Open Journal of Molecular and Integrative Physiology
Vol.3 No.3(2013), Article ID:36335,9 pages DOI:10.4236/ojmip.2013.33018
Novel genomic biomarkers for acute gentamicin nephrotoxicity in dog
![]()
1Drug Safety Sciences, Janssen Pharmaceutical Research & Development LLC, San Diego, USA
2CREATe-Genomic Technologies, Janssen Pharmaceutical Research & Development LLC, San Diego, USA
3Drug Safety Sciences, Janssen Pharmaceutical Research & Development LLC, Spring House, USA
4Safety Assessment, AstraZeneca Pharmaceuticals, Watham, USA
Email: *jmcduffi@its.jnj.com
Copyright © 2013 James Eric McDuffie et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received 20 June 2013; revised 11 July 2013; accepted 22 July 2013
Keywords: Biomarker; Gentamicin; Nephrotoxicity; Dog
ABSTRACT
Objectives: Novel biomarkers indicative of drug-induced kidney injury (DIKI) in dogs would have significant application in preclinical drug development. We conducted a feasibility study to identify genomic expression profiles for monitoring progressive, acute DIKI in dogs. Materials and Methods: Animals were intramuscularly administered either 0.9% physiological saline or gentamicin (40 mg/kg/day) for 10 consecutive days and euthanized on day 11. Serum and urine samples were collected at various time points and kidney samples were collected at necropsy for biomarker measurements. Results: Acute gentamicininduced renal histopathology changes were localized to the proximal convoluted tubules and characterized as slight-to-marked, diffuse cortical-medullary tubular epithelial degeneration/necrosis. Serum creatinine (sCr) and blood urea nitrogen (BUN) elevations suggestive of mild renal dysfunction were first observed on days 7 to 8. Gentamicin-induced increased urinary kidney injury molecule-1 (KIM-1) mRNA was observed on day 6 preceding detectable elevations of sCr and/or BUN. Increased urinary KIM-1 mRNA correlated with multifocal KIM-1 immunostaining in the corticomedullary tubular epithelial cells. Microarray analysis revealed changes in additional mRNA expression products detected in urine and/or kidney that should be further investigated for use as potential biomarkers for acute gentamicin related nephrotoxicity in dogs. Conclusion: These findings suggested the utility of non-invasive urinary genomic parameters for monitoring acute DIKI in dogs.
1. INTRODUCTION
Novel urinary protein biomarkers of renal injury, including albumin, clusterin, kidney injury molecule-1 (KIM- 1), total protein, cystatin C, and trefoil factor 3 (TFF3), β2-microglobulin and renal papillary antigen-1 (RPA-1) have been qualified in rats [1]. The utility of these biomarkers in dogs has not been well-defined. In dogs, non-invasive monitoring for renal dysfunction is generally based on elevated serum creatinine (sCr) measurements; yet, sCr elevations are not specific for renal tissue injury and often occur late in the progression of drug inducing kidney injury (DIKI) [2].
Beagle dog was chosen as the model for this study based on the common use of this model in preclinical toxicology studies to identify potential target organs for toxicity and safety parameters for subsequent clinical monitoring. The relevant kidney nephrotoxicant related to histopathology findings observed in this species are anticipated to be monitorable in humans and warrants further identification and characterization of novel, noninvasive urinary genomic parameters for monitoring drug-induced kidney injury in dogs. Novel sensitive and selective biomarkers indicative of DIKI in dogs would have significant application in preclinical drug development.
The aminoglycoside antibiotic gentamicin was selected as the test article in this study as it is known to induce acute kidney injury in dogs, a phenomenon which matches the gentamicin related to human pathology and clinical scenario [2]. Gentamicin induces nephrotoxicity in dogs by inhibiting protein synthesis in renal cells. Lesions ascribed to gentamicin nephrotoxicity include proximal tubular degeneration/necrosis [3,4]. Here, we provide data relative to novel genomic markers for acute gentamicin nephrotoxicity in dogs.
2. MATERIALS AND METHODS
2.1. Canine Gentamicin Nephrotoxicity Model
All animal care and use procedures were conducted in an Association for Assessment and Accreditation of Laboratory Animal Care International‑accredited facility under an Institutional Animal Care and Use Committee-approved protocol. This study included two male beagle dogs at approximately 6 years of age and mean pre-dose (day 1) body weight of 12.5 kg (Marshall Bio Resources, North Rose, NY). Animals were individually housed in stainless steel cages. Physical examinations were conducted on both dogs prior to assignment as control (vehicle)-treated and test article (gentamicin)-treated animals. Based on the most recently recorded pre-dose body weight, one dog received vehicle (0.9% physiologic saline USP) and the other gentamicin (Gentamicin solution for injection, TW Medical, Lago Vista, TX, 40 mg/kg/ day). Gentamicin or vehicle was administered via the intramuscular (IM) route in the biceps femoris muscle once daily for 10 consecutive days. In this study, predose is referred to as day 0. Individual whole body weights were recorded at pre-dose, on days 2, 6, 8, and 10, and immediately prior to necropsy. Cage side examinations were conducted at least thrice daily: at pre-dose (between approximately 7:00 and 7:30 am), at approximately 3 hours post dose, and in the afternoon (between approximately 3 and 4 pm) for signs of toxicity, moribundity, and mortality. Individual food and water consumption were assessed daily by visual inspection in the afternoon (between approximately 3:00 and 4:00 pm). Housed animals were provided water ad libitum via a water bowl. Subsequent to daily the cage side examinations at approximately 3 hours post dose, approximately 125 g of Adult dry food mixed with approximately 125 g of Adult canned wet food was provided for ad libitum consumption via a food bowl. Except for day 4, where no biological samples were collected, whole blood was collected (to process to serum) from both animals at approximately 2 hours prior to dosing, from the gentamicin-treated dog at approximately every 24 hours after the first dose and from the control animal on days 9 and 10. Urine was collected via catheter at pre-dose and every 24 to 48 hours after the first dose up to day 10. Urine (up to 10 mL per animal) was stored in individual tubes at each collection time point. Serum and urine aliquots were stored at −80˚C until time of analysis. Serum chemistry parameters (serum creatinine and urea nitrogen, sCr and BUN) were measured using an automated clinical analyzer at a contract research organization laboratory. Monitoring of serum chemistry parameter changes continued until the time point when the sCr level in the gentamicin treated dog measured at ≥10-fold the initial pre-dose value. Increased sCr indicative of gentamicin related renal dysfunction was based on a quantitative score (Table 1).
