Open Journal of Medical Microbiology
Vol.4 No.2(2014), Article
ID:46694,8
pages
DOI:10.4236/ojmm.2014.42014
Isolation and Identification of Multi-Drug Resistant Strains of Non-Lactose Fermenting Bacteria from Clinical Refuses in Major Hospitals of Khyber Pakhtunkhwa, Pakistan
Muhammad Zahid1*, Minhas Akbar1, Aftab Alam Sthanadar1,2, Pir Asmat Ali1, Mudassir Shah1,3, Iram Alam Sthanadar1, Muhammad Kaleem1, Muhammad Aslam1, Khayyam1, Nasrullah4, Muhammad Ismail Khan1, Syeda Mahreen Ul Hassan5, Noor Jehan Begum1
1Department of Zoology, Islamia College University, Peshawar, Pakistan
2Department of Zoology and Animal Sciences, Post Graduate College Dargai, Malakand, Pakistan
3Government Degree College Dara Adam Kheil, FR Kohat, Pakistan
4Punjab Institute of Cardiology, Lahore, Pakistan
5Department of Microbiology Shaheed Benazir Bhutto Women University Peshawar, Peshawar, Pakistan
Email: *mzahidsafi75@yahoo.com
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 29 April 2014; revised 29 May 2014; accepted 5 June 2014
ABSTRACT
Purpose: The present studied was performed in order to investigate the drug resistance of different non-lactose fermenting gram negative bacteria from clinical isolates. The bacteria were morphologically characterized through culturing and gram staining techniques were used for the identification of different bacterial strains. Methods: A total of 324 samples were collected from patients, after they were diagnosed by physicians at different hospitals at district Peshawar. Samples were morphologically identified by blood agar, MacConkey agar and Eosine Methylene Blue, identified by gram staining techniques. Modified Kirby-Bauer Disc diffusion method was used to test the in-vitro susceptibility of the identified isolates to different antibiotics. Results: The nonlactose fermenting gram negative bacteria were isolated from samples of blood (33.30%), pus/ wound (33.30%), urine (23.30%) and from ascetic/pleural fluids (10.20%). The study revealed that Pseudomonas aeroginosa showed high resistance against Gentamicin (74%) and Aztreonam (74%), followed by Ciprofloxacin (59.20%) and Amikacin (33.30). Tazocin was active as low resistance (18.50%) is shown. More resistance was seen in Morganella morganii against Aztreonam (77.7%) followed by Gentamicin (62.90%), Ciprofloxacin (40.70%). Tazocin show low resistance (3.70%). Multidrug resistant Proteus mirabillis was highly resistance to Gentamicin (66.60%), followed by Aztreonam (62.90%), Amikacin (55.50%), Ciprofloxacin (40.20%) and low resistance to Tazocin was (22.20%). Salmonella typhi demonstrated high resistance against Amikacin (62.90%), followed by Aztreonam (48.10%), Tazocin (40.70%). Gentamicin showed low resistance (29.60%), and hence it is more active against S. typhi. Conclusions: It can be concluded from the present study that different species of non-lactose fermenting gram negative bacteria have shown a different resistivity pattern. This study is a gate way for better and suitable management strategy for the infections caused by non-Lactose fermenting bacteria in the sampling region.
Keywords:Multidrug Resistant, Non-Lactose Fermenting Gram Negative Bacteria, Disc Diffusion Technique

1. Introduction
To develop anti-biotic resistance is quite natural and common in microbes. It is a natural process, which cannot be ceased and checked due to constantly evolving nature of microbes to the chemicals around them. The phenomenon is very important regarding its practical and economic implications. It is the same anti-biotic resistance issue that people cannot be effectively treated and have to stay ill for longer periods of time. The development of tolerance in microbes to more than one drug is commonly coined as multi drug resistance as per definition criteria (MDR) [1] . Among different bacteria, non-fermenters are gram-negative bacteria that cannot ferment sugars to produced energy for their physiology of life. Such Gram negative non-fermenting bacteria (NFGNB) were isolated from different clinical specimens, which are usually discarded at hospital and clinical levels [2] . Because of the extreme multidrug resistance problems, species of this group offer serious challenges for healthcare management. As mostly, non-fermenting (gram-negative) bacteria are of niche pathogens that readily cause serious and considerable infections in unhealthy individuals. Contrarily, on the other side, they are primarily healthcare-associated pathogens so they rarely cause infection in healthy persons [3] .
