Advances in Microbiology
Vol. 2 No. 2 (2012) , Article ID: 19667 , 8 pages DOI:10.4236/aim.2012.22010
Bacterial Colonization of the Equine Gut; Comparison of Mare and Foal Pairs by PCR-DGGE*
1Department of Animal and Food Sciences, University of Kentucky, Lexington, USA
2Agricultural Research Service, Forage-Animal Production Research Unit, USDA, Lexington, USA
Email: #michael.flythe@ars.usda.gov
Received January 31, 2012; revised March 20, 2012; accepted April 8, 2012
Keywords: Colonization; development; microbial ecology; neonates
ABSTRACT
Horses, like all animals, are born without the symbiotic microbes that occupy the gastrointestinal tracts of mature animals. As grazing animals, horses rely on these microbes to fully utilize the grasses and other cellulosic feeds that they consume. Thus, colonization of the foal’s gastrointestinal tract must occur between birth and weaning. The feces of nine mare and foal pairs were sampled from the day of parturition until 12 weeks of age, and the samples were analyzed by polymerase chain reaction amplification of the bacterial 16S rRNA gene and denaturing gradient gel electrophoresis (PCR-DGGE). The gels from feces of day (d) 0 foals had no or very few ( = 3, n = 6) bands, which indicates that species richness was low. The number of bands increased during the first 4 days of life, and by d 14 the foals and mares had similar numbers of bands (
= 3, n = 6) bands, which indicates that species richness was low. The number of bands increased during the first 4 days of life, and by d 14 the foals and mares had similar numbers of bands (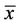 = 28, n = 23). Some bands were present in young foals, but not in mares or in foals on d 42 or d 84, which indicated succession of bacterial species. When the PCR-DGGE profiles were compared with Dice’s algorithm, all mare-foal pairwise similarities on d 14 and later were as great as the pairwise similarities between mares. These results are consistent with the idea that foals are born with a sterile gut, colonization proceeds rapidly, and a mature microbial community is present in the first few weeks of life.
= 28, n = 23). Some bands were present in young foals, but not in mares or in foals on d 42 or d 84, which indicated succession of bacterial species. When the PCR-DGGE profiles were compared with Dice’s algorithm, all mare-foal pairwise similarities on d 14 and later were as great as the pairwise similarities between mares. These results are consistent with the idea that foals are born with a sterile gut, colonization proceeds rapidly, and a mature microbial community is present in the first few weeks of life.
1. Introduction
It has long been held that neonates are born sterile, and that the symbiotic microbiome is acquired by postpartum colonization [1,2]. The placenta provides a barrier to both pathogens and commensal microbes that are present in the mother, and the first exposure to bacteria occurs during birth. Recent studies of human infants support the hypothesis that the initial inoculum comes from the vagina of the mother during birth [3,4]. Colonization occurs rapidly, but the predominant species depend on the diet [5]. In the case of humans, succession appears to correspond to changes in diet. The community stabilizes as the baby approaches 1 y, but studies of older children are not available. Recent evidence indicates that the placenta [6-8] and even the meconium [9] can contain the DNA sequences of commensal bacteria. Nevertheless, most documented cases of in utero exposure to bacteria are best characterized as infection. Listeria monocytogenes, Fusobacterium necrophorum, and Coxiella burnetii have all been associated with placental infection in mammalian species [7,10,11].
The problems associated with in utero exposure to bacteria must be considered in light of the metabolic contributions of the gut microflora to the host. That is, the normal microflora are beneficial, and the ability to maintain these symbionts is adaptive. This beneficial symbiosis is especially important in herbivores. Herbivores rely on the microbes in their gastrointestinal tracts to break down the fibrous materials that they consume [12]. Ruminants have a pre-gastric fermentation that occurs in the rumen, but non-ruminant herbivores also require symbiotic fermentation to effectively utilize fiber. In the case of horses, this fermentation occurs in the large intestine. The volatile fatty acids produced in the large intestine represent the majority of the energy absorbed by horses under some dietary circumstances [13]. Given these observations, it seems likely that the normal flora must be present prior to weaning.
