Open Journal of Genetics
Vol.4 No.5(2014), Article
ID:48990,8
pages
DOI:10.4236/ojgen.2014.45034
Detection of Contamination and Analysis of Vertical Transmission of BmNPV in Eggs and Moths of Bombyx mori
Cláudia Regina das Neves Saez1, Roxelle Ethienne Ferreira Munhoz1, Naiara Climas Pereira1, Thaís Souto Bignotto1, Verônica Aureliana Fassina1, Graziele Milani Pessini1, Laura Beatriz Garay2, Lucinéia de Fátima Chasko Ribeiro3, Rose Meire Costa Brancalhão3, Maria Aparecida Fernandez1
1Departamento de Biotecnologia, Genética e Biologia Celular, Universidade Estadual de Maringá, Maringá, Brasil
2Seda y Fibras, Hernandárias, Paraguay
3Universidade Estadual do Oeste do Paraná, Cascavel, Brasil
Email: aparecidafernandez@gmail.com
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 26 June 2014; revised 25 July 2014; accepted 18 August 2014
ABSTRACT
This study reports the molecular detection of Bombyx mori nucleopolyhedrovirus (BmNPV) in silkworm strains of the Universidade Estadual de Maringá Brazilian Germplasm Bank (UBGB). DNA extraction was carried out by using six Bombyx mori female moths of each strain, followed by PCR amplification. A pair of primers was designed based on a specific sequence of the baculovirus genome related to the BmNPV ORF 14. Another pair of primers was used to amplify the silkworm Actin A3 gene segment, which was used as positive control. Twenty gene pools were analyzed, and fifteen revealed a fragment of 443 base pairs (bp), which indicated the presence of the BmNPV. The frequency of contaminated moths was as following: 100% for silkworm strains M18-2, M12-2 and J1; 83% for C25, C75 and C24 strains; 66% for KR01; 50% for M11-A; 33% for AS3, B106, M8 and M11 and 16% for C211, E8 and Hindu strains. These are promising results for the identification of contaminated B. mori moths by BmNPV, which may prevent virus proliferation in subsequent generations. We also analyzed DNA samples extracted from B. mori eggs, but the results were not conclusive regarding the detection of the fragments of the expected size (443 bp). The difficulty in detecting BmNPV contamination in B. mori eggs may be due to the low concentration of virus in samples.
Keywords:Silkworm Diseases, Sericulture, Viruses in Insects, BmNPV, Bombyx mori

1. Introduction
Bombyx mori Linneaus is a domesticated lepidopteron and its cocoons are used to produce silk yarn. The field production of the larvae until cocoons is called sericulture. This insect has four morphological stages during its life cycle: egg, larvae, pupa and moth [1] [2] .
There are more than 3000 strains of B. mori [3] which are maintained in germplasm banks around the world in order to preserve this genetic collection. Universidade Estadual de Maringá Brazilian Germplasm Bank (UBGB) is the only public germplasm bank of this species in Brazil.
The main problems in sericulture are caused by diseases that can have several causes, such as protozoa, virus and fungi, which can infect silkworm during all life stages [4] . In this context, diseases caused by virus can affect B. mori in different stages of its life cycle, representing a serious problem to the global sericulture [5] [6] . Bombyx mori nucleopolyhedrovirus (BmNPV) belongs to the gender Alphabaculovirus [7] and it is a highly infective pathogen, because it has polyhedral occlusion bodies, which contain several virions inside.
This virus can be inherited and this characteristic is very important to the viral dominance, but it still requires more studies. Likewise, settling the inheritability of BmNPV through the generations is a very important aspect to be studied [8] . This fact is one of the most important conditions for the sericulture industry, and it is called vertical transmission; in other words, the parents can transmit the virus to its progeny, becoming unfavorable to use the surviving moths to production of eggs [9] -[16] .
This work aimed to standardize a protocol to identify the contamination of BmNPV among B. mori strains of the UBGB and analyze the vertical transmission of this baculovirus between the generations.