There are many reasons which indicate that why bacterial resistance should be an important concern for physicians. Bacterial resistance often results in treatment failure, which can pose considerable economic costs, especially in critically ill patients. For the management and treatment of infections in hospitalized patients, multidrug resistant bacteria required new approaches [4] . The major problem at clinic levels is emergence of multiple resistances. That is why the physician must know that the sensitivity level of pathogen before treatment could be started [5] . Antimicrobial resistance has become a major health problem worldwide, affecting every country to some degree. Because of the significant problem of increasing antimicrobial resistance, the global scientific community has tried to find new solutions; one of the most important ones is the evaluation and use of old antibiotic compounds. This study will help physicians, clinicians and consultants to decide its use in the most appropriate clinical infections. Microbial drug resistance is an unavoidable outcome of the use of antimicrobial agents [6] . The present study identified the various non-fermenters by staining techniques. The non-fermenting bacteria were isolated from various clinical specimens from patients admitted at Services Hospital Peshawar. The study was conducted, to check out their clinical importance, patterns of susceptibility of anti-microbes and to recognize the different health care related infections they cause [2] .
Drug resistance also leads to prolonged epidemics, and consequently there is a greater risk of infection to others. Self-medication and improper unauthorized use of antibiotics can also lead to drug resistance. Drug resistance is a serious issue in Pakistan. Pakistan is a developing country and because of drug resistance, an enough chunk of revenue is annually allocated for the import of new drugs which could be alternatively used on other developmental programs as well. Patients are put on new drugs, and stay for a longer period in hospital. Thus more resources and time are consumed due to the problem of drug resistance in pathogenic microbes.
2. Materials and Methods
2.1. Sample Collection
A total 350 clinical samples were included in this study. Samples were collected from patients’ diagnosed by physicians, at their visit to different major hospitals at Peshawar. Sampling was made from 1st March to end of October 2012, for a period of 6 months. Clinical isolates were collected from pus/wound, urine, ascetic/plural fluids and blood.
2.2. Isolation of Clinical Specimens
The clinical samples isolation of were carried out according to the protocol [7] . The collected samples from pus/ wound, blood, urine and ascetic/plural fluids were spread on blood, MacConkey and Eosine Methyline Blue (EMB) agar plates and incubated at 37˚C for 24 - 48 hours.
2.3. Gram Staining
Gram staining was carried out as early described [8] to identify the NFGNB bacteria.
2.4. Antibiotic Sensitivity Test
The Kirby-Bauer Disc Diffusion Method was used to test the in vitro susceptibility of the identified isolates to Ceftazidime (30 µg), Cefoperazone (75 µg), Ceftriaxone (30 µg), Cefepime (30 µg), Imipenem (10 µg). Pseudomonas aeroginosa colonies were picked up from the culture plate with the help of a sterile platinum wire loop and emulsified in 4 ml of sterile peptone water to match with 0.5 McFarland turbidity standards (1.5 × 108 cfu/ml). The surface of Mueller Hinton Agar (Oxoid, Basingstoke, UK) in a Petri dish was inoculated evenly through a sterile swab and for 10 minutes was allowed the agar to dry. A multichannel disc dispenser (Oxoid, Basingstoke, UK) was used to deposit the antibiotics discs onto the surface of the inoculated medium. The plate was then incubated at 37˚C for 24 hours. With measuring scale the diameters of zone of inhibition were measured in millimeters after 24 hours of period of incubation [9] . The above procedure was repeated thrice for each P. mirabillis, M. morganii and for S. typhi isolates.
