Journal of Biosciences and Medicines
Vol.03 No.11(2015), Article ID:60920,9 pages
10.4236/jbm.2015.311004
Holter ECG-Based Apnea Hypopnea Index to Screen Obstructive Sleep Apnea: A New Proposal and Evaluation of Feasibility
Motoki Sakai1, Daming Wei2
1Faculty of Policy Informatics, Chiba University of Commerce, Chiba, Japan
2Tohoku University, Graduate School of Medicine, Miyagi, Japan

Copyright © 2015 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).


Received 5 August 2015; accepted 3 November 2015; published 6 November 2015

ABSTRACT
Obstructive sleep apnea syndrome (OSAS) is a common sleep disorder. It has been reported that approximately 40% of patients with moderate or severe OSAS die within the first eight years of disease. In hospitals, OSAS is inspected using polysomnography, which uses a number of sensors. Because of the cumbersome nature of this polysomnography, an initial OSAS screening is usually conducted. In recent years, OSAS screening techniques using Holter electrocardiogram (ECG) have been reported. However, the techniques so far reported cannot perform an OSAS severity assess- ment. The present study presents a new method to distinguish the obstructive sleep apnea (OSA) and non-OSA epochs at one-second intervals based on the Apnea Hypopnea Index assessment, defined as the duration of continuous apnea. In the proposed method, the time-frequency components of the heart rate variability and three ECG-derived respiration signals calculated by the com- plex Morlet wavelet transformation are adopted as features. A support vector machine is employed for classification. The proposed method is evaluated using three eight-hour ECG recordings containing OSA episodes from three subjects. As a result, the sensitivity and specificity of classification are found to reach approximately 90%, a level suitable for OSAS screening in clinical settings.
Keywords:
Obstructive Sleep Apnea Syndrome, Heart Rate Variability, ECG-Derived Respiration, Complex Morlet Wavelet Transformation, Support Vector Machine

1. Introduction
Obstructive sleep apnea syndrome (OSAS), which is caused by repetitive occlusions of the upper airways, is a common sleep disorder. It has been reported that approximately 80% of sleep apnea cases are OSAS. OSAS itself is not necessarily fatal, but is a cause of hypertension, arrhythmia, cardiac arrest, diabetes, and dyslipidemia, which bring further risks of brain infarct or cardiac infarct [1] . Reports indicate that approximately 40% of patients with moderate or severe OSAS die within the first eight years of disease [2] . Thus, early detection and treatment of OSAS are important.
To assess OSAS, a polysomnogram (PSG) is used. This adopts the electroencephalogram (EEG), electrocardiogram (ECG), peripheral capillary oxygen saturation (SpO2), pressure transducers, and nasal cannula to obtain information related to sleep stages, heart rate, and respiratory variation. Because of the cumbersome nature of PSG inspection, a screening examination using fewer sensors is conducted for patients suspected of having OSAS. The most convenient form of this screening is to use only the ECG.
The Holter ECG-based OSAS screening uses features calculated from the heart rate variability (HRV) and ECG-derived respiration (EDR). The HRV analysis is effective for OSAS detection because the heart rate changes rapidly after an episode of obstructive sleep apnea (OSA). In previous research [3] [4] , the time- frequency components of HRV time series were used to screen OSAS. Hayano et al. presented indices to detect OSAS in the time domain [5] [6] . The EDR is a respiratory waveform estimated from the ECG signal. In many cases, the estimated respiratory waveform is obtained from the R wave amplitude or beat-to-beat fluctuations [7] - [10] . These fluctuations of the R wave are effective in estimating the respiratory waveform because the R wave amplitude is influenced by respiratory fluctuations. As for HRV, features of the EDR signal are computed in the time, frequency, or time-frequency domain.
Two approaches are generally used: data mining algorithms such as the support vector machine (SVM), AdaBoost, or linear discriminant analysis [11] - [15] ; and empirical algorithms that screen OSAS by defining discrimination conditions from empirical data such as the cycle variation of heart rate method [6] .
In previous research using Holter ECG-based OSAS screening, the goal has been to assess the existence or absence of an OSA episode. In other words, existing OSAS screening methods cannot determine the severity of OSAS. Generally, OSAS severity is determined using the Apnea Hypopnea Index (AHI), whereby the severity of OSAS is defined by the total number of apnea and hypopnea per hour of sleep [1] , as shown in Table 1.