Several studies have cited the early appearance of microbes in newborn ruminants, such as lambs and calves [14-20]. By 6 weeks, calves have some of the same bacteria as adults, and by 9 - 13 weeks, the bacteria isolated from calves were typical of those found in mature cattle [16,19]. Less is known about the colonization of equine gastrointestinal tract. Foal meconium was reported to be free of bacteria, which is consistent with the premise of a sterile gastrointestinal tract [21]. Lactobacilli, enterococci, and clostridia were isolated from feces as early as 3 d post parturition. Enrichments of cellulolytic and lactic acid bacteria indicated that these guilds were present in feces at levels similar to mature horses by 12 weeks [22].
The two aforementioned studies tested the foal microflora with culture-based techniques. Culture-based evaluation has distinct strengths. However, diversity is not adequately assessed by cultivation because of the selectivity of the media that are employed. Indeed, most microbial clades were unknown until the advent of molecular microbial ecology [23]. The purpose of the current study was to observe the colonization of the foal gastrointestinal tract with a molecular community “fingerprinting” method. The method selected was PCR amplification of a portion of the 16S rRNA gene subunit, followed by denaturing gradient gel electrophoresis (PCR-DGGE). The hypotheses were: 1) bacteria are not initially detectable in newborn foals; 2) species richness (i.e. band number) increases over time; and 3) the microbiomes of foals are more similar to their own dams than to other foals by 12 weeks of age. The methods are presented below with a table of samples. Then the results are presented with three accompanying figures. A figure also included in the discussion, which pays special attention to the most complete set of samples that were collected from a single mare-foal pair. Finally, conclusions are made.
2. Materials and Methods
2.1. Animal Management
All procedures were approved by the Institutional Animal Care and Use Committee at the University of Kentucky (Protocol No. 2008-0311). Nine mare-foal pairs of Thoroughbred or Quarter Horse breeding were used for the study. During late gestation mares were housed outdoors in cool-season grass pastures and fed supplemental hay and concentrate. As mares approached parturition they were kept in box stalls at night and in pastures during the day. After parturition mares and foals were kept in stalls or small paddocks for a few days and then returned to pasture, except on days when samples were collected. Mares and foals were house primarily outdoors, where they had ad libitum access to cool season grass pasture. Mares were provided a pelleted concentrate twice a day to meet their nutrient requirements. While the foals were not specifically fed the concentrate, they did have access to their dam's feed bucket.
2.2. Collection
Fecal samples were collected from mare-foal pairs on day 0 (day of birth), day 1, day 4, day 14, day 42 and day 84. Foals were born in the night or early morning. Day 0 samples were obtained during the day following the night or morning of parturition (0 to 24 hours after birth). Samples were collected from the mare and foal of a given pair on the same day. Due to the difficulty in obtaining foal fecal material, not all mare-foal pairs are represented at each time point. Some unpaired animals are included to expand the data set. On collection days, the mare-foal pair was placed in a 4.25 m × 4.25 m box stall and monitored so that samples could be collected immediately following defecation. A fecal collection harness was sometimes used in the collection process for foals. The harnesses were made out of cloth and lined with plastic. In the case of the very young foals (<2 weeks), the entire defecation was often retained; otherwise a subsample (approximately 100 - 200 g) of the feces was collected. In older foals and mares, several subsamples were collected from within the fecal pile and compiled. Fecal samples were placed in plastic bags, excess air was removed, and samples were immediately frozen in a −20 degree Celsius freezer.