Animals were euthanized on day 11. At necropsy, representative kidney samples were flash frozen and stored at ‑80ºC until time of analysis. Additional samples were fixed in 10% neutral-buffered formalin, processed, embedded in paraffin, sectioned (4 mm), and stained with hematoxylin and eosin (H&E). Histologic slides were evaluated in a blinded fashion using a semi-quantitative scale. Renal tubule injury was characterized as tubular degeneration/necrosis, tubular regeneration, tubular hypertrophy, moderate hyaline casts, intratubular proteinaceous debris, and interstitial inflammation. The distribution and increased number of cells affected were assigned a quantitative severity score: 0 = no abnormality noted, 1 = slight (minimal, <25%), 2 = mild (25% - 50%), 3 = moderate (>50%), and 4 = severe (>75%). Representative photomicrograph images were captured using a Nikon E800 light microscope (Nikon, Melville, NY) equipped with Q imaging digital camera and Image Pro Plus 7.0 software (Media Cybermetrics, Bethesda, MD). Slides were scanned at 20× magnification using the NanoZoomer 2.0 HT Digital Pathology system scanner (Hamamatsu Photonics, Bridgewater, NJ).
2.2. Urine and Kidney mRNA Extraction
Total RNA was extracted from urine using the ZR Urine RNA Isolation Kit (Zymo Research, Irvine, CA). Total RNA was extracted from kidney by first disrupting the tissue through homogenization, followed by purification using an RNeasy Mini Kit (Qiagen, Valencia, CA).
2.3. Microarray Analysis
GeneChip Canine Genome 2.0 Arrays were purchased from Affymetrix (Santa Clara, CA) and used according to instructions provided by the vendor. The microarray analysis and the gene annotations were provided by Affymetrix. The updated gene annotations were obtained from various sources [5-25].
2.4. Urinary and Renal Kim-1 Analysis
Custom TaqMan assays were designed to measure canine
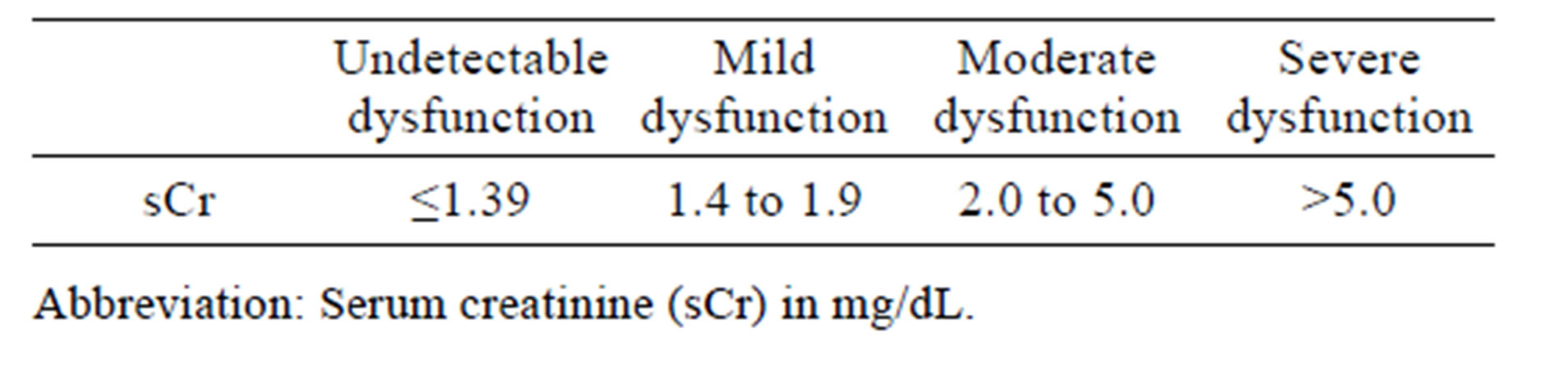
Table 1. Renal dysfunction severity scale.
urinary and renal levels of KIM-1 mRNA and β-actin mRNA expression using quantitative polymerase chain reaction (qPCR). The assays were made by Applied Biosystems Inc., Foster City, CA. For KIM-1, the forward primer sequence 5’-CGGCCCAGACTGACAGT-3’, reverse primer sequence 5’-CCACCCCCTGTGCTCAA-3’, and probe sequence 5’-CACGGCAACAATACTG‑3’ were used. β-actin was used as an endogenous control whereby the forward primer sequence 5’-CCACTTTCCTGTCTTACCCAATGTT-3’, reverse primer sequence 5’-CCGTTTTCCAACTTCAAGGCAATTA-3’, and probe sequence 5’‑CACCCACGGTGTTCTG-3’ were employed. For qPCR analysis, 1 μg of representative kidney total RNA or 100 ng of urine total RNA was reverse transcribed to cDNA using the High-Capacity cDNA Archive Kit. Water was used as a negative control. Triplicate samples were subjected to TaqMan assay on the 7300 Real-Time PCR System (Applied Biosystems, Inc.). KIM-1 (HACVR1) gene expression was determined by calculating the difference in CT from the reference gene β-actin. The average CT values for target genes were subtracted from the average housekeeping gene CT values to yield the ΔCT. Results were expressed as relative quantification of KIM-1 mRNA expression.