2. Material and Methods
2.1. Biological Material
Twenty B. mori strains of Chinese, Japanese and Indian geographical origin were obtained at the UBGB: C211, C24, C25, AS3, C75, C36 and KR01 (Chinese strains); B82, Hindu and B106 (Indian strains); and M11, M11-A, M11-2, M12-2, M18-2, J1, F6, M8, M18 and E8 (Japanese strains).
2.2. Genomic DNA Extraction of Bombyx mori Moths
Genomic DNA of six female moths, of each strain, was isolated according to [17] . After isolation, DNA was purified with phenol and chloroform (1:1) and precipitated with 2 volumes of ethanol. Quantification of DNA was carried out at the spectrophotometer UV-1650PC UV (Shimadzu). DNA was then diluted to the concentration of 60 ng/μL, which was used to the PCR (Polymerase Chain Reaction) reactions.
One of the aims of this work was to investigate the event of vertical transmission of BmNPV in B. mori. Therefore, DNA was isolated from female and male moths that were previously identified and mating with each other. DNA isolation of the resulting eggs was also performed.
2.3. DNA Extraction of B. mori Eggs
The eggs were identified according to their parents and the DNA was extracted using the KOH method, with modifications [18] [19] . Twenty eggs were transferred to microtubes containing 500 µL of KOH 60%. Samples were incubated for 1 hour and 30 minutes in room temperature until the color of the eggs was purplish to reddish. Subsequently, KOH was removed and the eggs were washed twice with 500 µL of distilled water. Then, 500 µL of HCl 1 M was added to avoid the formation of neutralizing salts. Again, eggs were washed twice with distillated water to remove HCl. Then, the digestion buffer for Proteinase K (1 M TrisHCl pH 9.0; 0.5 M EDTA pH 8.0; SDS 20% e Sacarose 16%) were added and the eggs were ground inside the microtubes using a pestle. Samples were then centrifuged for 10 minutes. The supernatant phase was recovered and 1.5 µL of proteinase K was added. Samples were incubated for 2 hours in 50˚C. DNA was purified with phenol-chloroform, precipitated with ethanol, suspended with TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) and stored at −20˚C.
2.4. Genomic DNA Extraction of BmNPV
Viral DNA was obtained according to the methodology previously described [20] . The DNA was extracted from geographic isolated BmNPV, which was conceded by R. C. Brancalhão and L. F. C. Ribeiro, from Universidade Estadual do Oeste do Paraná (Unioeste). This DNA was used as a positive control in the PCR reactions.
2.5. DNA Amplification of BmNPV Nucleopolyhedrovirus
A pair of primers was constructed using a known sequence from the baculovirus genome, corresponding to the Open Reading Frame (ORF) 14 [21] . These primers were used to amplify the BmNPV DNA. According to Santos et al. [20] , the ORF 14 codes a protein from NPV viral envelope and it is associated with stress response, such as the ones caused by balucovirus infection in B. mori. The primers ORF14 Forward (5’ATG GAC GGT GTA AAG TTG CTG G3’) and ORF14 Reverse (5’TCA AAA TCA ACG CCG TCG TC3’) amplify a fragment of 443 base pairs from BmNPV genome.
One pair of primers for the B. mori Actin A3 gene (Actin Forward 5’AGA TGA CCA GAT CAT GTT CG3’; Actin Reverse 5’GAG ATC CAC ATC TGT TGG AAG3’), which amplified a fragment of 721 base pairs, was used as a positive control in the PCR reactions.
Six samples of DNA from B. mori females were used in the PCR amplifications. The mix reaction contained Taq polymerase buffer 1× (Biolabs), 1.5 mM MgCl2, 0.2 mM of each dNTP, 1 U of Taq polymerase (Biolabs), 20 ng of genomic DNA and 1.2 µM of each primer. PCR reactions were conducted in a Thermal Cycler (Eppendorf) and were repeated at least twice. The amplification conditions consisted of 35 cycles: denaturation at 94˚C for 1 minute, annealing at 61˚C for 1 minute and extension at 72˚C for 1 minute, followed by a final cycle of extension of 72˚C for 30 minutes.