2.3. PCR-DGGE Analysis
Three sub-samples were extracted from each fecal sample, and then combined prior to PCR in an attempt to obtain a representative sample. DNA was extracted from individual fecal sub-samples using the QiaAMP DNA Mini Stool Kit (Qiagen, Valencia, CA). The DNA template was amplified using PCR. PureTaq Ready—ToGoÔ PCR beads were used according to the manufacturers instructions (GE Healthcare; Piscataway, NJ). The thermal cycler was an Eppendorf Mastercycler (Hauppauge, NY). The PCR cycles were: 94˚C for 5 min, 10 cycles of 94˚C for 0.5 min, 65˚C for 0.5 min, 72˚C for 0.75 min, 25 cycles of 94˚C for 0.5 min, 60˚C for 0.5 min, 72˚C for 0.75 min, final extension step at 72˚C for 5 min. 341F-GC and 901R primers [24] were used for the amplification process. PCR products were stored at −20˚C. Amplicons within the PCR products were separated using denaturing gradient gel electrophoresis (6% polyacrylamide, 40% to 60% denaturing gradient, 56˚C, 69 V, 17 h). The gels were stained with Gel Red (Phenix Research Products, Candler, NC). The Kodak Gel Logic System (1D Image Analysis Software V3.6.5; Rochester, NY) was used for photography. Each sample was subjected to PCR and separation at least twice.
The photographs were imported into BioNumerics v. 6.0 (Applied Maths, Austin, TX). The images were normalized during data import. The standards used for normalization were the PCR product of two pure cultures (Clostridium acetobutylicum and Peptostreptococcus anaerobius). Preliminary experiments demonstrated that these bacteria yielded five distinct bands that were distributed throughout the lane. The standards were run in 4 of the 14 lanes used on each gel. Band categories were assigned to each 1% of the 10 cm gel. The minimum profiling value was set at 5% of the maximum value of a lane. No bands that were identified by the software were removed. Obvious bands that the software did not identify were manually identified. Comparisons were made using Dice’s algorithm [25] and reported as percent similarity. UPGMA (Unweighted Pair Group Method with Arithmetic Mean) dendrograms were constructed from Dice’s analysis in BioNumerics with the algorithm. Cartesian plots were made in KaleidaGraph v. 4.1 (Synergy Software; Reading, PA) with data generated in BioNumerics. The significant differences in positive band categories were determined by Student’s T-test, assuming unpaired data and unequal variance.
3. Results
Fifty-three samples were collected from mares and foals, including 46 same-day samples from mare-foal pairs (Table 1). When the positive band categories (i.e. a 1 mm length of the lane) in the DGGE lanes were counted, the mares had 23 to 41 bands in a given sample (Figure 1). Three foal samples from 0 d (first 24 h post parturition) had no bands. The other three d 0 samples had 2, 4 and 12 bands, respectively. The mean foal band number was 19 by d 1 (24 to 48 h post parturition), and 23 by 2 d. The difference in the number of positive band categories between the mares and the foals in the first 24 h post parturition was significant (P < 0.001), but band number differences were not significant in subsequent samples.
In addition to the number of bands, the gel lanes were analyzed based on the position of positive band categories. The majority of bands occurred in categories between 35 and 95 mm of the gel (Figure 2). However, some foals that were d 14 or younger had one or more bands that occurred before 35 mm Bands located in this region of the gel (<35 mm) were not observed in older foals. Only one mare sample (G, day 1) had bands before 35 mm. The positive and negative band categories of all the samples were quantitatively compared and a dendrogram was constructed (Figure 2). The within-mare similarity ranged from 44.9% to 81.4%. Primarily because of the low number of positive band categories, the d 0 foals did not group with the mares. Three of the five d 1 foals were most similar to another 1 d foal or to a d 0 foal. The exceptions were foal (a), which was most similar to mare
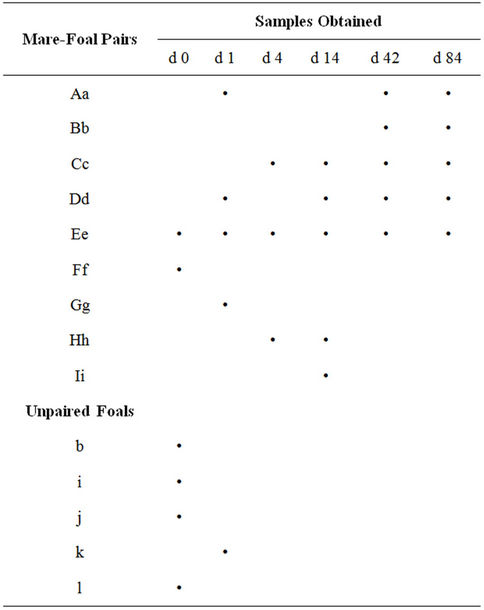
Table 1. Fecal samples collected from mare-foal pairs or individual foals were collected. The samples were collected on 0 to 84 days post parturition as indicated. Majuscule and miniscule letters are used to represent the mare and foal, respectively.
