolated from healthy and diarrhoeic swine through the use of toxin gene profiling.
2. Materials and Methods
2.1. Samples
One hundred and ten swine samples (all faeces) were collected, using a stratified random sampling, from different farms in the area of Parma and Reggio Emilia provinces (Italy) during the period beginning of 2008 to end of 2011. The faecal samples were from sixty-seven healthy animals and 43 with gastrointestinal tract disease.
All faecal specimens were naturally voided. Assays were performed on faeces within 3 hours from the collection, after which they were immediately stored at −20˚C.
2.2. Faecal Sample Culture
All faecal samples were cultured onto pre-reduced Schaedler agar plates (Oxoid, Basingstoke, Hampshire, England), and at the same time inoculated into cooked meat broth (Oxoid, England). Samples were also streaked onto pre-reduced selective medium containing cycloserinecefoxitin-fructose agar (CCFA) for C. difficile isolation. Plates were incubated anaerobically at 37˚C for 48 - 72 hours. After 3 days of incubation into cooked meat broth, the samples were subjected to heat shock for spore selection at 100˚C for 5 min., followed by subculture onto Schaedler agar and/or CCFA plates.
Preliminary identification of C. difficile was based on colony morphology, odor (horse manure), lack of aerotolerance and cellular morphology following Gram staining. Species identity was confirmed through the rapid latex slide agglutination test (C. difficile, Oxoid, England) and Rapid ID32A (bioMérieux SA, Marcy-l’Etoile, France).
Isolates which were anaerobic, Gram-positive, rodshaped, and produced a double zone of haemolysis on blood were preliminarily considered to be C. perfringens. Reverse Christie-Atkins-Munch-Peterson (CAMP) testing [16] was performed on colonies accompanied by positive controls (Streptococcus agalactiae ATCC 27956 and C. perfringens internal control of Istituto Zooprofilattico Sperimentale della Lombardia e dell’Emilia RomagnaSezione di Parma, Italy).
All isolates were stored on cryopreservation beads (MAST Diagnostics, D.I.D, Diagnostic International Distribution S.p.A., Italy) at −70˚C.
2.3. Reference Strains for PCRs
C. difficile VPI 10463 and 51377 were used as C. difficile tcdA+/tcdB+ and cdtA+/cdtB+ controls, respectively. C. perfringens ATCC 12917 cpa+/cpe+ was utilized as positive control for duplex and multiplex PCRs. C. perfringens NCTC 8346, ATCC 373, and ATCC 27324 were used as cpa+/etx+, cpa+/cpb+/cpb2+ and cpa+/iap+ /cpe+/cpb2+ controls, respectively, for multiplex PCR.
2.4. Rapid Immunoassays
The, in vitro, toxin production by C. difficile was detected by two distinct immunological tests (ProSpecT Clostridium difficile Toxin A/B, Remel, USA, and C. diff Quik Chek CompleteTM, TechLab, Princeton, USA) on isolate following 3 and 5 days of anaerobic growth into cooked meat broth. C. difficile VPI 10463 was used as TcdA+/TcdB+ positive control.
2.5. Extraction of C. difficile and C. perfringens DNA
For each C. difficile C. perfringens or strain, a 100 ml suspension of cells in sterile water was vortexed, incubated at 100˚C for 5 and 10 min., respectively, and centrifuged at 12,000 g (Microliter Centrifuge, Hermle Z 233 M-2, Delchimica Scientific Glassware s.r.l.) for 2 min. Five ml of this preparation were used as the DNA template for all PCR assays. All PCRs were performed with a Techne TC-32 thermal cycler (Barloworld Scientific Ltd, Milano, Italy).
2.6. Duplex PCRs for the C. difficile TcdA/B and Binary Toxin Encoding Genes
All C. difficile isolates and the reference strains were PCR-screened for the presence of (a) TcdA/B-encoding genes (624-bp tcdA and 412-bp tcdB gene fragments), as previously described by Spigaglia and Mastrantonio [17], and (b) binary toxin genes (375-bp cdtA and 510-bp cdtB gene fragments), as described by Stubbs et al. [18]. The reaction products were subjected to 1.5% agarose gel electrophoresis (120 V, 1 h) and visualized by ethidium bromide staining and ultraviolet light exposure.