In the AHI-based assessment, the duration of an OSA episode must be measured. However, no previous studies record the duration of OSA episodes. In most studies, the existence or absence of an OSA episode is evaluated at one-minute intervals, as in the PhysioNet/CinC challenge (in the PhysioNet database, the existence or non-existence of OSA episodes is annotated every minute).
In this study, we propose a method that detects OSA episodes at one-second intervals to quantitatively assess the OSAS severity. For this purpose, the time-frequency components of EDR and HRV signals are adopted as features of OSA and non-OSA, meaning that relatively few beats are needed. Our motivation is simply to realize a practical and quantitative method for OSAS severity screening, and to contribute to the early detection and treatment of OSAS.
2. Method
In this paper, the time-frequency components of the EDR and HRV computed with the complex Morlet wavelet transformation (CMORWT) are used as classification features for OSA and non-OSA epochs, and an SVM is used as the classifier. The process of the overall methodology is shown in Figure 1.
2.1. Preprocessing
In previous studies [11] [15] , median filtering of the raw ECG signals was performed at widths of 200 ms and
Table 1. AHI-based definition of OSAS severity.
Figure 1. Processing flow of OSA episode detection method.
600 ms before future extractions, because the baseline wander decreases the accuracy of the EDR series. After median filtering, R wave detection can be performed using the wavelet transformation modulus maxima (WTMM) method [16] .
2.2. Calculation of EDR and HRV Signals
On the basis of previous research regarding Holter ECG-based OSAS screening, we adopt the HRV and EDR signals calculated from the area of the R wave in this research. In addition, two further EDR methods are proposed in this research.
・ HRV
HRV is the cycle variation over the duration of the heartbeat, which is defined as the time interval from one R wave to the next. This is also called the R-R interval time series, and is computed as follows:
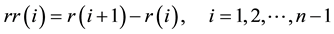 (1)
(1)
where 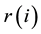 is the i-th R wave peak time. Furthermore, the R-R interval time series is resampled into 1 Hz periods using a cubic spline for the following time-frequency analysis.
is the i-th R wave peak time. Furthermore, the R-R interval time series is resampled into 1 Hz periods using a cubic spline for the following time-frequency analysis.
・ EDR1: Area of R wave
For the first EDR, an area of the R wave is computed [3] . This is the most typical EDR method. The onset and offset of the R wave are initially detected by searching for the lowest absolute values within 100 ms (before and after) of the minimum value of an R wave peak. Next, the preprocessed ECG signal between onset and offset is integrated to obtain the QRS complex area (Figure 2(a)). Areas are computed in all QRS complexes, and the resulting EDR series is then resampled. (This resampling is also performed in the EDR2 and EDR3 methods.)
・ EDR2: QRST integration
The second EDR method involves QRST integration, as shown in Figure 2(b). Generally, a length of the QT- segment changes with variations in heart rate, and it is clear that the heart rate is also influenced by changes in
Figure 2. Range of ECG area calculations: (a) EDR1; (b) EDR2; (c) EDR3.
respiration. In fact, it may be that the QRST integration is more sensitive to changes in respiration than the area of the R wave. In this study, the onset and offset of each elementary wave (i.e., Q and T waves) is also detected by the WTMM method [16] .
・ EDR3: Area between two R waves
Although EDR1 and EDR2 are promising for OSAS detection, they have the practical disadvantage that the onset or offset of each elementary wave cannot necessarily be detected in a Holter ECG recording. The detection of an R wave is more robust than for other elementary waves. Therefore, we propose the area between two R waves (Figure 2(c)) as a third EDR method, because this only requires the detection of R waves.
2.3. CMORWT-Based Feature Extraction of Time-Frequency Area
Next, the time-frequency components of the calculated EDR and HRV signals are computed using the wavelet transformation. In this research, we adopt the CMORWT, because it can relate scale levels to actual frequencies [17] , which enables us to relate the time-frequency components to common frequency features such as HF (0.20 - 0.35 Hz) or LF (0.05 - 0.2 Hz).
The mother wavelet of CMORWT is defined as:
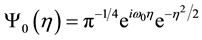 (2)
(2)
where 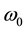 is the characteristic eigen-frequency. The scaled and shifted mother wavelet
is the characteristic eigen-frequency. The scaled and shifted mother wavelet 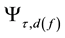 is computed with the control factor
is computed with the control factor 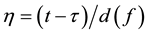 (f is the main receptive frequency).