Besides the amplification using ORF 14 or actin primers separately, we also carried out some multiplex PCR with both pairs of primers at the same reaction. ORF 14 primers were used to confirm the infection by BmNPV and the primers for the Actin gene were used as a control of the reaction. Positive control (DNA virus samples) and negative control (B. mori DNA sample free of contamination) were used to validate the results.
2.6. Electrophoresis
The products of simple and multiplex PCR reaction were visualized in 1.5% agarose gel, in a horizontal system with TBE 0.5× (45 mM Tris-borato, 1 mM EDTA, pH 8.0) running buffer. Gel was photographed under ultraviolet light and documented using UVP Biolmaging System. Results were analyzed according to the presence or absence of DNA fragments.
2.7. Cloning and Sequencing
In order to confirm if the fragment of 443 bp obtained in the PCR reactions was from BmNPV, the PCR amplified DNA fragment of the strain C25 was cloned in the pGEM-T Easy Vector System (Promega) vector. Sequencing was performed as described by Silva et al. [22] , using MegaBACE 1000 (Amersham Biosciences) automatic sequencer. The alignment of the nucleotide sequence and the verification of homology and identity were obtained with the online tools EMBL and Blastn from NCBI.
3. Results and Discussion
3.1. Molecular Identification of Silkworm Strains from the UEM Germplasm Bank Contaminated with the BmNPV
PCR amplifications from specific segments of Bombyx mori genome and/or BmNPV led to estimation of the number of contaminated moths in each of the 20 silkworm strains from the UEM germplasm bank. Six moths were analyzed of each silkworm strain. The results showed that the strains B82, M11-2, M18, C36 and F6 presented no fragment of 443 bp, which indicates absence of the baculovirus and, therefore, we assumed that they were not contaminated with BmNPV.
Moths of silkworm strains that were identified as free of contamination revealed only one fragment of 721 bp, which refers to the amplification of Bombyx mori actin A3 gene. Of the twenty strains analyzed, fifteen revealed a fragment of 443 bp, indicating the presence of the BmNPV virus in these strains. The frequency of contaminated moths was investigated by individual PCR amplifications of each moth from the strains. Results revealed that the frequency of moths contaminated with BmNPV was: 100% for M18-2, M12-2 and J1 strains; 83% for C25, C75 and C24; 66% for KR01; 50% for M11-A; 33% for AS3, B106, M8 and M11; and 16% for C211, E8 and Hindu (Table 1). Moths contaminated with BmNPV showed two fragments in the multiplex PCR analyses, one of 721 bp and one of 443 bp (Figure 1).
Results revealed high rate of contamination by this pathogen among the strains of UBGB. The high levels of contamination by BmNPV can be explained by one or more reasons, such as, lack of adequate disinfection of silkworm eggs and/or farming buildings with formol. Besides, the improper silkworm larvae management can cause vertical transmission during the processes of maintenance and multiplication of silkworm genetic resources.
The detection of BmNPV contamination in every developmental stage of B. mori is crucial to avoid the dissemination of the disease. Detection of BmNPV in silkworm moths was performed to verify the level of virus infection between the strains from UBGB and, consequently, prevent vertical transmissions of the baculovirus. Eggs laid by silkworm moths that were identified as infected by BmNPV were disposed, avoiding future contamination of the strains maintained in the UBGB.
Other articles also detected the infection of silkworm moths by BmNPV, using PCR with specific primers for the detection of this baculovirus, which corroborates our results [15] and [23] .
3.2. Cloning and Sequencing of ORF 14
The nucleotide sequence of BmNPV, ORF 14, isolated from the strain C25 was sequenced and compared to other sequences from Genbank, using Basic Local Alignment Search Tool, BLAST [24] . ClustalW alignment [25] revealed 100% of nucleotide similarity with BmNPV isolate T3 (Figure 2) and 99% of similarity with Autographa californica MNPV PRF 23, whose product is related to a protein of the viral envelope [19] .
3.3. Investigation of the BmNPV Vertical Transmission
Occasionally, PCR amplifications of eggs laid by silkworm moths contaminated with BmNPV revealed that vertical transmission may be occurred. However, results obtained in the present work were not consistent and conclusive regarding the occurrence of vertical transmission of BmNPV in silkworms from UBGB. Nevertheless, BmNPV contamination of B. mori eggs was detected by PCR with specific primers for the polyhedrin gene, as reported by Kaewwises [26] .