2.7. Duplex PCR for the C. perfringens Phospholipase C (PLC) and CPE Encoding Genes
All C. perfringens isolates and the ATCC 12917 reference strain were PCR-screened for the presence of PLC and CPE-encoding genes as previously described by Fach and Popoff [19]. Amplified products were subjected to agarose gel electrophoresis as above mentioned.
2.8. Multiplex PCR for the C. perfringens Toxins Encoding Genes
All C. perfringens isolates, along with the four reference strains, were PCR-subjected for the detection of α (cpa), β (cpb), ε (etx), CPE (cpe), ι (iap), and β2 (cpb2) toxin encoding genes, as described by Baums et al. [20]. The reaction products were subjected to agarose gel electrophoresis as above.
3. Results
Sixty-five of the 110 faecal samples (59.1%) resulted positive: 38 for C. difficile and 17 for C. perfringens (15.4% = 17 of 110). One sample tested C. perfringens and C. difficile-positive, at the same time: on the whole, 39 C. difficile isolates (35.4% = 39 of 110). Thirty-eight of the 39 C. difficile-positive samples belonged to healthy swine and the strains resulted tcdA/tcdB and cdtA/cdtBnegative by PCRs and toxins A/B-negative by immunological tests.
On the contrary, the C. difficile strain isolated, at the same time, from a C. perfringens cpb2-positive diarrhoeic sample was tcdA/tcdB-positive (Figure 1) and cdtA/cdtBpositive by PCRs, and toxins A/B-positive by immunoassays.
Out of the 17 C. perfringens strains, 10 (58.8%) were from diarrhoeic swine. All C. perfringens isolates were type A; eight of them (47.1%), belonging to diarrhoeic animals, were also cpb2-positive by multiplex PCR (Figure 2).
4. Discussion
Clostridium difficile is ubiquitous in the environment. In addition to humans, C. difficile has also been found in calves, ostriches, chickens, elephants, dogs, horses, and pigs, but its role in infection and its pathogenesis in ani-
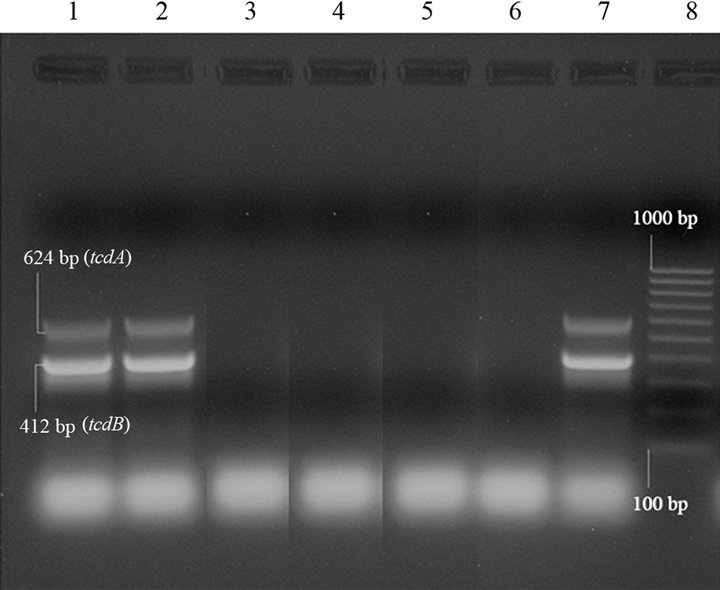
Lanes 1 and 2: C. difficile tcdA+/tcdB+strain, amplified in duplicate; lanes 3 - 5: C. difficile tcdA-/tcdB-strains; lane 6: negative controls (“0 DNA”); lane 7: C difficile positive control (tcdA+/tcdB+); lane 8: molecular size markers (100 bp Molecular Ruler, Biorad, Italy).
Figure 1. Duplex PCR for tcdA and tcdB genes of Clostridium difficile isolates from swine.