2.5. Renal KIM-1 Immunostaining
Kidney paraffin blocks were sectioned at 4 mm, mounted on glass slides, deparaffinized, and hydrated in PBS. Endogenous peroxide was blocked using 3% hydrogen peroxide solution. To avoid nonspecific reaction with primary antibody, slides were pretreated with 10% normal donkey serum. Slides were incubated for 30 minutes at 4˚C with 10% normal rabbit serum before incubation for 2 hours at 4˚C with primary rabbit monoclonal antibody to canine KIM‑1/TIM-1/HACVR1 (Sino Biological, Inc., 70001-R202) at 1:50. Normal rabbit IgG (Millipore, Billerica, MA, 12 - 370) was used as negative control. Slides were incubated for 30 minutes at room temperature with donkey anti-goat IgG SP-biotin conjugates, species absorbed (Millipore, AP180B) which was used as secondary antibody at 1:2000. The immunoreactivities were visualized VECTASTAIN Elite ABC Systems reagents (Vector Labs, Inc., Burlingame, CA, PK6100) and Liquid DAB+ Substrate Chromagen System reagents (DAKO North America, Inc., Carpinteria, CA, K3468) followed by counterstaining with hematoxylin. Slides were scanned at 20× magnification as previously described in Section 2.1.
3. RESULTS
3.1. Traditional Renal Biomarkers
In this study, gentamicin-induced DIKI was defined by temporal measurements of elevated sCr and confirmed by histopathology. Increased sCr measurements (1.7 and 4.7 mg/dL), suggestive of gentamicin-induced mild and moderate renal dysfunction were observed on days 7 and 9, respectively (Table 2).
Gentamicin related increased sCr at >10-fold the initial pre-dose value and indicative of severe renal dysfunction was observed on day 10. Mild increased BUN in the gentamicin treated dog was first observed on day 7.
3.2. Renal Histopathology
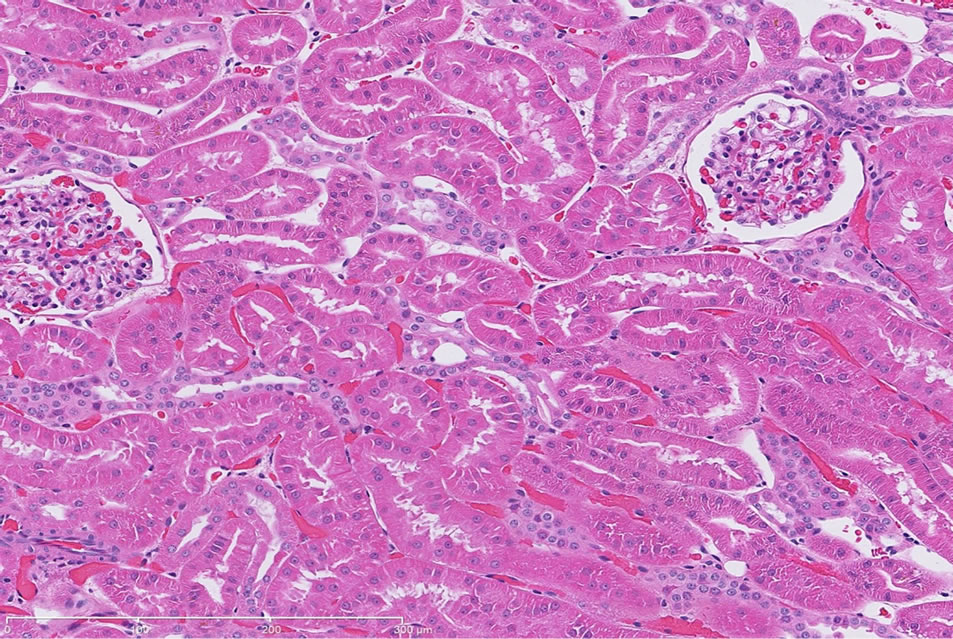
There were no significant treatment related renal histopathology findings in the control dog following repeat administration of saline (Figure 1(a)).
Gentamicin-induced renal injury had three distinct histologic features (in order of decreasing severity): marked, diffuse corticomedullary tubular epithelial degeneration/ necrosis; moderate tubular regeneration and hypertrophy; and moderate, diffuse pelvic granular and/or hyaline casts (Figure 1(b) and Table 3). Additional histologic features were noted, however, these findings could not be definitively considered as gentamicin treatment related due to the incidence of these findings commonly observed in naïve dogs.
3.3. Novel Genomic Renal Biomarkers in Dogs
Increased gene expression ≥2-fold (above baseline in the same animal) following administration of gentamicin and literature-based annotations (renal distribution and/or biological processes) were the primary inclusion criteria for the candidate genomic biomarker transcripts. Microarray analysis revealed 1000 upregulated genes in both kidney and urine following gentamicin administration. Of these genes, 350 were increased at least 5-fold in kidney and 2-fold in urine. Gentamicin induced a unique, urinary gene expression biomarker profile between days 2 and 6 prior to elevations in traditional nephrotoxicity biomarkers indicative of gentamicin-induced renal dysfunction which first occurred on Days 7 to 8 (Table 4).
The biological relevance of the genes identified remains inconclusive as representative serial tissue (kidney and/or other relevant target organ) sampling was not included in this feasibility study. Based on references in the public domain, likely annotations (renal distribution and/or biological processes) for the gene ontologies observed in the gentamicin treated dog in this study have been summarized; yet, warrants further investigation (Table 5).
As shown in the heatmap, variations in gene expression levels were detected for 18 different genes in urine collected on day 10 and kidneys collected at study termination on day 11. These observations suggested both a plausible biological relevance and an invasive (kidney)

Table 2. sCr and BUN measurements.