3. Results
A total of 324 drug resistant bacteria were isolated from 350 clinical samples processed at Microbiology Laboratory of Services Hospital Lahore. The identification of bacterial strains was then identified on the basis of morphological cultural, and biochemical analysis (Table 1). The occurrence of isolates was lower in males (47%, n = 324) as compared to females (53%, n = 324) while highest frequency of MDR-NFGNB was observed among young patients of age from 11 - 30 years 54.6%). As there were 171 cases out of 324 total study sample. As compared to rest of the age groups, the frequency of MDR-NFGNB was 23.1% in age group of 31 - 60 years age. Similarly it was 22.2% in case of age group from 61 - 74 years. The distribution of clinical isolates among different age group is showed in Table1 Overall infection rate was higher in young individuals (11 - 30 years) but P. aeroginosa infection was higher in old age (61 - 74), shown in Table1
The Distribution of MDR-NFGNB P. mirabilis, M. morganii, P. aeroginosa, and S. typhi in different clinical specimens (blood, pus/wound, urine, ascetic/pleural fluids) are shown in Table2 The results were presented on simple cylindrical bar graph in Figure 1. It was observed that Pseudomonas aeriginosa was found to be 77.00%, 62.90% resistant against Aztreonam and Gentamicin respectively. The resistance found in P. aeroginosa against other antibiotics included Ciprofloxacin (40.70%); Amikacin (18.50%); Tazocin (03.70%) as shown in Table3
MDR-Patteren in P. mirabilis was of a higher degree of resistance to Gentamicin (66.60%) was detected in P. mirabilis isolates. The frequency of resistance against was Aztreonam (62.90%); Amikacin (55.50%); Ciprofloxacin (40.03%); Tazocin (22.20%) (Table 3). As Tazocin showed lower resistance to P. mirabilis.
MDR-Patteren in Salmonella typhi isolates exhibited high resistance against Amikacin (62.90%). Resistance to other antibiotics included Aztreonam (48.10%); Tazocin (40.70%); Ciprofloxacin (37.03%); Gentamicin (29.60%) and thus a lower resistance to Cefepime were observed (Table 3). For the sake of convenience, results in Table 3 were presented on simple horizontal bar graph in Figure 2.
4. Discussion
Multi-drug resistance in bacteria is of very much concern to clinicians as of development of resistance to multi drugs in microorganisms [10] . In microorganisms, antibiotic resistant pathogens causes financial burden, in
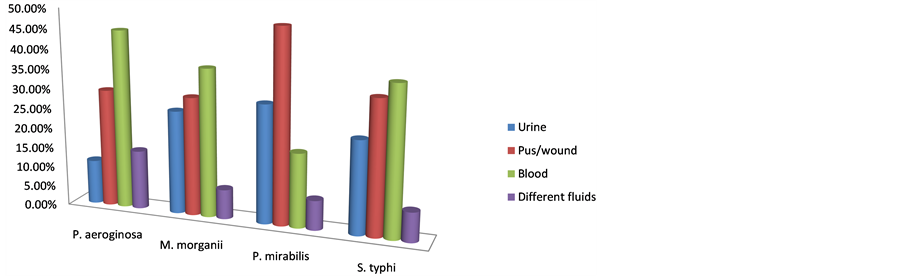
Figure 1. MRD-Bacteria percent isolated samples from different parts of Patients’ bodies.
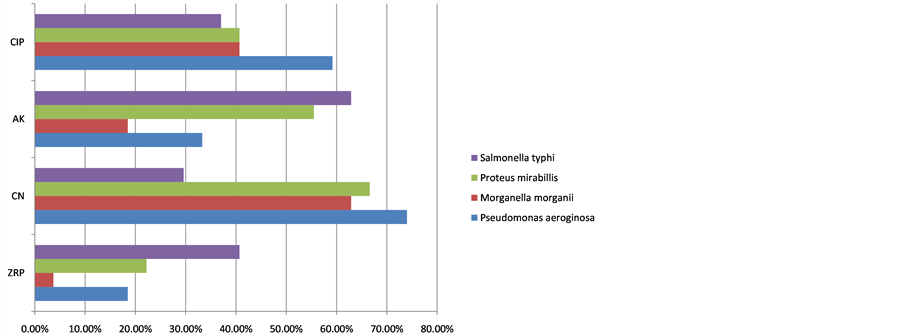
Figure 2. Antibiotics percent resistance in Multi Drug Resistant Bacteria from isolated samples.
creases chances of treatment failure and readily spread from person to other person/patients [11] . In the last fifty years, due to the widespread use of antibiotics, non-fermentative gram negative bacilli have emerged important health care-associated pathogens. Recently studies were conducted on important areas like identification of nonfermentative gram negative bacilli and monitoring their susceptibility patterns for the appropriate management of its infections. Further to make clear the fact that it is important to establish the clinical relevance of the isolated non fermentative gram negative bacilli, before they are considered as pathogens. This would prevent unnecessary usage of antibiotics and the rise of drug-resistant strains [12] . A very limited data on the prevalence and resistance pattern of NFGNB is available especially in the sampling area. The present study was aimed to highlight the issues.