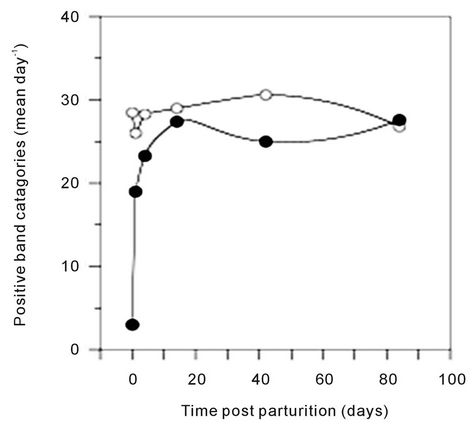
Figure 1. Average number of 16S DGGE bands found in the feces of mares (open circles) or foals (filled circles) on each sample day post parturition.
(G) on d 1, and foal (g), which grouped with mares, and older foals. Mares and foals that were d 42 or d 84 post parturition did not form distinct groups. The similarity
Figure 2. Similarity analysis of the 16S DGGE profiles from all mare and foal fecal samples. Similarity was determined by Dice’s analysis of the positive and negative band categories. The similarity (%) is indicated on the scale at the top of the UPGMA dendrogram. Majuscule (mare) and miniscule (foal) letters represent pairs. The sample day post parturition is indicated.
(pairwise, Dice’s analysis) of each mare sample was compared to its corresponding foal sample on each day (Figure 3). The similarity was initially low, but rapidly increased in the first few days of life. By d 84, the similarity ranged from 54% to 87%. The mare/foal pair with 87% similarity was (Ee). They represent the most complete data subset in the study (Figure 4). The foal gel lanes (Figure 4(a)), the foal and mare band number (Figure 4(b)), and the pairwise similarity (Figure 4(c)) are shown.
4. Discussion
Figure 4 shows the PCR-DGGE analysis of the feces from a single foal (e) from his birthday to 12 weeks of age. We conjecture that the curves shown for band number (Figure 4(b)) and similarity to his dam (Figure 4(c)) provide an example of the rate and extent of colonization of equine gastrointestinal tract. Fewer samples were collected from any other mare/foal pair, but the data from these samples are consistent.
When the feces collected from foal (e) on the day of his birth was analyzed, no DGGE bands were evident. Five other foals were sampled on d 0. Two of these had no bands, and three had small numbers of bands. The d 0 fecal samples were never available immediately after parturition, but rather at any time within the first 24 h. One interpretation is that the foals were born sterile, and colonization began in the first 24 h. However, DNA sequences from commensal bacteria were recently found in the placentae of humans and mice [6-8]. It is possible that bacteria were present in the foals at numbers below
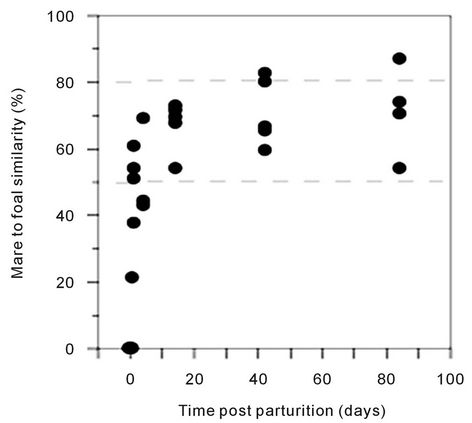
Figure 3. Pairwise similarity of the 16S DGGE profiles of mare-foal pairs on each sample day post parturition. Similarity was determined by Dice’s analysis of the positive and negative band categories. The dashed lines represent the lowest and highest similarity values within the mare group.
the limit of detection for the touchdown PCR procedure that was used.