 (a)
(a) (b)
(b)
Figure 1. (a) Representative H&E-stained kidney section from a dog following repeat dosing of 0.9% physiologic saline. 20× magnification. (b) Representative H&E-stained kidney section from a dog following repeat dosing of gentamicin (40 mg/kg/day). Renal injury characterized as marked, diffuse corticomedullary tubular epithelial degeneration/necrosis; moderate tubular regeneration and hypertrophy; and moderate, diffuse pelvic granular and/or hyaline casts. Mild intratubular proteinaceous debris and slight interstitial inflammation were also observed. 20× magnification.
versus non-invasive (urine) relationship for the treatment related gene expression changes observed in this feasibility study in dogs (Figure 2).

Table 3. Gentamicin related renal histopathology.

Table 4. Gentamicin related genomic urinary renal biomarkers.
3.4. Urinary and Renal KIM-1 mRNA
Gentamicin-induced urinary KIM-1 mRNA changes were first observed on Day 6, prior to detection of elevated levels of sCr and/or BUN on day 7 (Table 6). Re-

Table 5. Novel urinary genomic biomarker annotations.

Figure 2. Heatmap of gene expression changes in urine collected on day 10 and kidneys collected at termination on Day 11 from dogs following 9-day repeat dose administration of either 0.9% physiologic saline or gentamicin (40 mg/kg/day).
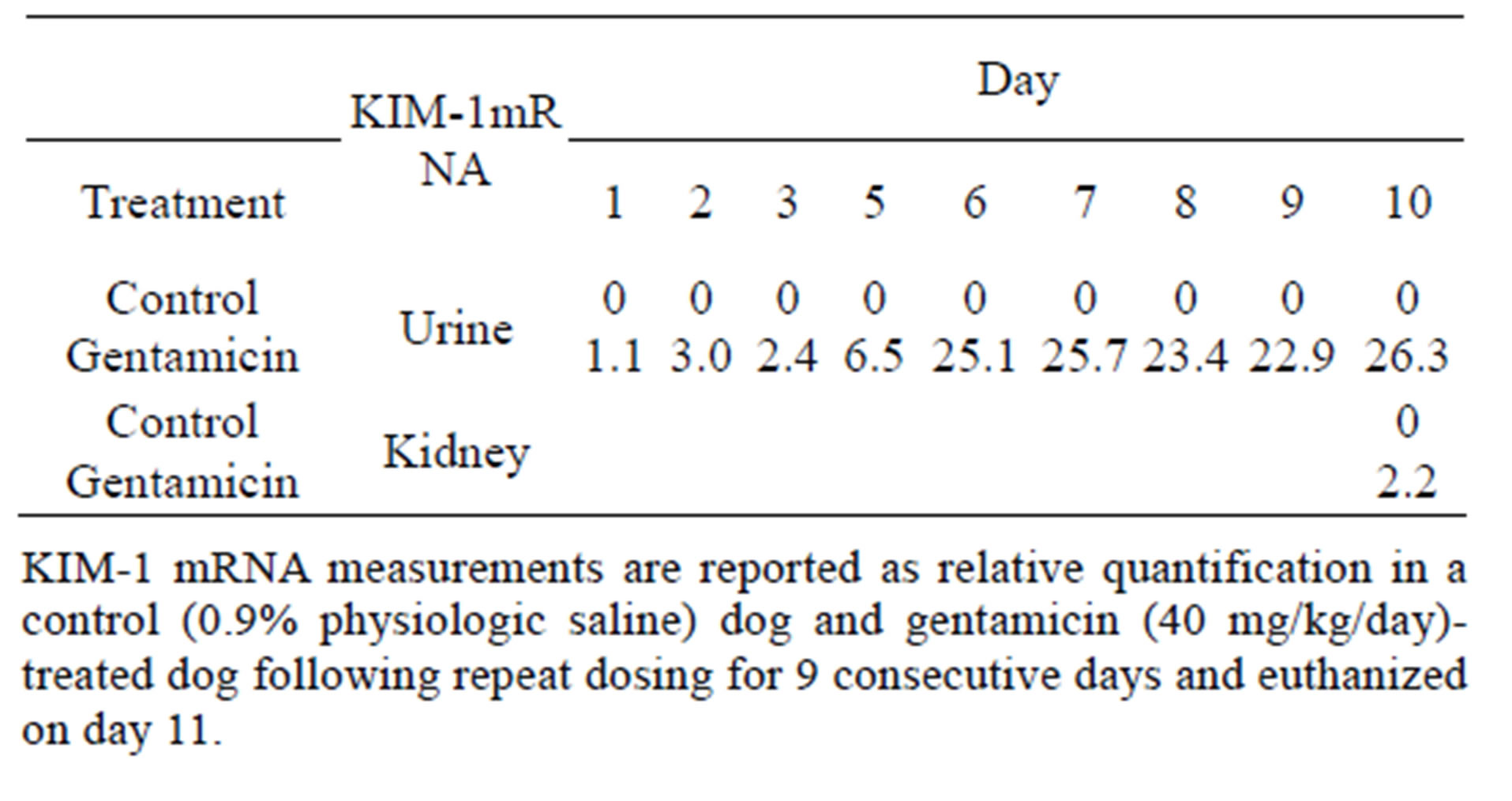
Table 6. Quantification of urinary and renal KIM-1.
nal KIM-1 mRNA was detected in the gentamicin-treated dog kidney with microscopic evidence of DIKI.
3.5. Renal Kim-1 Immunostaining
Immunostaining of KIM-1 protein expression was localized in the corticomedullary tubular epithelial cells (more apical) in the gentamicin‑treated dog, however, KIM-1 immunostaining was not detected in the control dog following repeat dose administration of saline (Figures 3(a)-(d)).
4. DISCUSSION
As observed in other species, sCr elevation is not specific for renal tissue injury in canines and is often detectable only after significant injury has occurred. In dogs, gentamicin inducing nephrotoxicity is primarily characterized histologically as tubular degeneration/necrosis of cells in the proximal tubules [3,4]. Temporal sCr and BUN changes and histologic evidence observed in this study were consistent with previous reports of gentamicin inducing renal injury. Increased sCr and BUN levels indicative of progressive DIKI were first observed on day 7 to 8. Novel genomic biomarkers indicative of DIKI in dogs may prove to be value-added in preclinical drug development, particularly if they outperform or are added to the value of concurrent sCr and/or BUN measurements.