MDR-NFGNB was isolated from suspected patients of different age groups (11 - 74). The highest number of MDR-NFGNB was isolated from age group of 11 - 30 years, followed by age group of 31 - 60 and 61 - 74 years. People were susceptible to infection at the early and middle age (11 - 30 years) because at this age the people are more active, have more social contacts, so will have more chances of infection. Chances of infection depend on number of exposure to the injurious microbes. With increasing age the social contact of an individual decreases so have less chances of developing infectin.
In the present study, Gender wise distribution of MDR-NFGNB was isolated both from males and females and highest numbers were recorded in females. This could be due to the social activity of females in their life in developing countries like Pakistan. Where females are much ignored as compared to male so their food cleanliness is not good as males, as a result their immune system is weak. Furthermore, females give birth child’s so admitted frequently to hospitals and thus has more chances of infection.
Different non-lactose fermenting bacteria P. aeroginosa, M. morganii, P. mirabillis and S. typhi were isolated and identified from blood, pus/wound, urine and ascetic fluids. The present results were similar to the results
*n for numbers: % for percentage.
reported by other investigators [2] [13] [14] for P. aeroginosa, Anjum et al. [15] , Javeed et al. [16] , for M. morganii, further Singla et al. [17] and Lee et al. [18] . A higher frequency of P. mirabillis was observed in pus (48.2%) as compared that reported by Saito et al. [19] and Jombo et al. [20] . Previous study of (Nagshtly et al. [21] ; Hassan et al. [22] and unique in the sense as it identified S. typhi from urine, blood, pus/wound, ascetic and pleural fluids 29.60%, 37.03%, 25.90%, 7.40% respectively.
In-vitro MDR-Patteren in P. aeroginosa showed 74.00%, 74.00%, and 18.50% resistance against Aztreonam, Gentamicin and Tazocin respectively in the current study. These results are consistent with the findings of some earlier studies carried out by Satti et al. [23] , Rajat et al. [24] and Romao et al. [25] reported the same results in Morganella morganii in-vitro MDR-Patteren showed 3.7% against Tazocin. These findings are quite in line with the findings of Falagas et al. [26] and Xiao et al. [27] . Our findings are well supported by their work [26] [27] . The findings of Lee et al. [18] and Falagas et al. [26] reported 9% resistance against Aztreonam, 8% against Ciprofloxacin, 4% against Amikacin. Our work is not in line with their work. All such findings are beyond the range and quite against our findings. This difference may be due to geographical differences as antibiotic resistance differs from region to region and from time to time.
In our present investigations, the in-vitro MDR―P. mirabillis showed 22.20%, 55.50% and 66.60% resistance against Tazocin, Amikacin, Gentamicin which are in clear contradiction with the findings of Falagas et al. [26] , who reported 100%, 100% and 0.3% resistance for the same antibiotics. The present study in-vitro MDR-Patteren S. Typhis showed 29.60% resistance against Gentamicin, and 37.03% resistance against Ciprofloxacin. These results are in agreement with the work of Akinyemi et al. [28] who identified 30% resistance against
Table 2 . Clinical isolates of gram negative non-fermenters bacteria from different sites in hospitals.
*n for numbers: % for percentage.
Table 3. Antibiotic resistance percentage of gram negative non fermented bacteria (n = 81) isolated from different clinical samples.
Gentamicin. Islam et al. [29] recoded 30% resistance against Ciprofloxacin in S. typhi. Likely, Pokhare et al. [30] reported 43% resistance against Ciprofloxacin. Salmonella typhi resistance against Tazocin, Aztreonam and Amikacin has been recoded for first time in this region with figures, i.e. 40.70%, 48.10% and 62.90% respectively.
Acknowledgements
The authors acknowledge with gratitude the invaluable support and assistance provided by Professor Dr. Abdul Hamid Jan. We are also thankful to Microbiology laboratories and its technical staff for extending us lab facilities.
References
- White, D.G., Hudson, C., Maurer, J.J. and Ayers, S. (2000) Characterization of Chloramphenicol and Florfenical Resistance in Escherichia coli Associated with Bovine Diarrhea. Journal of Clinical Microbiology, 38, 4593-4598.