By d 1, the sample from foal (e) yielded 19 bands (Figures 4(a) and (b)), which indicates that bacteria were present in the feces. The ways that normal microflora are acquired has been examined in other species. For example, gastrointestinal tracts of vaginally delivered human babies are dominated by the mother’s vaginal and intestinal microflora, but babies delivered by caesarian section primarily carry environmental microbes [3]. It is reasonable to assume that a foal is also colonized by microbes from the dam’s vagina, as well as from grooming and other maternal contact. Newborn foals often exhibit searching behavior prior to nursing and may suckle various parts of the mare’s body in this process. Once nursing in established, the foal may suckle the udder more than 100 times a day in the neonatal period [26]. The exterior of the mare’s body, including the udder, is not sterile and thus could provide a source of ingested microbes for the foal. Additionally, foals, like many juvenile herbivores, are coprophagic [26]. Establishment of the normal microflora may be an important function of this behavior. It has been shown that foals usually consume fresh, rather than dry feces. Compared to dried feces, fresh feces would be more likely to contain viable microbes that could successfully colonize the gastrointestinal tract. In addition, foals preferentially consume the feces of their dam, even when fresh feces from another horse are also present [27]. The propensity to consume the feces of their dam may explain the relatively high level of microbial similarity in mare-foal pairs.
Foal (e) had bands in the first 35 mm of the gel on d 1, d 4, and d 14, but they were not present in the two later samples. These “early bands” were common among the young foals, but on only one band before 35 mm was observed in a single mare sample. It is possible that these bands represented transient bacteria that persist for a limited time in the foal’s gastrointestinal tract. In primary succession, the first organisms that colonize new habitat are called pioneer species, and the hypothesis that these transient bands correspond to microbial pioneer species in the foal gastrointestinal tract is consistent with either the facilitation or the inhibition models of succession [28]. Bacterial succession was observed in two human babies using PCR-DGGE [2]. However, if these bands represent pioneer species, it is uncertain how they affect the developing microbiome or the host.
When foal (e) was sampled on d 42, it was observed that he had more positive band categories than his dam. The similarity of his sample to hers was 60%. Likewise, all of the other mare/foal pairwise similarities were greater than 50% in the 42 d and 84 d samples. Correspondingly, the mares and older foals did not form distinct groups when analyzed. These data indicate that the
 (a)
(a) (b)(c)
(b)(c)
Figure 4. Colonization of a foal’s gastrointestinal tract by bacteria. The 16S DGGE profiles from the feces are shown on each sample day (a). Part (b) shows the number of positive band categories from the foal (filled circles) and the mare (open circles), and the pairwise similarity is shown in part (c). Similarity was determined by Dice’s analysis of the positive and negative band categories.
species richness in the foals’ gastrointestinal tracts was similar to that of mature horses by the sixth week of life. The similarity of the older foals to the mares and the apparent decrease in the acquisition of new species suggests that a climax community was achieved by approximately 6 weeks.
It is important to acknowledge the limitations of PCRDGGE and similar methods. Traditional measures of ecological diversity, such as the Shannon Index, require the number and size of the populations in a community [29,30]. PCR-DGGE results are not indicative of population size, and quantitative biases associated with similar methods have been demonstrated [31]. However, these methods can be used to estimate species richness, or the number of species in a habitat [2,6,19,24,32]. Furthermore, results from culture-based and physiological studies are consistent with the hypothesis that a foal can possess a gastrointestinal microbial community in the climax state by 6 - 12 weeks post parturition. In vitro fiber digestion by the fecal microbes of 1 d foals was approximately half that of mares, but by 4 weeks of age there was no difference between the foals and mares [33].