Table 1. Number of moths contaminated in silkworm strains belonging to Universidade Estadual de Maringá Brazilian Germplasm Bank (UBGB).
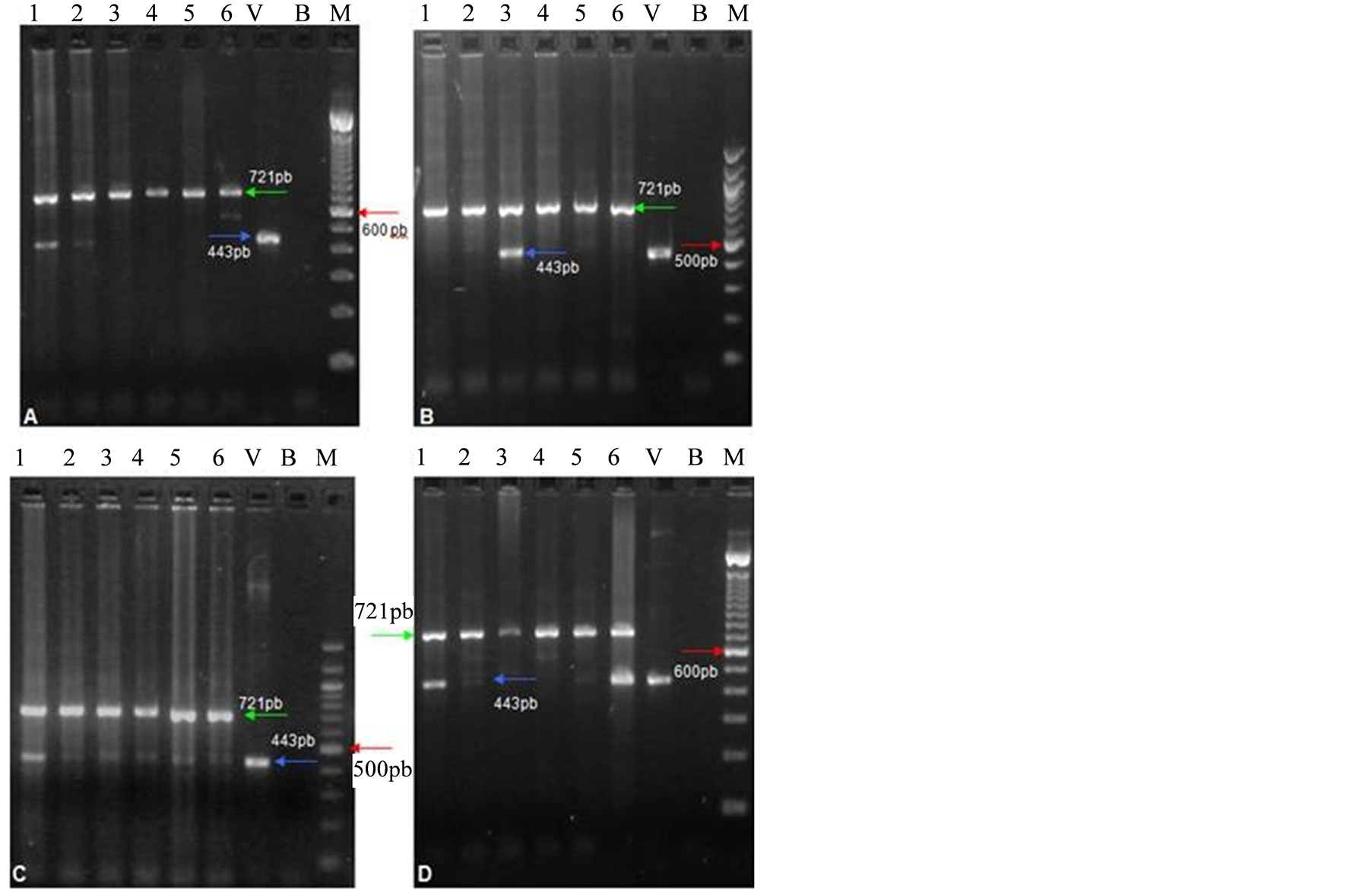
Figure 1. Multiplex PCR of Bombyx mori moths contaminated with BmNPV virus. M—Molecular marker 100 bp (Invitrogen in (A) and (D); Biolabs in (B) and (C)). V—Positive control (DNA of BmNPV virus). B—Negative control. (A) Samples 1 to 6: M8 strain; BmNPV contamination can be visualized in samples 1 and 2; (B) Samples 1 to 6: C211 strain; BmNPV contamination can be visualized in sample 3; (C) Samples 1 to 6: J1 strain; every sample is contaminated; (D) Samples 1 to 6: KR01 strain; BmNPV contamination can be visualized in samples 1, 2, 5 and 6.