The correlation of gentamicin related to DIKI and associated with gene expression in dogs had not been previously reported. In this study, microarray analysis indicated that approximately 1000 genes were upregulated following gentamicin administration to dog, of which 20 genes had been previously related to DIKI in various rodents, canines and/or humans [6-27]. We identified 18 genes which were increased in both kidney and urine of the gentamicin-treated animal following repeat dosing suggesting the potential utility of these parameters as biomarkers for monitoring DIKI in dogs. Urinary mRNA changes observed on day 2 or 3 indicated potential for biomarkers of early DIKI. Other mRNA changes were not observed until day 5; and these transcripts were then
 (a)
(a)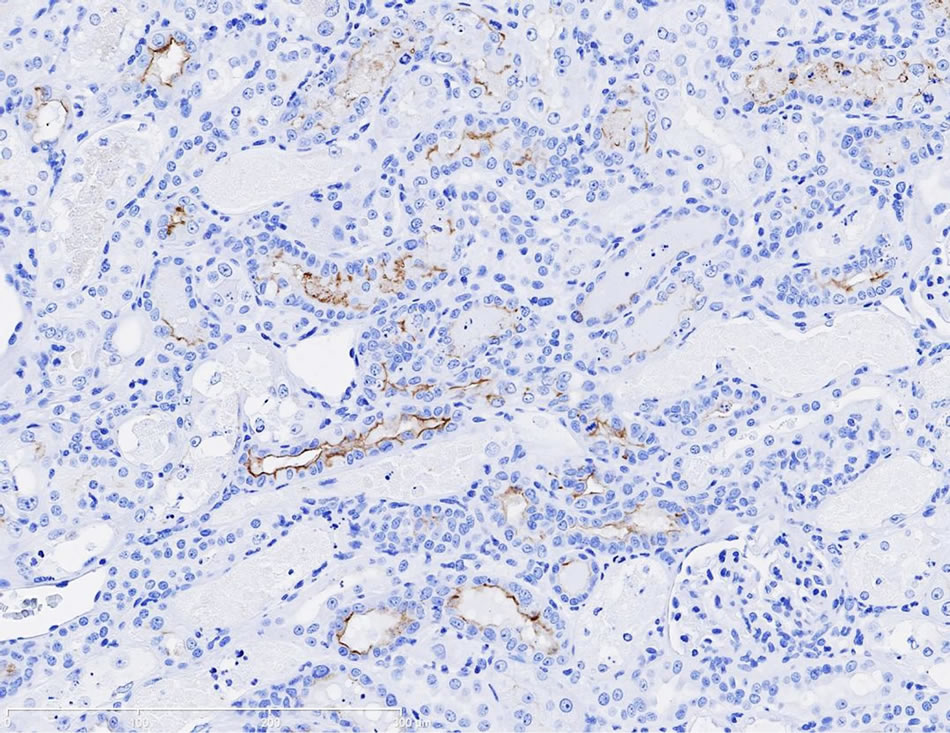 (b)
(b)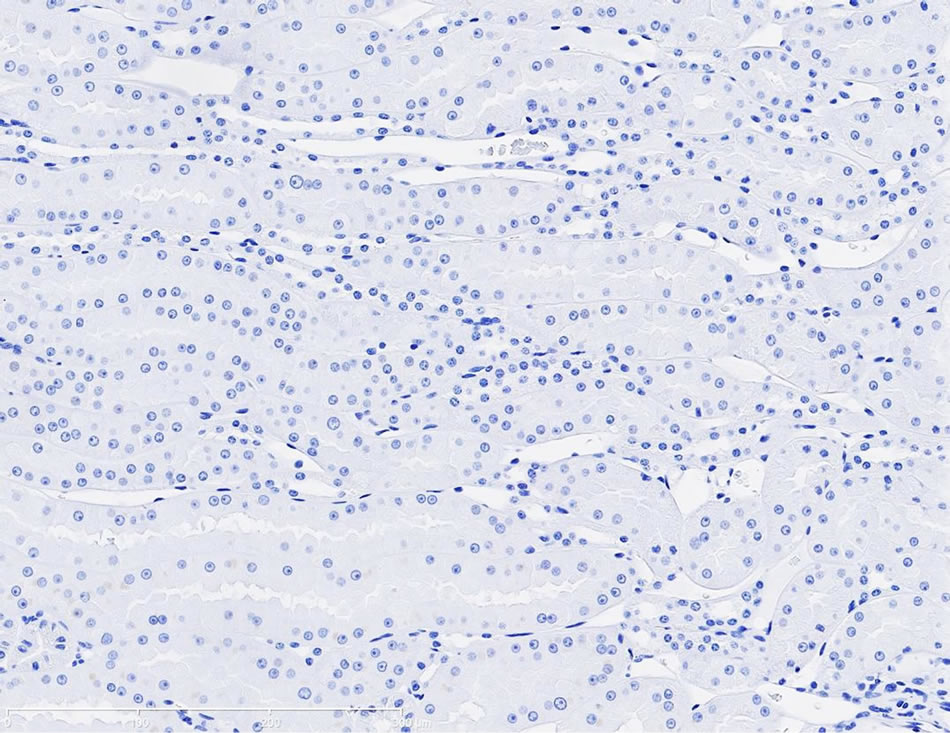 (c)
(c)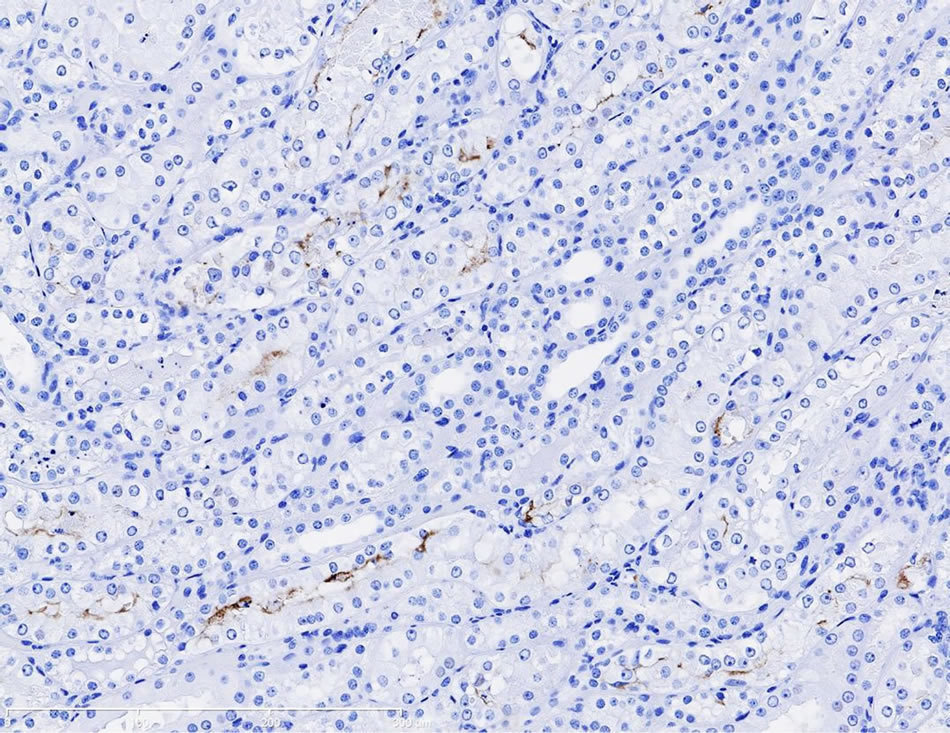 (d)
(d)
Figure 3. (a) Photomicrographs of representative KIM-1 immunostaining in the renal cortex in a dog following 9-day repeat dosing of saline. KIM-1 protein expression was not detected. 20× magnification; (b) Photomicrographs of representative KIM-1 immunostaining in the renal cortex (epithelial cells) in a dog following 9-day repeat dosing of gentamicin (40 mg/kg/day). 20× magnification; (c) Photomicrographs of representative KIM-1 immunostaining in the renal medulla in a dog following 9-day repeat dosing of saline. KIM-1 protein expression was not detected. 20× magnification; (d) Photomicrographs of representative KIM-1 immunostaining the renal medulla (epithelial cells) in a dog following 9-day repeat dosing of gentamicin (40 mg/kg/day). 20× magnification.
sustained through the remainder of the observational period suggestive of potential biomarkers of progressive DIKI.
Urinary KIM-1 is a qualified renal biomarker for monitoring DIKI in rats [1]. Robust KIM-1 renal gene expression following gentamicin treatment in rats has been previously reported [24,25]. In the present feasibility study in dog, a custom TaqMan assay was designed to evaluate canine KIM-1 gene induction following gentamicin administration. Specifically, we measured urinary and renal levels of KIM-1 mRNA expression using qPCR because unlike the Affymetrix arrays specific for rats and humans, the canine microarray used in this study for canine gene expression profiling did not include KIM-1. Increased urinary renal KIM-1 mRNA was observed prior to increased sCr or BUN, which are two