Likewise, cellulolytic and lactic acid bacteria were found in similar numbers in 12 week old foals and mares [22].
Weaning is the greatest dietary change in the life of a mammal. Even in human nutrition, we have not successfully imitated milk. This is evidenced by significant compositional differences in the intestinal microflora of human infants that are raised on formulas versus human milk [34]. We conjecture that the transition from milk to feed is even greater for herbivores that rely on fermentative microbes to provide metabolic access to that feed. Although neonatal foals obtain most of their nutrients from milk, they may consume pasture, hay or grain as early as 1 day of age. By 5 weeks of age foals spend more than 20% of their time grazing or eating non-milk foods [27]. Overall, the timeline of the microbial establishment of the gastrointestinal tract is consistent with the time at which plant-based feeds become important in diet of the foal.
5. Conclusion
To the best of our knowledge, this is the first attempt to observe colonization of the equine gastrointestinal tract using molecular methods. The data provide evidence that the beneficial symbiosis between the microbiome and the horse begins early. It appears that foals are born without bacteria in the gastrointestinal tract, but colonization begins during the first day. Colonization proceeds rapidly, there is some evidence of succession, and a mature microbial community is present by the sixth week of life.
6. Acknowledgements
This work was funded by the United States Department of Agriculture, Agricultural Research Service, and the Kentucky Agricultural Experiment Station. We would like to thank Dr. Kim Cook for her assistance in implementing PCR-DGGE in our laboratory.
REFERENCES
- H. Tissier, “Recherches sur la Flore Intestinale des Nourrissons (état Normal et Pathologique),” G Carre and C Naud, Paris, 1900.
- C. F. Favier, E. E. Vaughan, W. M. De Vos and A. D. L. Akkermans, “Molecular Monitoring of Succession of Bacterial Communities in Human Neonates,” Applied and Environmental Microbiology, Vol. 68, No. 1, 2002, pp. 219-226. doi:10.1128/AEM.68.1.219-226.2002
- G. Biasucci, M. Rubini, S. Riboni, L. Morelli, E. Bessi and C. Retetangos, “Mode of Delivery Affects the Bacterial Community in the Newborn Gut,” Early Human Development, Vol. 86, Suppl. 1, 2010, pp. 13-15. doi:10.1016/j.earlhumdev.2010.01.004
- M. G. Dominguez-Belloa, E. K. Costellob, M. Contrerasc, M. Magrisd, G. Hidalgod, N. Fierere and R. Knight, “Delivery Mode Shapes the Acquisition and Structure of the Initial Microbiota across Multiple Body Habitats in Newborns,” Proceedings of the National Academy of Sciences, Vol. 107, No. 26, 2010, pp. 11971-11975. doi:10.1073/pnas.1002601107
- C. F. Favier, W. M. De Vos and A. D. L. Akkermans, “Development of Bacterial and Bifidobacterial Communities in Feces of Newborn Babies,” Anaerobe, Vol. 9, No. 5, 2003, pp. 219-229. doi:10.1016/j.anaerobe.2003.07.001
- R. Satokari, T. Groenroos, K. Laitinen, S. Salminen and E. Isolauri, “Bifidobacterium and Lactobacillus DNA in the Human Placenta,” Letters in Applied Microbiology, Vol. 48, No. 1, 2009, pp. 8-12. doi:10.1111/j.1472-765X.2008.02475.x
- A. B. Onderdonk, M. L. Delaney, A. M. DuBois, E. N. Allred and A. Leviton, “Detection of Bacteria in Placental Tissues Obtained from Extremely Low Gestational Age Neonates,” American Journal of Obstetrics and Gynecology, Vol. 198, No. 1, 2008, pp. 110.e111-110.e117.