Figure 2. Partial nucleotide sequence alignment of Bombyx mori ORF 14 and BmNPV, isolate T3 (GenBank Accession number L33180.1).
In the present work, we carried out a screening of silkworm moths from UBGB that could be naturally contaminated by BmNPV. Silkworm moths were primarily analyzed regarding contamination by BmNPV. In the cases in which the results were positive for BmNPV contamination, the respective eggs of the contaminated moths had their DNA isolated and, subsequently, PCR amplified with primers for ORF 14 and Actin A3. This process was carried out to verify the contamination by BmNPV in the silkworm eggs.
Previously was verified that the dispersion of baculovirus among silkworm moths may occur after vertical transmission to the progeny through the inner or surfaces of the eggs [10] and [11] . The vertical transmission of NPV in B. mori occurs by oral inoculation of occlusion bodies in 5th instar [15] .
Some results were presented in this present work; however, they were not consistent with the vertical transmission hypothesis. We could not detect the presence of BmNPV in some silkworm eggs laid by moths that were previously identified as contaminated by the baculovirus. The eggs of contaminated moths did not reveal the fragment of 443 bp that indicates the presence of BmNPV, maybe due to the low concentration of the virus in our samples. In the present work, we employed the methodology of the conventional PCR for detection of BmNPV. However, a technique that allows the quantification of viral load could enable the detection of the baculovirus.
Two hypotheses are consistent with the observations of the not contaminated silkworm eggs laid by BmNPV contaminated moths. First, the detection of BmNPV was impossible due to the reduced viral load in the silkworm eggs. Second, maybe there is some anti-viral mechanism during the larvae stage in which BmNPV infection can be blocked.
Vertical transmission was previously investigated by several researches. For instance, employing the PCRbased technique for detection of BmNPV in silkworm eggs laid by contaminated female moths the authors [14] concluded that BmNPV can be detected even when samples were as reduced as a unique silkworm egg.
Khumnoi [16] also identified vertical transmission of BmNPV in a Thai B. mori strain, detecting viral DNA in PCR reactions with primers for the polyhedrin gene of BmNPV. Results obtained by [16] were similar to those found in the present work and confirmed that some larvae infected by BmNPV survived to the virus and that the silkworm moths derived from these larvae could vertically transmit the BmNPV to their progeny. Therefore, PCR-based techniques can be employed to detect BmNPV in eggs and in every developmental stage of B. mori.
The knowledge of infection by BmNPV in silkworm moths will be usefull from UEM germplasm bank to detect the presence of the virus by sampling analyses. For instance, egg-laying identified as contaminated by BmNPV will be evaluated and decisions will be taken regarding the permanence of this egg-laying, with the purpose of eliminating contaminated larvae in the next generations.
The detection technique based on the conventional PCR methodology used in the present work provides an effective, fast, reliable and practical way for detecting BmNPV in moths of B. mori and should also be optimized for the detection in silkworm eggs. This methodology proved to be adequate to promote a common practice for governmental institutions and private sectors involved in sericulture. Besides, it can be routinely used to monitor and protect the proliferation of BmNPV in silkworm rearing. However, further studies on the sensitivity of PCR will provide even more efficient BmNPV detections, especially in B. mori eggs, preventing losses in the sericulture production. Future works using techniques such as real-time PCR may be carried out to quantitatively determine the baculovirus and thus promote better detection of BmNPV in eggs.