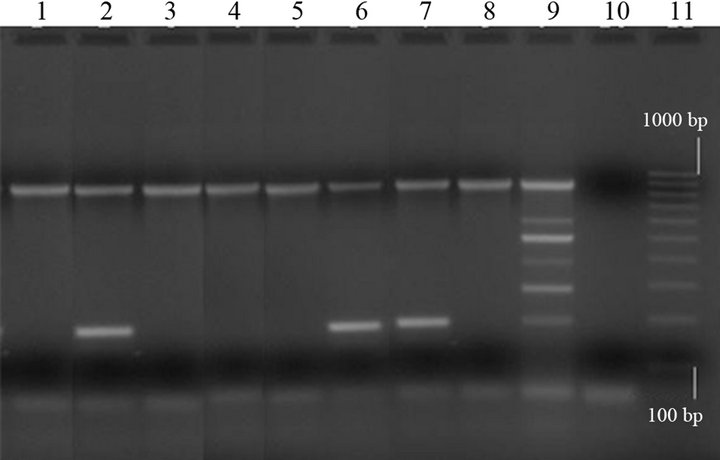
Lanes 1, 3, 4, 5 and 8: type A strains (cpa+); lanes 2, 6 and 7: type A strains (cpa+/cpb2+); lane 9: C. perfringens positive control (cpa+/cpb+/cpe+/etx+ /iap+/cpb2+); lane 10: negative control (“0 DNA”); lane 11: molecular size markers (100 bp Molecular Ruler, Biorad, Italy).
Figure 2. Multiplex PCR of Clostridium perfringens isolates from swine.
mals are largely poorly understood and possibly underestimated [21-23].
This bacterium is an important cause of enteric disease in humans. It is the most commonly diagnosed cause of hospital-and antimicrobial agent-associated diarrhoea in people. Similarly, any association between antibiotic usage and C. difficile colonization or diarrhoea in animals is less well documented than that in humans, although the acquisition of C. difficile in dogs and cats during hospitalization in an intensive-care unit was associated with the development of diarrhoea [24].
Clostridium difficile has been reported as an agent of neonatal swine enteritis and represents a significant concern to the pork industry [25,26].
Recent evidence suggests that it may be emerging as an important community-associated pathogen. In fact, this organism has also been found in retail meat, and concerns about the role of food in the epidemiology of community-associated C. difficile infection (CDI) have been expressed [22].
Clostridium perfringens may be one of the most widespread pathogen. It is commonly found in terrestrial and marine environments and is also readily found in intestinal contents of healthy humans and other animals [27, 28].
This organism can cause gas gangrene and food poisoning in humans; necrotic enteritis in poultry; enterotoxaemia in lambs and calves; and enteritis in pigs, cattle, dogs, and horses [3,10].
Clostridium perfringens type A is the most common of all the C. perfringens types. This bacterium produces alpha toxin (CPA) as well as other non-typing toxins, such as enterotoxin (CPE) and β2 (CPB2) [29-31].
Enterotoxin has been associated with diarrhoeal disease in some animal species, and, more importantly, with food poisoning in humans [3,15]. The β2-toxin has been associated with the onset of enteritis in pigs, horses, and cattle [11,14].
Type A strain, that produce only CPA among the major toxins, is a member of the normal flora of warmblooded animals and is recovered from environment contaminated by faeces. However, when properly equipped genetically and placed in opportune situations, the organism can cause gas gangrene, food poisoning, and gastrointestinal illness in humans, necrotic enteritis in chickens, necrotizing enteritis in piglets, and abomasitis, tympany, and hemorrhagic enteritis in calves [3,28,32].
Although C. perfringens type A has been linked to abomasal ulcers and inflammation, as well as necrotic enteritis, in calves and cows, and CPAand CPB2-encoding genes have been detected in some of these cases, the bacteria have also been isolated from the intestinal content of healthy animals. Therefore, its role as an intestinal pathogen is still unclear [31].
In this study, the 38 C. difficile-positive swine samples belonged to healthy animals and these isolates were nontoxigenic (tcdA/tcdB and cdtA/cdtB-negative by PCRs and toxins A/B-negative by immunological tests); in one C. perfringens cpb2-positive diarrhoeic sample, a toxigenic C. difficile strain (2.56% = 1 of 39 isolates) was also isolated. It tested tcdA/tcdB and cdtA/cdtB-positive by PCRs and toxins A/B-positive by immunoassays (0.9% = 1 isolate of 110 samples).
There was 100% correlation between the results of PCRs and the toxin phenotype.
We found a higher isolation percentage (34.5% = 38 of 110 samples) of C. difficile non-toxigenic strains in swine than in other studies [33]. Really, C. difficile readily colonizes the large intestines of neonates of most species mammals [26].