- H. E. Jones, K. A. Harris, M. Azizia, L. Bank, B. Carpenter, J. C. Hartley, N. Klein and D. Peebles, “Differing Prevalence and Diversity of Bacterial Species in Fetal Membranes from Very Preterm and Term Labor,” PLoS ONE, Vol. 4, No. 12, 2009, p. e8205. doi:10.1371/journal.pone.0008205
- E. Jimenez, M. L. Marin, R. Martin, J. M. Odriozola, M. Olivares, J. Xaus, L. Fernandez and J. M. Rodriguez, “Is Meconium from Healthy Newborns Actually Sterile?” Research in Microbiology, Vol. 159, No. 3, 2008, pp. 187- 193. doi:10.1016/j.resmic.2007.12.007
- J. S. Agerholm, M. Boye and B. Aalbæk, “Ovine Fetal Necrobacillosis,” Journal of Comparative Pathology, Vol. 136, No. 4, 2007, pp. 213-221. doi:10.1016/j.jcpa.2007.01.012
- R. M. Jones, D. F. Twomey, S. Hannon, J. Errington, G. C. Pritchard and J. Sawyer, “Detection of Coxiella burnetii in Placenta and Abortion Samples from British Ruminants Using Real-Time PCR,” Veterinary Record, Vol. 167, No. 25, 2010, pp. 965-967. doi:10.1136/vr.c4040
- R. E. Hungate, “Studies on Cellulose Fermentation: III. The Culture and Isolation for Cellulose-Decomposing Bacteria from the Rumen of Cattle,” Journal of Bacteriology, Vol. 53, No. 5, 1947, pp. 631-645.
- M. J. Glinsky, R. M. Smith, H. R. Spires and C. L. Davis, “Measurement of Volatile Fatty Acid Production Rates in the Cecum of the Pony,” Journal of Animal Science, Vol. 42, No. 6, 1976, pp. 1465-1470.
- K. L. Anderson, T. G. Nagaraja, J. L. Morrill, T. B. Avery, S. J. Galitzer and J. E. Boyer, “Ruminal Microbial Development in Conventionally or Early Weaned Calves,” Journal of Animal Science, Vol. 64, No. 4, 1987, pp. 1215-1226.
- G. Fonty, P. Gouet, J. P. Jouany and J. Senaud, “Establishment of the Microflora and Anaerobic Fungi in the Rumen of Lambs,” Journal of General Microbiology, Vol. 133, No. 7, 1984, pp. 1835-1843.
- P. Gouet, J. M. Nebout, G. Fonty and J. P. Jouany, “Cellulolytic Bacteria Establishment and Rumen Digestion in Lambs Isolated after Birth,” Canadian Journal of Animal Science, Vol. 64, No. 5, 1984, pp. 163-164. doi:10.4141/cjas84-205
- H. Minato, M. Otsuka, S. Shirasaka, H. Itabashi and M. Mitsumori, “Colonization of Microorganisms in the Rumen of Young Calves,” Journal of General and Applied Microbiology, Vol. 38, No. 5, 1992, pp. 447-456. doi:10.2323/jgam.38.447
- F. W. Lengemann and N. N. Allen, “Development of Rumen Function in the Dairy Calf. II. Effect of Diet upon Characteristics of the Rumen Flora and Fauna of Young Calves,” Journal of Dairy Science, Vol. 42, No. 7, 1959, pp. 1171-1181. doi:10.3168/jds.S0022-0302(59)90709-X
- R. E. Mueller, E. L. Iannotti and J. M. Asplund, “Isolation and Identification of Adherent Epimural Bacteria during Succession in Young Lambs,” Applied and Environmental Microbiology, Vol. 47, No. 4, 1984, pp. 724- 730.
- A. Zoilecki and C. A. E. Briggs, “The Microflora of the Rumen of the Young Calf: II. Source, Nature, and Development,” Applied and Environmental Microbiology, Vol. 24, No. 2, 1961, pp. 148-163.