We advise for the need to investigate the rate of silkworm moths contaminated in germplasm banks, because the absence of disease symptoms could not signal absence of baculovirus infection.
4. Conclusion
The results obtained in this study are promising for detection of moths contaminated with BmNPV, using simple techniques, such as PCR, avoiding the proliferation of the baculovirus in subsequent generations, and assisting in a proper management of B. mori strains, in order to protect the genetic pool of this specie in the UEM germplasm bank.
Acknowledgements
This work was supported by CAPES, CNPq, FINEP/Fundação Araucária and Secretaria de Estado da Ciência, Tecnologia e Ensino Superior—FUNDO PARANÁ.
References
- Hanada, Y. and Watanabe, J.K. (1986) Manual de criação do bicho-da-seda. Cocamar, Curitiba.
- Nagaraju, J. and Goldsmith, M.R. (2002) Silkworm Genomics—Progress and Prospects. Current Science, 83, 411-425.
- Yamamoto, T. (2000) Silkworm Strains. In: Nakatsuij, N., Ed., Strains Maintenance and Databank for Life Science, Kyoritsu Shuppan, Tokyo, 45-49.
- Zanetti, R. (2005) Doenças e pragas do bicho-da-seda, notas de aula de ENT 100—Sericicultura. http://www.den.ufla.br/siteantigo/Professores/Ronald/Disciplinas/Notas%20Aula/Sericicultura%20doencasbicho.pdf
- Sengupta, K., Kumar, P. and Baig, M. (1990) Handbook on Pest and Disease Control of Mulberry and Silkworm. Economic and Social Commission for Asia and Pacific, UNESCAP (United Nations Economic and Social Commission for Asia and the Pacific), Bangkok.
- Brancalhão, R.M.C. (2002) Vírus entomopatogênicos no bicho-da-seda: Taxonomia e citopatologia causada por nucleopolyhedrovirus em células de Bombyxmori. Biotecnologia Ciência e Desenvolvimento, 24, 54-58.
- Jehle, J.A., Blissard, G.W., Bonning, B.C., Cory, J.S., Herniou, E.A., Rohrmann, G.F., Theilmann, D.A., Thiem, S.M. and Vlak, J.M. (2006) On the Classification and Nomenclature of Baculoviruses: A Proposal for Revision. Archives of Virology, 151, 1257-1266. http://dx.doi.org/10.1007/s00705-006-0763-6
- Beukema, S.J. (1992) Towards More Realistic Models of Disease and Insect Population Dynamics: Virus-Tent Caterpillar Interactions. Doctoral Thesis, University of British Columbia, Vancouver.
- Neelgund, Y.F. and Mathad, S.B. (1978) Transmission of Nuclear Polyhedrosis Virus in Laboratory Population of the Armyworm, Mythimna (Pseudaletia) separata. Journal of Invertebrate Pathology, 31, 143-147. http://dx.doi.org/10.1016/0022-2011(78)90001-0
- Ridhards, A., Matthews, M. and Christain, P. (1998) Ecological Considerations for the Environmental Impact Evaluation of Recombinant Baculovirus Insecticides. Annual Review of Entomology, 43, 493-517. http://dx.doi.org/10.1146/annurev.ento.43.1.493
- Kukan, B. (1999) Vertical Transmission of Nucleopolyhedrovirus in Insects. Journal of Invertebrate Pathology, 74, 103-111. http://dx.doi.org/10.1006/jipa.1999.4873
- Fuxa, J.R., Weidner, E.H. and Richter, A.R. (1992) Polyhedra without Virions in a Vertically Transmitted Nuclear Polyhedrosis Virus. Journal of Invertebrate Pathology, 60, 53-58. http://dx.doi.org/10.1016/0022-2011(92)90153-U
- Fuxa, J.R., Richter, A.R., Ameen, A.O. and Hammock, B.D. (2002) Vertical Transmission of TnSNPV, TnCPV, AcMNPV and Possibly Recombinant NPV in Trichoplusia ni. Journal of Invertebrate Pathology, 79, 44-50. http://dx.doi.org/10.1016/S0022-2011(02)00003-4
- Ikuno, A.A., Margatho, L.F.F., Harakava, R., Akamatsu, M.A., Martins, E.M.F., Porto, A.J. and Ferreira, V.C.A. (2004) Direct Application of the New PCR Protocol for Evaluation and Monitoring of Bombyx mori Infection by Nucleopolyhedrovirus. Arquivos do Instituto Biologico, 71, 309-315.