Out of the 17 C. perfringens type A swine isolates (15.4% = 17 of 110), 10 (58.8%) were from diarrhoeic swine and eight of them (80.0%) were also cpb2-positive. Percentages of positive cultures were different in diarrhoeic and healthy swine (23.2% = 10 of 43, versus 10.4% = 7 of 67). However, this difference was not statistically significant (two-tailed Fisher’s P = 0.103). Probably, the high rate of occurrence of cpb2-positivity among swine strains isolated from animals with enteritis could be consistent with the contention that CPB2 plays a role in pathogenesis of the disease [34,35]. On the contrary, the detection of strains harbouring cpb2 in healthy animals is not a necessary risk in itself, although β2-toxigenic C. perfringens can become an emerging health threat if circumstances appear which provoke enteric dybiosis or immunosuppression [14].
We could conclude that, since C. difficile and C. perfringens, in particular non-toxigenic strains, can be found in healthy pigs, as commonly in the colon of clinically normal animals, their isolation may have little diagnostic relevance.
The molecular genotyping/toxinotyping should be applied to establish a final diagnosis and to assess properly the full implications and the epidemiological impact of these findings in particular in samples of healthy animals and aid in the development of effective intervention methods for controlling clostridial disease outbreaks.
5. Acknowledgements
The authors wish to thank Mrs. Cinzia Reverberi and Mr. Roberto Lurisi for their technical support, and the Istituto Zooprofilattico Sperimentale della Lombardia e dell’Emilia Romagna-Sezione di Parma (Italy) for kindly providing some C. perfringens-positive strains.
REFERENCES
- X. Song, J. G. Bartlett, K. Speck, A. Naegeli, K. Carroll and T. M. Perl, “Rising Economic Impact of Clostridium difficile-Associated Disease in Adult Hospitalized Patient Population,” Infection Control and Hospital Epidemiology, Vol. 29, No. 9, 2008, pp. 823-828. doi:10.1086/588756
- M. K. Keel and J. G. Songer, “The Comparative Pathology of Clostridium difficile-Associated Disease,” Veterinary Pathology, Vol. 43, No. 3, 2006, pp. 225-240. doi:10.1354/vp.43-3-225
- J. G. Songer, “Clostridial Enteric Diseases of Domestic Animals,” Clinical Microbiology Reviews, Vol. 9, No. 2, 1996, pp. 216-234.
- B. E. Waggett, B. C. McGorum, U. Wernery, D. J. Shaw and R. S. Pirie, “Prevalence of Clostridium perfringens in Faeces and Ileal Contents from Grass Sickness Affected Horses: Comparisons with 3 Control Populations,” Equine Veterinary Journal, Vol. 42, No. 6, 2010, pp. 494- 499. doi:10.1111/j.2042-3306.2010.00105.x
- A. A. Baker, E. Davis, T. Rehberger and D. Rosener, “Prevalence and Diversity of Toxigenic Clostridium perfringens and Clostridium difficile among Swine Herds in the Midwest,” Applied and Environmental Microbiology, Vol. 76, No. 9, 2010, pp. 2961-2967. doi:10.1128/AEM.02459-09
- J. G. Songer, K. W. Post, D. J Larson., B. H. Jost and R. D. Glock, “Infection of Neonatal Swine with Clostridium difficile,” Swine Health and Production, Vol. 8, No. 4, 2000, pp. 185-189.
- M. C. Hammitt, D. M., Bueschel M. K. Keel, R. D. Glock, P. Cuneo, D. W. DeYoung, C. Reggiardo, H. T. Trinh and J. G. Songer, “A Possible Role for Clostridium difficile in the Etiology of Calf Enteritis,” Veterinary Microbiology, Vol. 127, No. 3-4, 2008, pp. 343-352. doi:10.1016/j.vetmic.2007.09.002
- A. Rodriguez-Palacios, H. R. Stämpfli, T. Duffield, A. S. Peregrine, L. A. Trotz-Williams, L. G. Arroyo, J. S. Brazier and J. S. Weese, “Clostridium difficile PCR Ribotypes in Calves, Canada,” Emerging Infectious Diseases, Vol. 12, No. 11, 2006, pp. 1730-1736. doi:10.3201/eid1211.051581
- F. Van Immerseel, J. de Buck, F. Pasmans, G. Huyghebaert, F. Haesebrouck and R. Ducatelle, “Clostridium perfringens in Poultry: An Emerging Threat for Animal and Public Health,” Avian Pathology, Vol. 33, No. 6, 2004, pp. 537-549. doi:10.1080/03079450400013162
- L. Petit, M. Gilbert and M. R. Popoff, “Clostridium perfringens: Toxinotype and Genotype,” Trends in Microbiology, Vol. 7, No. 3, 1999, pp. 104-110. doi:10.1016/S0966-842X(98)01430-9
- D. Slavić, P. Boerlin, M. Fabri, K. C. Klotins, J. K. Zoethout, P. E. Weir and D. Bateman, “Antimicrobial Susceptibility of Clostridium perfringens Isolates of Bovine, Chicken, Porcine, and Turkey Origin from Ontario,” Canadian Journal of Veterinary Research, Vol. 75, No. 2, 2011, pp. 89-97.