- Y. Sakaitani, N. Yuki, F. Nakajima, S. Nakanishi, H. Tanaka, R. Tanaka and M. Morotomi, “Colonization of Intestinal Microflora in Newborn Foals,” Journal of Intestinal Microbiology, Vol. 13, No. 1, 1999, pp. 9-14.
- V. Julliand, A. De Vaux, L. Villard and Y. Richard, “Preliminary Studies on the Bacterial Flora of Faeces Taken from Foals, from Birth to Twelve Weeks. Effect of the Oral Administration of a Commercial Colostrum Replacer,” Pferdeheilkunde, Vol. 12, No. 3, 1996, pp. 209-212.
- N. R. Pace, “A Molecular View of Microbial Diversity and the Biosphere,” Science, Vol. 276, No. 5313, 1997, pp. 734-740. doi:10.1126/science.276.5313.734
- E. O. Casamayor, H. Schäfer, L. Bañeras, C. Pedrós-Alió- and G. Muyzer, “Identification of and Spatio-Temporal Differences between Microbial Assemblages from Two Neighboring Sulfurous Lakes: Comparison by Microscopy and Denaturing Gradient Gel Electrophoresis,” Applied and Environmental Microbiology, Vol. 66, No. 2, 2000, pp. 499-508. doi:10.1128/AEM.66.2.499-508.2000
- L. R. Dice, “Measures of the Amount of Ecologic Association between Species,” Ecology, Vol. 26, No. 3, 1945, pp. 297-302. doi:10.2307/1932409
- S. Crowell-Davis, “Normal Behavior and Behavioral Problems,” In: C. Kobluk, T. Ames, R. Geor, Eds., The Horse: Diseases and Clinical Management, W.B. Saunders, Philadelphia, 1995, pp. 1-20.
- S. L. Crowell-Davis, K. Houpt and J. Carnevale, “Feeding and Drinking Behavior of Mares and Foals with Free Access to Pasture and Water,” Journal of Animal Science, Vol. 60, No. 4, 1985, pp. 883-889.
- J. H. Connell and R. O. Slayter, “Mechanisms of Succession in Natural Communities and Their Role in Community Stability and Organization,” American Naturalist, Vol. 111, No. 982, 1977, pp. 1119-1144. doi:10.1086/283241
- C. E. Shannon, “A Mathematical Theory of Communication,” Bell System Technical Journal, Vol. 27, 1948, pp. 379-423.
- C. J. Krebs, “Ecological Methodology,” Addison Wesley Longman, Menlo Park, 1999.
- J. C. Frey, E. R. Angert and A. N. Pell, “Assessment of Biases Associated with Profiling Simple, Model Communities Using Terminal-Restriction Fragment Length Polymorphism-Based Analyses,” Journal of Microbiological Methods, Vol. 67, No. 1, 2006, pp. 9-19. doi:10.1016/j.mimet.2006.02.011
- G. Muyzer, E. C. De Waal and A. G. Uitterlinden, “Profiling of Complex Microbial Populations by Denaturing Gradient Gel Electrophoresis Analysis of Polymerase Chain Reaction-Amplified Genes Coding for 16S rRNA,” Applied and Environmental Microbiology, Vol. 59, No. 3, 1993, pp. 695-700.
- S. Hayes, H. Werner and L. M. Lawrence, “In Vitro Assessment of Fiber Digestion Capacity in Foals,” Proceedings of the 18th Equine Nutrition and Physiology Symposium, East Lansing, 4-7 June 2003, pp. 273-274.
- H. Yoshioka, K. Iseki and K. Fujita, “Development and Differences of Intestinal Flora in the Neonatal Period in Breast-Fed and Bottle-Fed Infants,” Pediatrics, Vol. 72, No. 3, 1983, pp. 317-321.
NOTES
*Mandatory Disclaimer: Proprietary or brand names are necessary to report factually on available data; however, the USDA neither guarantees nor warrants the standard of the product, and the use of the name by the USDA implies no approval of the product, nor exclusion of others that may be suitable.
#Corresponding author.