- Khurad, A.M., Mahulikar, A., Rathod, M.K., Rai, M.M., Kanginakudru, S. and Nagaraju, J. (2004) Vertical Transmission of Nucleopolyhedrovirus in the Silkworm, Bombyx mori L. Journal of Invertebrate Pathology, 87, 8-15. http://dx.doi.org/10.1016/j.jip.2004.05.008
- Khumnoi, S. (2007) Vertical Transmission of Nucleopolyhedrovirus in Thai Mulberry Silkworm, Bombyx mori L. (Lepidoptera: Bombycidae). Master of Science (Agricultural Biotechnology), Kasetsart University, Bangkok.
- Pereira, N.C., Munhoz, R.E.F., Bignotto, T.S., Bespalhuk, R., Garay, L.B., Saez, C.R.N., Fassina, V.A., Nembri, A. and Fernandez, M.A. (2013) Biological and Molecular Characterization of Silkworm Strains from the Brazilian Germplasm Bank of Bombyx mori. Genetics and Molecular Research, 12, 2138-2147. http://dx.doi.org/10.4238/2013.June.28.1
- TakahashiI, K., Takaku, M. and Hasegawa, M. (1925) A Experiment of Dissection and Transparent Method with Silkworm Egg. Sanshishinpou, 33, 47-59.
- Hatakeyama, Y. and Hayasaka, S. (2003) A New Method of Pebrine Inspection of Silkworm Egg Using Multiprimer PCR. Journal of Invertebrate Pathology, 82, 148-151. http://dx.doi.org/10.1016/S0022-2011(03)00019-3
- Santos, S.A., Silva, J.L.C., Balani, V.A., Seixas, F.A.V. and Fernandez, M.A. (2010) Conserved Baculoviral ORFs 10 and 14 from Bombyx mori Multiple Nucleopolyhedrovirus. Genetics and Molecular Research, 9, 457-470. http://dx.doi.org/10.4238/vol9-1gmr723
- Gomi, S., Majima, K. and Maeda, S. (1999) Sequence Analysis of the Genome of Bombyx mori Nucleopolyhedrovirus. Journal of General Virology, 80, 1323-1337.
- Silva Jr., W.A., Costa, M.C., Valente, V., Sousa, J.F., Paço-Larson, M.L., Espreafico, E.M., Camargo, S.S., Monteiro, E., Holanda, A.J., Zago, M.A., Simpson, A.J. and Neto, E.D. (2001) PCR Template Preparation for Capillary DNA Sequencing. Biotechniques, 30, 537-542.
- Kaewwises, M., Attathom, T., Chaeychomsri, S. and Chowpongpang, S. (2004) Detection of Nucleopolyhedrovirus of Mulberry Silkworm, Bombyx mori by PCR Technique. Proceedings of the 42nd Kasetsart University Annual Conference, Kasetsart, 3-6 February 2004, 174-182.
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W. and Lipman, D.J. (1990) Basic Local Alignment Search Tool. Journal of Molecular Biology, 215, 403-410. http://dx.doi.org/10.1006/jmbi.1990.9999
- Thompson, J.D., Higgins, D.G. and Gibson, T.J. (1994) CLUSTAL W: Improving the Sensitivity of Progressive Multiple Alignment through Sequence Weighting, Positions-Specific Gap Penalties and Weight Matrix Choice. Nucleic Acids Research, 22, 4673-4680.
- Kaewwises, M. (2006) Potential Application of PCR-Based Method for Early Detection Osgrasserie Disease of Silkworm Bombyx mori. Doctoral Thesis, Kasetsart University, Bangkok.