- M. Lebrun, J. G. Mainil and A. Linden, “Cattle Enterotoxaemia and Clostridium perfringens: Description, Diagnosis and Prophylaxis,” Veterinary Record, Vol. 167, No. 1, 2010, pp. 13-22. doi:10.1136/vr.167.1.12
- B. Kreft, K. Dalhoff and K. Sack, “Necrotizing Enterocolitis: A Historical and Current Review,” Medizinische Klinik (Munich), Vol. 95, No. 8, 2000, pp. 435-441. doi:10.1007/s000630050003
- U. Schotte, U. Truyen and H. Neubauer, “Significance of β2-Toxigenic Clostridium perfringens Infections in Animals and Their Predisposing Factors—A Review,” Journal of Veterinary Medicine, Vol. 51, No. 10, 2004, pp. 423-426. doi:10.1111/j.1439-0450.2004.00802.x
- P. Lahti, A. Heikinheimo, T. Johansson and H. Korkeala, “Clostridium perfringens Type A Strains Carrying a Plasmid-Borne Enterotoxin Gene (Genotype IS1151-cpe or IS1470-like-cpe) as a Common Cause of Food Poisoning,” Journal of Clinical Microbiology, Vol. 46, No. 1, 2008, pp. 371-373. doi:10.1128/JCM.01650-07
- A. G. Buchanan, “Clinical Laboratory Evaluation of a Reverse CAMP Test for Presumptive Identification of Clostridium perfringens,” Journal of Clinical Microbiology, Vol. 16, No. 4, 1982, pp. 761-762.
- P. Spigaglia and P. Mastrantonio, “Molecular Analysis of the Pathogenicity Locus and Polymorphism in the Putative Negative Regulator of Toxin Production (TcdC) among Clostridium difficile Clinical Isolates,” Journal of Clinical Microbiology, Vol. 40, No. 9, 2002, pp. 3470- 3475. doi:10.1128/JCM.40.9.3470-3475.2002
- S. Stubbs, M. Rupnik, M. Gilbert, J. Brazier, B. Duerden and M. Popoff, “Production of Actin-Specific ADP-Ribosyltransferase (Binary Toxin) by Strains of Clostridium difficile,” FEMS Microbiology Letters, Vol. 186, No. 2, 2000, pp. 307-312. doi:10.1111/j.1574-6968.2000.tb09122.x
- P. Fach and M. R. Popoff, “Detection of Enterotoxigenic Clostridium perfringens in Food and Faecal Samples with a Duplex PCR and the Slide Latex Agglutination Test,” Applied and Environmental Microbiology, Vol. 63, No. 11, 1997, pp. 4232-4236.