- Gokale, S.K. and Metgud, S.C. (2010) Characterization and Antibiotic Sensitivity Pattern of Non-Fermenting Gram Negative Bacilli from Various Clinical Samples in a Tertiary Care Hospital, Belgaum. Journal of Pharmaceutical and Biomedical Science, 17, 2230-7885.
- Gowan Jr., Mc. and John, E. (2006) Resistance in Non-Fermenting Gram-Negative Bacteria: Multidrug Resistance to the Maximum. The American Journal of Medicine, 119, 29-36. http://dx.doi.org/10.1016/j.amjmed.2006.03.014
- Moon, A.H. and Kamble, V.A. (2012) Comparative Studies on: Drug Resistance Patterns of Common Clinical Bacterial Isolates against Essential Oils and Commercial Antibiotics. Asiatic Journal of Biotechnology Resources, 3, 1447- 1454.
- Zehra, A., Naqvi, B.S., Bushra, R. and Ali, S.Q. (2010) Comparative Study on Resistance Pattern of Different Pathogens against Cefixime and Cefepime. Jordan Journal of Pharmaceutical Science, 3, 145-156.
- Masood, S.H. and Aslam, N. (2010) In Vitro Susceptibility Test of Different Clinical Isolates against Ceftriaxone. The Open Museum Journal, 25, 199-202. http://dx.doi.org/10.5001/omj.2010.56
- Cheesbrough, M. (2000) District Laboratory Practice Manual in Tropical Countries Part 2. Cambridge University Press, Cambridge, 178-179.
- Harley, J. and Prescott, L. (1990) Laboratory Exercises in Microbiology. Wm. C. Brown Publishers, 49-53.
- Gloria, A., Cheryl, B., John, E., Richard, F., Joan, S.K., Tanja, P., Joy, W. and Scott, F.D. (2003) Manual for the Laboratory Identification and Antimicrobial Susceptibility Testing of Bacterial Pathogens of Public Health Importance in the Developing World. Centers for Disease Control and Prevention, Atlanta, and World Health Organization, Department of Communicable Disease Surveillance and Response, 103-118.
- Roshan, M., Ikram, A., Mirza, I.A., Malik, N., Abbasi, S.A. and Alizai, S.A. (2011) Susceptibility Pattern of Extended Spectrum B-Lactamase Producing Isolates in Various Clinical Specimens. Journal of the College of Physicians and Surgeons, 21, 342-346.
- Mahmod, A. (2000) Bacteriology of Surgical Site Infections and Antibiotic Susceptibility Pattern of the Isolates at a Tertiary Care Hospital in Karachi. Journal of Pakistan Medical Association, 50, 256-259.
- Upgade, A., Prabhu, N., Gopi, V. and Soundararajan, N. (2012) Current Status of Antibiotic Resistant Non-Fermentative Gram Negative Bacilli among Nosocomial Infections. Advances in Applied Science Research, 3, 738-742.
- Bhargavi, P.S., Gopala, R.T.V., Mukkanti, K., Dinesh, K.B. and Krishna, T.P. (2010) Increasing Emergence of Antibacterial Resistance Mainly in Uropathogens: Southeast Part of India. International Journal of Microbio Research, 2, 1-6.
- Bhongle, N.N., Nagdeo, N.V. and Thombare, V.R. (2012) The Prevalence of Metallo β-Lactamases in the Clinical Isolates of Pseudomonas aeruginosa in a Tertiary Care Hospital: An Alarming Threat. Journal of Clinical Diagnose Research, 6, 1200-1202.
- Anjum, F. and Mir, A. (2010) Susceptibility Pattern of Pseudomonas aeruginosa against Various Antibiotics. African Journal of Microbiology Research, 4, 1005-1012.
- Javeed, I., Hafeez, R. and Anwar, M.S. (2011) Antibiotic Susceptibility Pattern of Bacterial Isolates from Patients Admitted to a Tertiary Care Hospital in Lahore. D Biomedica, 27, 19-23.
- Singla, N., Kaistha, N., Gulati, N. and Chander, J. (2010) Morganella morganii Could Be an Important Intensive Care Unit Pathogen. Indian Journal of Critical Care Medicine, 14, 154-155. http://dx.doi.org/10.4103/0972-5229.74176
- Lee, I.K. and Liu, J.W. (2006) Clinical Characteristics and Risk Factors for Mortality in Morganella morganii Bacteremia. Journal of Microbial Immunological Infection, 39, 328-334.