traditional renal biomarkers. We recently reported on changes in urinary KIM-1 mRNA and protein in dogs following cisplatin administration [26]. In this study, KIM-1 mRNA as measured by qPCR was detected in kidney from the Gentamicin-treated dog; and there was no lower detectable limit in the control animal. Gentamicin-related increased urinary KIM-1 mRNA levels occurred on day 6. In the gentamicin treated dog, increased KIM-1 immunostaining was detected and localized to the epithelial cells in the corticomedullary tubules. These findings suggested that a unique genomic signature for DIKI is monitorable in both kidney and urine in dogs. Urinary KIM-1 mRNA appeared as a useful noninvasive parameter for monitoring DIKI in dogs; and this characteristic lends to its potential use in monitoring DIKI across multiple species [27]. Similar to the observation observed in this study in dog relative to KIM-1, upregulated C2, clusterin, osteopontin, Cxcl10 [24,25], and proliferating cell nuclear antigen (PCNA) and glutathione S-transferase mu (GST mu) [25] gene expression in kidney from gentamicin-treated rat have been previously reported. The correlation between gentamicininduced KIM-1 expression changes observed in this study and subsequent changes in KIM-1 protein requires further investigation in dog. The availability of fit-forpurpose validated canine specific immunoassays would support the possibility of testing whether or not urinary KIM-1 and/or other proteins that are readily detectable in biological fluids may serve as non-invasive biomarkers for monitoring DIKI in dog which have been previously qualified for specific use(s) in rat preclinical studies.
Qualification of Canine Renal Biomarkers
The identification of genomic biomarkers for monitoring DIKI in dogs is needed because of the non-specificity and late occurrence of sCr elevations. Reagent development must be primarily based on the unique molecular characteristics of these next generation biomarkers; and canine-specific antibodies and ELISAs intended for measuring novel renal biomarkers have been recently marketed. For example, the Dog NGAL ELISA kit (BIOPORTO® Diagnostics, Gentofte, Denmark) has been used to demonstration neutrophil gelatinase-associated lipocalin (NGAL) in urine as a sensitive and predictive biomarker for acute DIKI in gentamicin-treated dogs [28]. Similar utility of urinary NGAL has been shown in gentamicin-treated rats [28]. NGAL has not been qualified in rats to date.