- C. G. Baums, U. Shotte, G. Amtsberg and R. Goethe, “Diagnostic Multiplex PCR for Toxin Genotyping of Clostridium perfringens Isolates,” Veterinary Microbiology, Vol. 100, No. 1-2, 2004, pp. 11-16. doi:10.1016/S0378-1135(03)00126-3
- M. Rupnik, “Is Clostridium difficile-Associated Infection a Potentially Zoonotic and Foodborne Disease?” Clinical Microbiology and Infection, Vol. 13, No. 5, 2007, pp. 457- 459. doi:10.1111/j.1469-0691.2007.01687.x
- J. S. Weese, B. P. Avery, J. Rousseau and R. J. ReidSmith, “Detection and Enumeration of Clostridium difficile Spores in Retail Beef and Pork,” Applied and Environmental Microbiology, Vol. 75, No. 15, 2009, pp. 5009- 5011. doi:10.1128/AEM.00480-09
- J. Freeman, M. P. Bauer, S. D. Baines, J. Corver, W. N. Fawley, B. Goorhuis, E. J. Kuijper and M. H. Wilcox, “The Changing Epidemiology of Clostridium difficile Infections,” Clinical Microbiology Reviews, Vol. 23, No. 3, 2010, pp. 529-549. doi:10.1128/CMR.00082-09
- J. Cloten, S. Kruth, L. Arroyo and J. S. Weese, “Prevalence and Risk Factors for Clostridium difficile Colonization in Dogs and Cats Hospitalized in an Intensive Care Unit,” Veterinary Microbiology, Vol. 129, No. 1-2, 2008, pp. 209-214. doi:10.1016/j.vetmic.2007.11.013
- K. W. Post, B. H. Jost and J. G. Songer, “Evaluation of a Test for Clostridium difficile Toxins A and B for the Diagnosis of Neonatal Swine Enteritis,” Journal of Veterinary Diagnostic Investigation, Vol. 14, No. 3, 2002, pp. 258-259. doi:10.1177/104063870201400314
- M. K. Keel and J. G. Songer, “The Distribution and Density of Clostridium difficile Toxin Receptors on the Intestinal Mucosa of Neonatal Pigs,” Veterinary Pathology, Vol. 44, No. 6, 2007, pp. 814-822. doi:10.1354/vp.44-6-814
- S. L. Marks and E. J. Kather, “Bacterial-Associated Diarrhea in the Dog: A Critical Appraisal,” Veterinary Clinics: Small Animal Practice, Vol. 33, No. 5, 2003, pp. 1029-1060. doi:10.1016/S0195-5616(03)00091-3
- M. C. Ferrarezi, T. C. Cardoso and I. S. Dutra, “Genotyping of Clostridium perfringens Isolated from Calves with Neonatal Diarrhea,” Anaerobe, Vol. 14, No. 6, 2008, pp. 328-331. doi:10.1016/j.anaerobe.2008.12.001
- L. Ceci, P. Paradies, M. Sasanelli, et al., “Haemorrhagic Bowel Syndrome in Dairy Cattle: Possible Role of Clostridium perfringens Type A in the Disease Complex,” Journal of Veterinary Medicine Series A, Vol. 53, No. 10, 2006, pp. 518-523. doi:10.1111/j.1439-0442.2006.00884.x
- C. C. Brown, D. C. Baker and I. K. Barker, “Alimentary System,” In: M. G. Maxie, Ed., Jubb, Kennedy and Palmer’s pathology of domestic animals, 5th Edition, Saunders Elsevier, St. Louis, 2007, pp. 1-296.
- W. E. Morris, A. J. Venzano, A. Elizondo, D. A. Vilte, E. C. Mercado and M. E. Fernandez-Miyakawa, “Necrotic Enteritis in Young Calves,” Journal of Veterinary Diagnostic Investigation, Vol. 23, No. 2, 2011, pp. 254-259. doi:10.1177/104063871102300209
- Y. S. Sawires and J. G. Songer, “Clostridium perfringens: Insight into Virulence Evolution and Population Structure,” Anaerobe, Vol. 12, No. 1, 2006, pp. 23-43. doi:10.1016/j.anaerobe.2005.10.002
- J. G. Songer, R. Jones, M. A. Anderson, A. J. Barbara, K. W. Post and H. T. Trinh, “Prevention of Porcine Clostridium difficile-Associated Disease by Competitive Exclusion with Nontoxigenic Organisms,” Veterinary Microbiology, Vol. 124, No. 3-4, 2007, pp. 358-361. doi:10.1016/j.vetmic.2007.04.019
- S. Thiede, R. Goethe and G. Amtsberg, “Prevalence of β2 Toxin Gene of Clostridium perfringens Type A from Diarrhoeic Dogs,” Veterinary Record, Vol. 149, No. 9, 2001, pp. 273-274. doi:10.1136/vr.149.9.273
- D. M. Bueschel, B. H. Jost, S. J. Billington, H. T. Trinh and J. G. Songer, “Prevalence of cpb2, Encoding β2 Toxin, in Clostridium perfringens Field Isolates: Correlation of Genotype with Phenotype,” Veterinary Microbiology, Vol. 94, No. 2, 2003, pp. 121-129. doi:10.1016/S0378-1135(03)00081-6
NOTES
*Corresponding author.