- Saito, R., Okugawa, S., Kumita, W., Sato, K., Chida, T., Okamura, N., Moriya, K. and Koike, K. (2007) Clinical Epidemiology of Ciprofloxacin Resistant Proteus Mirabilis Isolated from Urine Samples of Hospitalized Patients. Clinical Microbiology and Infection, 13, 1204-1206. http://dx.doi.org/10.1111/j.1469-0691.2007.01826.x
- Jombo, G.T.A., Emanghe, U.E., Amefule, E.N. and Damen, J.G. (2012) Nosocomial and Community Acquired Uropathogenic Isolates of Proteus Mirabilis and Antimicrobial Susceptibility Profiles at a University Hospital in Sub-Saharan Africa. Asian Pacific Journal of Tropical Disease, 2, 7-11.
- Nagshetty, K., Shivannavar, T., Channappa and Gaddad, S.M. (2010) Antimicrobial Susceptibility of Salmonella typhi in India. Journal of Infection in Developing Countries, 4, 70-73.
- Hasan, R., Zafar, A., Abbas, Z., Mahraj, V. and Malik, F. (2008) Zaidi A. Antibiotic Resistance among, Serovars Typhi and Paratyphi A in Pakistan (2001-2006). Journal of Infection in Developing Countries, 2, 289-294. http://dx.doi.org/10.3855/jidc.224
- Satti, L., Abbasi, S., Qumar, T.A., Khan, M.S. and Hashmi, Z.A. (2010) In Vitro Efficacy of Cefepime against MultiDrug Resistant Pseudomonas aeruginosa, an Alarming Situation in our Setup. The Open Drug Resistance Journal, 1, 12-16. http://dx.doi.org/10.2174/1876521101101010012
- Rajat, R.M., Ninama, G.L., Kalpesh, M., Rosy, P., Kanu, P. and Vegad, M.M. (2012) Antibiotic Resistance Pattern in Pseudomonas aeruginosa Species Isolated at a Tertiary Care Hospital, Ahmadabad. National Journal of Medical Research, 2, 156-159.
- Romão, C.M.C.P.A., Faria, Y.N.D., Pereira, L.R. and Asensi, M.D. (2006) Susceptibility of Clinical Isolates of Multi Resistant Pseudomonas aeruginosa to a Hospital Disinfectant and Molecular Typing. Mem Inst Oswaldo Cruz, Rio de Janeiro, 100, 541-548.
- Falagas, M.E., Kavvadia, P.K., Mantadakis, E., Kofteridis, D.P., Bliziotis, I.A., Saloustros. E., Maraki, S. and Samonis, G. (2005) Morganella morganii Infections in a General Tertiary Hospital. Infection, 34, 315-321.
- Xiao-Bo, M., Xiao-Ju, L., Li-Yuan, M., Hui-Li, C., Wen-Zhao, C., Xiao-Jingo, G., Chuan-Min, T. and Zhi-Xing, C. (2008) Antimicrobial Susceptibility Analysis of Ninety-One Clinical Isolates of Morganella morganii. Chinese Journal of Antibiotics.
- Akinyemi, K.O., Smith, S.I., Oyefolu, A.O.B. and Coker, A.O. (2005) Multidrug Resistance in Salmonella enterica serovar typhi Isolated from Patients with Typhoid Fever Complications in Lagos, Nigeria. Public Health, 119, 321-327. http://dx.doi.org/10.1016/j.puhe.2004.04.009
- Islam, M.J., Das, K.K., Sharmin, N., Hasan, M.N. and Azad, A.K. (2008) Antimicrobial Susceptibility of Salmonella Serovars Isolated from Blood. Journal of Innovative Development Strategy, 2, 22-27.
- Pokhare, B.M., Koirala, J., Daha, R.K., Mishra, S.K., Khadga, P.K. and Tuladhar, N.R. (2006) Multidrug-Resistant and Extended-Spectrum Beta-Lactamase (ESBL)-Producing Salmonella enterica (Serotypes Typhi and Paratyphi A) from Blood Isolates in Nepal: Surveillance of Resistance and a Search for Newer Alternatives. International Journal of Infectious Diseases, 10, 434-443. http://dx.doi.org/10.1016/j.ijid.2006.07.001
NOTES

*Corresponding author.