To this end, considering the recent availability of canine specific reagents, the constitutive nature and secretory characteristics of selected genes upregulated in both kidney and urine following the administration of gentamicin in dogs, the relevance of these parameters may be further investigated which could lead to the qualification of novel sensitive and selective biomarkers indicative of DIKI in dogs. Furthermore, selected novel renal biomarker gene/protein sequences are sufficiently conserved across humans, canines, non-human primates, and rats. Taken together, novel canine renal biomarkers would support opportunities to identify DIKI in preclinical studies in dogs that to date remain challenging or unlikely monitorable in order to delineate the relevance of such findings to other preclinical studies and to the clinical setting.
5. AUTHOR CONTRIBUTIONS
JEM: Prepared manuscript. JG: Performed the gene expression analyses. AB: Performed gene expression data reporting. JM: Provided histopathology analysis. MS: Contributed to the data interpretation. DL: Served as the principal investigator. MW: Contributed to the data interpretation and design for confirmatory studies. SS: Participated in the research design and data interpretation.
6. ACKNOWLEDGEMENTS
We would like to thank the Renal Integrated Safety Assessment team within Janssen Pharmaceutical Research & Development, LLC for their ongoing support to ascertain novel renal biomarkers within specific contexts of use to enable proprietary drug development programs to progress safely and faster.
REFERENCES
- Dieterle, F., Sistare, F., Goodsaid, F., Papaluca, M., Ozer, J.S., Webb, C.P., Baer, W., Senagore, A., Schipper, M.J., Vonderscher, J., Sultana, S., Gerhold, D.L., Phillips, J.A., Maurer, G., Carl, K., Laurie, D., Harpur, E., Sonee, M., Ennulat, D., Holder, D., Andrews-Cleavenger, D., Gu, Y.Z., Thompson, K.L., Goering, P.L., Vidal, J.M., Abadie, E., Maciulaitis, R., Jacobson-Kram, D., Defelice, A.F., Hausner, E.A., Blank, M., Thompson, A., Harlow, P., Throckmorton, D., Xiao, S., Xu, N., Taylor, W., Vamvakas, S., Flamion, B., Lima, B.S., Kasper, P., Pasanen, M., Prasad, K., Troth, S., Bounous, D., Robinson-Gravatt, D., Betton, G., Davis, M.A., Akunda, J., McDuffie, J.E., Suter, L., Obert, L., Guffroy, M., Pinches, M., Jayadev, S., Blomme, E.A., Beushausen, S.A., Barlow, V.G., Collins, N., Waring, J., Honor, D., Snook, S., Lee, J., Rossi, P., Walker, E. and Mattes, W. (2010) Renal biomarker quailfication submission: A dialog between the FDA-EMEA and predictive safety testing consortium. Nature Biotechnology, 28, 455-462. doi:10.1038/nbt.1625
- Bellomo, R., Ronco, C., Kellum, J.A., Mehta, R.L. and Palevsky, P. (2004) Acute renal failure—Definition, outcome measures, animal models, fluid therapy and information technology needs: The second international consensus conference of the acute dialysis quality initiative (ADWI) group. Critical Care, 8, R204-R212. doi:10.1186/cc2872
- Powell, H.S., Thompson, W.L., Luthe, M.A., Stern, R.C., Grossniklaus, D.A., Bloxham, D.D., Groden, D.L., Jacobs, M.R., DiScenna, A.O., Cash, H.A. and Klinger, D.J. (1983) Once-daily vs. continuous amino glycoside dosing: Efficacy and toxicity in animal and clinical studies of gentamicin, netilmicin, and tobramycin. The Journal of Infectious Diseases, 147, 918-932. doi:10.1093/infdis/147.5.918
- Reiner, N.E., Bloxham, D.D. and Thompson, W.L. (1978) Nephrotoxicity of gentamicin and tobramycin given once daily or continuously in dogs. Journal of Antimicrobial Chemotherapy, 4, 85-101. doi:10.1093/jac/4.suppl_A.85
- Affymetrix product support/technical. https://www.affymetrix.com/support/technical/byproduct.affx?product=canine
- Nishihara, K., Masuda, S., Shinke, H., Ozawa, A., Ichimura, T., Yonezawa, A., Nakagawa, S., Inui, K., Bonventre, J.V. and Matsubara, K. (2013) Urinary chemokine (C-C) motif ligand 2 (monocyte chemotactic protein-1) as a tubular injury marker for early detection of cisplatin nephrotoxicity. Biochemical Pharmacology, 85, 570-582. doi:10.1016/j.bcp.2012.12.019
- Seo, S.G., Lee, J.K., Yu, J.I., Yun, K.J., Chae, S.K. and Choi, S.C. (2010) Identification of the polymorphisms in IFITM3 gene and their association in a Korean population with ulcerative colitis. Experimental & Molecular Medicine, 42, 99-104. doi:10.3858/emm.2010.42.2.011
- Gröne, H.J., Weber, K., Gröne, E., Helmchen, U. and Osborn, M. (1987) Coexpression of keratin and vimentin in damaged and regenerating tubular epithelia of the kidney. The American Journal of Pathology, 129, 1-8.