(f is the main receptive frequency). 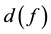 can be expressed as follows:
can be expressed as follows:
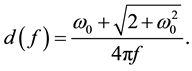 (3)
(3)
Let S(t) denote the EDR or HRV signal obtained from the ECG. Its wavelet component 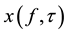 at time
at time 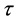 and frequency f is computed as:
and frequency f is computed as:
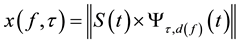 (4)
(4)
for f < {0.01, 0.02, …, 0.4} Hz.
2.4. Classification Using Support Vector Machine
In this research, we adopt an SVM [18] as a classifier to investigate OSA episodes. SVMs are effective for learning and separating datasets in high-dimensional feature space. However, we do not have to define explicit features, and can use several kernel functions that can be regarded as inner products in an unknown feature space.
In previous research [14] , the SVM was used with a linear kernel function, and this was shown to be effective. In this research, we also use a linear kernel function, and select the inner product as the kernel. The optimization step uses the gradient ascent algorithm.
The input data for the SVM classifier are the time-frequency components obtained by CMORWT at one- second intervals. We randomly select 50% of the measured ECG signals as learning data, and use the remaining data for the evaluation.
3. Experiment
To evaluate the proposed method, three subjects’ ECG recordings during sleep were used. All subjects were male, aged from 23 - 50. They may be OSAS candidates. The ECG signals were measured using a mobile OSAS monitor, the SAS-3200 produced by Nihon Kohden. ECG electrodes were placed on CM5, NASA, and CC5. (In this paper, CM5, NASA, and CC5 are called ch1, ch2, and ch3, respectively.) The ECG sampling rate was 1000 Hz. The recording length for each subject was approximately eight hours. Simultaneously, the actual respiration signals were measured using an air flow sensor with sampling rates of 40 Hz.
4. Evaluation
We labeled the measured ECG signal as an “OSA epoch” or “non-OSA epoch” according to respiratory waveform measured with the air flow sensor. In particular, when air flow was absent or decreased, its epoch was labeled as “OSA epoch”. Other epochs were labeled as “non-OSAS epoch”. These labeled epochs were used in the learning and evaluation steps.
To validate the effectiveness of the proposed method, the classification accuracy for OSA and non-OSA epochs was evaluated in each ECG channel. As evaluation indices for classification accuracy, we used the sensitivity (=TP/TP + FN) and specificity (=TP/TP + FP), where TP, TN, FN, and FP denote the true number of OSA epochs, true number of non-OSA epochs, false negative, and false positives, respectively.
5. Results and Discussion
5.1. Time-Frequency Components of Three EDRs and HRV
Figures 3-6 show the EDR1, EDR2, EDR3, and HRV spectrograms for one subject. These EDR spectrograms were obtained from the ECG signal measured through channel 3. The figures show the time-frequency components of OSA and non-OSA epochs. The colors in Figures 3-6 represent the energy intensity, with red indicating higher energy and blue indicating lower energy. From these figures, it can be seen that the time-frequency feature of the HRV series is most effective for the classification of OSA and non-OSA epochs. In particular, it can be seen that the energy difference between OSA and non-OSA epochs was large in the low-frequency region (0.01 - 0.05 Hz). Similarly, the effectiveness of the low-frequency region can be seen in the three EDR spectrograms. Among these, EDR3, which is calculated from the area between two R waves, seems to be more efficient than the other EDR methods.
Next, the distributions of low-frequency components of OSA and non-OSA epochs are presented to further confirm the effectiveness of EDR3 and HRV’s in this region. Figure 7 shows scatter diagrams of the 0.01 Hz and 0.05 Hz components for one subject in the time-frequency domain. In Figure 7, the time-frequency EDR3 characteristics obtained from channels 1, 2, and 3 are plotted. The scatter plot obtained from channel 2 illustrates some overlapping features of OSA and non-OSA epochs. In contrast, less overlap can be seen in the scatter diagrams obtained from channels 2 and 3 or that of HRV. These results imply that the time-frequency components of HRV, and EDR3 computed with channel 2 or 3 might be effective for classifying OSA and non-OSA epochs. In Section 5.2, we evaluate the classification accuracy based on these two time-frequency components.
5.2. Classification Accuracy
Table 2 lists the sensitivity and specificity of classifications performed using only the time-frequency components
Figure 3. Time-frequency representation of EDR1 signal.
Figure 4. Time-frequency representation of EDR2 signal.
of HRV and EDR3. The frequency range was from 0.01 - 0.05 Hz. Table 3 gives the sensitivity and specificity when classifications were performed with all time-frequency components. In both tables, the sensitivities and specificities are highest in channel 3, and those in channel 2 are lowest. This is consistent with the results in Figure 7. Moreover, this indicates that the accuracy level of the EDR signal is related to the classification.
In contrast, although Figures 3-7 suggest that low-frequency components are effective, Table 2 and Table 3 indicate that the classification accuracies increase with both low-frequency features and features in other frequency regions, because the sensitivities and specificities increase when all time-frequency components are adopted (the highest sensitivity and specificity are approximately 90% when the EDRs were computed from channel 3). From this, we can hypothesize that time-frequency components which are ineffective in the input space might contribute in the SVM-based mapping to the unknown feature space. However, effective features should have been selected using a feature selection method, such as a genetic algorithm. Adopting an efficient
Figure 5. Time-frequency representation of EDR3 signal.
Figure 6. Time-frequency representation of HRV signal.
Table 2. Classification accuracy obtained by features whose frequency region ranges from 0.01 Hz to 0.05 Hz.
feature selection method will increase the classification accuracy.
From these results, we can conclude that the classification of OSA and non-OSA epochs at one-second intervals is feasible for AHI-based assessment with time-frequency components of HRV and those of EDRs
Figure 7. Scatter diagrams of EDR3 and HRV time-frequency components (0.01 - 0.05 Hz) in each channel. (Triangles denote OSA epoch features in a one-second interval, asterisks denote those of non-OSA epochs.).
Table 3. Classification accuracy obtained by all features.
computed from a suitable channel. However, the number of subjects in this study was not significant. In future work, we will conduct experiments with a greater number of subjects to evaluate the validity of the proposed method.
6. Conclusion
This study proposed a new method to evaluate the AHI for OSAS screening using Holter ECG. The proposed method distinguishes OSA and non-OSA epochs every second. The time-frequency components of HRV and three EDR signals computed by CMORWT were used as features, and, classification was performed with SVM. Three eight-hour ECG recordings containing OSA episodes from three subjects were used to evaluate the proposed method. Our results showed that the sensitivity and specificity of classification reached approximately 90%, indicating the feasibility of this approach for clinical OSAS screening to enable early detection and treatment.
Cite this paper
MotokiSakai,DamingWei, (2015) Holter ECG-Based Apnea Hypopnea Index to Screen Obstructive Sleep Apnea: A New Proposal and Evaluation of Feasibility. Journal of Biosciences and Medicines,03,33-41. doi: 10.4236/jbm.2015.311004
References
- 1. He, J., Kryger, M.H., Zorick, F.J., Conway, W. and Roth, T. (1988) Mortality and Apnea Index in Obstructive Sleep Apnea. Experience in 385 Male Patients. Chest, 94, 9-14.
http://dx.doi.org/10.1378/chest.94.1.9 - 2. Yaggi, H.K., Concato, J., Kernan, W.N., Lichtman, J.H., Brass, L.M. and Mohsenin, V. (2005) Obstructive Sleep Apnea as a Risk Factor for Stroke and Death. New England Journal of Medicine, 353, 2034-2041.
http://dx.doi.org/10.1056/NEJMoa043104 - 3. Corthout, J., Van Huffel, S., Mendez, M.O., Bianchi, A.M., Penzel, T. and Cerutti, S. (2008) Automatic Screening of Obstructive Sleep Apnea from the ECG Based on Empirical Mode Decomposition and Wavelet Analysis. 30th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Vancouver, 20-25 August 2008, 3608-3611.
http://dx.doi.org/10.1109/iembs.2008.4649987 - 4. Penzel, T., McNames, J., Murray, A., de Chazal, P., Moody, G. and Raymond, B. (2002) Systematic Comparison of Different Algorithms for Apnoea Detection Based on Electrocardiogram Recordings. Medical & Biological Engineering & Computing, 4, 402-407.