- Baban, B., Liu, J.Y. and Mozafari, M.S. (2013) Endoplasmic reticulum stress response and inflammatory cytokines in type 2 diabetic nephropathy: Role of indoleamine 2,3-dioxygenase and programmed death-1. Experimental and Molecular Pathology, 94, 343-351. doi:10.1016/j.yexmp.2012.11.004
- Miki, Y., Nakata, T., Suzuki, T., Darnel, A.D., Moriya, T., Kaneko, C.Z., Hidaka, K., Shiotsu, Y., Kusaka, H. and Sasano, H. (2002) Systemic distribution of steroid sulfatase and estrogen sulfotransferase in human adult and fetal tissues. The Journal of Clinical Endocrinology & Metabolism, 87, 5760-5768. doi:10.1210/jc.2002-020670
- Theilig, F., Kriz, W., Jerichow, T., Schrade, P., Hӓhnel, B., Willnow, T., Le Hir, M. and Bachmann, S. (2007) Abrogation of protein uptake through megalin‑deficient proximal tubules does not safeguard against tubulointerstitial injury. Journal of the American Society of Nephrology, 18, 1824-1834. doi:10.1681/ASN.2006111266
- Gardberg, M., Talvinen, K., Kaipio, K., Iljin, K., Kampf, C., Uhlen, M. and Carpen, O. (2010) Characterization of diaphanous-related formin FMNL2 in human tissues. BMC Cell Biology, 11, 55-66. doi:10.1186/1471-2121-11-55
- Pluznick, J.L., Zou, D.-J., Zhang, X., Yan, Q., RodriguezGil, D.J., Eisner, C., Wells, E., Greer, C.A., Wang, T., Firestein, S., Schnermann, J. and Caplan, M.J. (2008) Functional expression of the olfactory signaling system in the kidney. Proceedings of the National Academy of Sciences, 106, 2059-2064. doi:10.1073/pnas.0812859106
- Ou, Z.L., Natori, Y. and Natori, Y. (2009) Transient and sequential expression of chemokine mRNA in glomeruli in puromycin aminonucleoside nephrosis. Nephron, 85, 254-257.
- Saito, A., Miyauchi, N., Hashimoto, T., Karasawa, T., Han, G.D., Kayaba, M., Sumi, T., Tomita, M., Ikezumi, Y., Suzuki, K., Koitabashi, Y., Shimizu, F. and Kawachi, H. (2011) Neurexin-1, a presynaptic adhesion molecule, localizes at the slit diaphragm of the glomerular podocytes in kidneys. American Journal of Physiology—Regulatory, Integrative and Comparative Physiology, 300, R340-R348. doi:10.1152/ajpregu.00640.2009
- Harper, S.J., Xing, C.Y., Whittle, C., Parry, R., Gillatt, D., Peat, D. and Mathieson, P.W. (2001) Expression of neuropilin-1 by human glomerular epithelial cells in vitro and in vivo. Clinical Science, 101, 439-446. doi:10.1042/CS20010025
- Hassoun, H.T., Grigoryev, D.N., Lie, M.L., Liu, M., Cheadle, C., Tuder, R.M. and Rabb, H. (2007) Ischemic acute kidney injury induces a distant organ functional and genomic response distinguishable from bilateral nephrectomy. Renal Physiology: American Journal of Physiology, 293, F30-F40. doi:10.1152/ajprenal.00023.2007
- Lagor, W.R., Brown, R.J., Toh, S-A., Millar, J.S., Fuki, I.V., de la Llera-Moya, M., Yuen, T., Rithblat, G., Billheimer, J.T. and Rader, D.J. (2009) Overexpression of apolipoprotein F reduces HDL cholesterol levels in vivo. Arteriosclerosis, Thrombosis, and Vascular Biology, 29, 40-46. doi:10.1161/ATVBAHA.108.177105
- Gaudreault, I., Guay, D. and Lebel, M. (2004) YB-1 promotes strand separation in vitro of duplex DNA containing either mispaired bases or cisplatin modifications, exhibits endonucleolytic activities and binds several DNA repair proteins. Nucleic Acids Research, 32, 316-327. doi:10.1093/nar/gkh170
- Hingorani, S., Molitoris, B.A. and Himmelfarb, J. (2009) Ironing out the pathogenesis of acute kidney injury. American Journal of Kidney Diseases, 53, 569-571. doi:10.1053/j.ajkd.2009.01.002
- Pizzorno, G., Cao, D., Leffert, J.J., Russell, R.L., Zhang, D. and Handschumacher, R.E. (2002) Homeostatic control of uridine and the role of uridine phosphorylase: A biological and clinical update. BBA-Molecular Basis of Disease, 2-3, 133-144.
- Holst, C.R., Bou-Reslan, H., Bryan B. Gore, B.B., Wong, K., Deanna Grant, D., Chalasani, S., Carano, R.A., Frantz, G.D., Tessier-Lavigne, M., Bolon, B., French, D.M. and Ashkenazi, A. (2007) Secreted sulfatases Sulf1 and Sulf2 have overlapping yet essential roles in mouse neonatal survival. PLoS ONE, 6, 1-14.
- Reule, S. and Gupta, S. (2011) Kidney regeneration and the role of stem cells. Organogenesis, 7, 135-139. doi:10.4161/org.7.2.16285
- Com, E., Boitier, E., Marchandeau, J.-P., Brandenburg, A., Schroeder, S., Hoffmann, D., Mally, A. and Gautier, J.-C. (2012) Integrated transcriptomic and proteomic evaluation of gentamicin nephrotoxicity in rats. Toxicology and Applied Pharmacology, 258, 124-133. doi:10.1016/j.taap.2011.10.015
- Ozaki, N., Matheis, K.A., Gamber, T., Feidl, T., Nolte, A., Kalkuhl, A. and Deschl, U. (2010) Identification of genes involved in gentamicin induced nephrotoxicity in rats—A toxicogenomic investigation. Experimental and Toxicologic Pathology, 61, 555-566. doi:10.1016/j.etp.2009.07.004
- Ward, P.D., La, D. and McDuffie, J.E. (2013) Renal transporters and biomarkers in safety assessment, new insights into toxicity and drug testing. In: S. Gowder, Ed., InTech. http://www.intechopen.com/books/new-insights-into-toxicity-and-drug-testing/renal-transporters-and-biomarkers-in-safety-assessment
- Bonventre, J.V. (2010) Next generation biomarkers for detecting kidney toxicity. Nature Biotechnology, 28, 336- 440. doi:10.1038/nbt0510-436
- Kai, K., Yamaguchi, T., Yoshimatsu, Y., Kinoshita, J., Teranishi, M. and Takasaki, W. (2013) Neutrophil gelatinase-associated lipocalin, a sensitive urinary biomarker of acute kidney injury in dogs receiving gentamicin. The Journal of Toxicological Sciences, 38, 269-277. doi:10.2131/jts.38.269
NOTES
*Corresponding author.