http://dx.doi.org/10.1007/BF02345072 - 5. Hayano, J., Tsukahara, T., Watanabe, E., Sasaki, F., Kawai, K., Sakakibara, H., Kodama, I., Nomiyama, T. and Fujimoto, K. (2013) Accuracy of ECG-Based Screening for Sleep-Disordered Breathing: A Survey of All Male Workers in a Transport Company. Sleep Breath, 17, 243-251.
http://dx.doi.org/10.1007/s11325-012-0681-7 - 6. Hayano, J., Watanabe, E., Saito, Y., Sasaki, F., Fujimoto, K., Nomiyama, T., Kawai, K., Kodama, I. and Sakakibara, H. (2011) Screening for Obstructive Sleep Apnea by Cyclic Variation of Heart Rate. Circulation: Arrhythmia and Electrophysiology, 4, 64-72.
http://dx.doi.org/10.1161/circep.110.958009 - 7. Travaglini, A., Lamberti, C., DeBie, J. and Ferri, M. (1998) Respiratory Signal Derived from Eight-Lead ECG. Proceeding of Computers in Cardiology, 13-16 September 1998, 65-68.
http://dx.doi.org/10.1109/cic.1998.731718 - 8. Ding, S., Zhu, X., Chena, W. and Wei, D. (2004) Derivation of Respiratory Signal from Single-Channel ECGs Based on Source Statistics. International Journal of Bioelectromagnetism, 6.
- 9. Bowers, E.J., Murray, A. and Langley, P. (2008) Respiratory Rate Derived from Principal Component Analysis of Single Lead Electrocardiogram. The Proceeding of Computers in Cardiology, Bologna, 14-17 September 2008, 437-440.
- 10. O’Brien, C. and Heneghan, C. (2007) A Comparison of Algorithms for Estimation of a Respiratory Signal from the Surface Electrocardiogram. Computers in Biology and Medicine, 37, 305-314.
http://dx.doi.org/10.1016/j.compbiomed.2006.02.002 - 11. Mendez, M.O., Bianchi, A.M., Matteucci, M., Cerutti, S. and Penzel, T. (2009) Sleep Apnea Screening by Autoregressive Models From a Single ECG Lead. IEEE Transactions on Biomedical Engineering, 56, 2838-2850.
http://dx.doi.org/10.1109/TBME.2009.2029563 - 12. de Chazal, P., Heneghan, C., Sheridan, E., Reilly, R., Nolan, P. and O’Malley, M. (2003) Automated Processing of the Single-Lead Electrocardiogram for the Detection of Obstructive Sleep Apnoea. IEEE Transactions on Biomedical Engineering, 50, 686-696.
http://dx.doi.org/10.1109/TBME.2003.812203 - 13. Kao, T.-P., Wang, J.-S., Lin, C.-W., Yang, Y.-T. and Juang, F.-C. (2012) Using Bootstrap AdaBoost with KNN for ECG-Based Automated Obstructive Sleep Apnea Detection. International Joint Conference on Neural Networks (IJCNN), Brisbane, 10-15 June 2012, 1-5.
- 14. Almazaydeh, L., Elleithy, K. and Faezipour, M. (2012) Obstructive Sleep Apnea Detection Using SVM-Based Classification of ECG Signal Features. Conference Proceedings—IEEE Engineering in Medicine and Biology Society, 4938-4941.
- 15. Kao, T.P., Wang, J.S., Lin, C.W., Yang, Y.T. and Juang, F.C. (2012) Using Bootstrap AdaBoost with KNN for ECG- Based Automated Obstructive Sleep Apnea Detection. Proceedings of WCCI 2012 IEEE World Congress on Computational Intelligence, 305-314.
- 16. Mallat, S. and Hwang, W.L. (1992) Singularity Detection and Processing with Wavelets. IEEE Transaction on Information Theory, 38, 617-643.
http://dx.doi.org/10.1109/18.119727 - 17. Lee, H., Cichocki, A. and Choi, S. (2006) Nonnegative Matrix Factorization for Motor Imagery EEG Classification. Lecture Notes in Computer Science, 4132, 250-259.
http://dx.doi.org/10.1007/11840930_26 - 18. Cristianini, N. and Shawe-Taylor, J. (2000) An Introduction to Support Vector Machines and Other Kernel-Based Learning Methods. Cambridge University Press, Cambridge.
http://dx.doi.org/10.1017/CBO9780511801389









