 Vol.3, No.4B, 36-48 (2013) Open Journal of Animal Sciences http://dx.doi.org/10.4236/ojas.2013.34A2005 A phylogenetic study of Drosophila splicing assembly chaperone RNP-4F associated U4-/U6-snRNA secondary structure Jack C. Vaughn*, Sushmita Ghosh, Jing Chen Department of Biology, Cell Molecular and Structural Biology Program, Miami University, Oxford, USA; *Corresponding Author: vaughnjc@MiamiOH.edu Received 15 August 2013; revised 23 September 2013; accepted 1 October 2013 Copyright © 2013 Jack C. Vaughn et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. ABSTRACT The rnp-4f gene in Drosophila melanogaster encodes nuclear protein RNP-4F. This encoded protein is represented by homologs in other eukaryotic species, where it has been shown to function as an intron splicing assembly factor. Here, RNP-4F is believed to initially bind to a recognition sequence on U6-snRNA, serving as a chaperone to facilitate its association with U4-snRNA by intermolecular hydrogen bonding. RNA conformations are a key factor in spli- ceosome function, so that elucidation of chang- in g seconda ry structures for interacti ng sn RNAs is a subject of considerable interest and impor- tance. Among the five snRNAs which participate in removal of spliceosomal introns, there is a growing consensus that U6-snRNA is the most structurally dynamic and may constitute the catalytic core. Previous studies by others have generated potential secondary structures for free U4- and U6-snRNAs, including the Y-shaped U4-/U6-snRNA model. These models were based on study of RNAs from relatively few species, and the popular Y-shaped model remains to be systematically re-examined with reference to the many new sequences generated by recent ge- nomic sequencing projects. We have utilized a comparative phylogenetic approach on 60 di- verse eukaryotic species, which resulted in a revised and improved U4-/U6-snRNA secondary structure. This general model is supported by observation of abundant compensatory base mutations in every stem, and incorporates more of the nucleotides into base-paired associations than in previous models, thus being more en- ergetically stable. We have extensively sampled the eukaryotic phylogenetic tree to its deepest roots, but did not find genes potentially encod- ing either U4- or U6-snRNA in the Giardia and Trichomonas data-bases. Our results support the hypothesis that nuclear introns in these most deeply rooted eukaryotes may represent evolutionary intermediates, sharing characteris- tics of both group II and spliceosomal introns. An unexpected result of this study was discov- ery of a potential competitive binding site for Drosophila splicing assembly factor RNP-4F to a 5’-UTR regulatory region within its own pre- mRNA, which may play a role in negative feed- back control. Keywords: RNP-4F; snRNA Secondary Structure; U4-/U6-snRNA Phylogeny; Spliceosome Evolution 1. INTRODUCTION Drosophila melanogaster, which is a cosmopolitan holometabolous insect found in all warm environments, has been an important model organism for genetic, mo- lecular, cellular and physiological studies for over a cen- tury. Its small size (usually 2 - 4 mm), short life cycle (10 - 14 days at 25˚C), high reproductive rate (an adult fe- male can lay 400 - 500 eggs in 10 days), completely se- quenced and largely annotated genome, well-developed techniques, and evolutionarily-conserved molecular path- ways all contribute to making Drosophila a research paradigm. It has been predicted that about 75% of human disease genes have clear homologs in D. melanogaster [1,2], an observation leading to the extensive use of Drosophila which has led to advances in the improve- ment of human health. The long-term objective of our research is to under- stand evolutionarily-conserved cellular, developmental, molecular and genetic mechanisms behind regulation of Copyright © 2013 SciRes. OPEN ACCESS  J. C. Vaughn et al. / Open Journal of Animal Sciences 3 (2013) 36-48 37 genes which encode intron splicing assembly factor pro- teins, a topic about which relatively little is known. The system which we are currently using to address these questions is the Drosophila rnp-4f gene, which encodes splicing assembly factor RNP-4F, and we are concen- trating on mechanisms of posttranscriptional level regu- lation [3-11]. This protein is believed to play a direct role during spliceosome assembly by acting as a chaperone to unwind U6-snRNA and thus facilitate its association with U4-snRNA via intermolecular h ydrogen bonding [12-1 6]. In the course of our work, we became interested in sec- ondary structure interactions within the Drosophila U4-/U6-s nRNA duplex. The major or U2-type molecular pathway for removal of spliceosomal in trons has been extensively studied [re- viewed in 17, 18], and shown to require direct participa- tion of five trans-acting small nuclear uracil-rich RNAs (snRNAs) termed U1, U2, U4, U5 and U6. These RNAs are each associated with specific sets of proteins to yield the corresponding biologically active snRNPs, which progressively interact with pre-mRNAs and with each other during the ensuing spliceosomal assembly. In addi- tion to these snRNAs, about 70 different snRNP proteins and more than 100 non- snRNP proteins have been shown to be spliceosomal components [reviewed in 19]. For example, the essential Saccharomyces cerevisiae pre- mRNA splicing protein Prp24, rep resented in Drosophila by its ortholog RNP-4F and in human by p110 [13,14] facilitates U4- and U6-snRNA pairing during spliceoso- mal assembly [16]. A succession of snRNA conformational changes ac- companies steps in the splicing pathway, which are es- sential in generation and function of the catalytic struc- ture. Elucidation of the changing secondary structures of the interacting snRNA molecules is therefore a subject of considerable interest and importance. The comparative phylogenetic approach [20,21] generates models in which existence of potential biologically significant stem-loops can be established by observation of com- pensatory base mutations in diverse species, and has proven to be a powerful technique. The original Y-shaped U4-/U6-snRNA duplex secondary structure model [12] was based on this methodology by comparing yeast, fruit-fly, plant and human sequences. Subsequent studies have shown that RNAs from various species can also be folded in accordance with this model [22-26]. However, no attempt has ever been made to systematically re-ex- amine the original model itself, utilizing the relative abundance of new sequences now available for analysis. 2. MATERIALS AND METHODS 2.1. Selection of U4- and U6-snRNA Sequences We began by utilizing the original Small RNA Data- base [27] as a source for sequences published early. We then carried out GenBank searches, followed by BLAST searches (http://www.ncbi.nlm.nih.gov/BLAST) in which bait sequences were derived from the major phylogenetic levels. Finally, the number of sequences available for study was further increased from early published work not submitted to GenBank. The BLAST search was more successful in finding U6-snRNAs, owing to their ex- tremely high sequence conservation. We did not use every sequence found, excluding for example those from eleven other Drosophila species [28] and also different species of Saccharomyces, since their inclusion would add little additional understand ing due to having v irtually identical sequences within a genus. This exercise (Table 1) yielded 42 U4- and 56 U6-snRNAs, of which 38 were both available in a given species and deemed optimal for our study. In total, sequences from some 60 different species were included in our study. 2.2. Alignment of U4- and U6-snRNA Sequences All sequences selected for this study were individually aligned with reference to the corresponding Drosophila genes using the ClustalW program (http://align.genome.jp), and the resulting alignment was further refined by eye. Finally, the alignment was adjusted using the emerging secondary structure results, to assure that homologous nucleotides would be compared for evidences of com- pensatory base mutations. The final alignments (not shown) included as few deletions (gaps) and insertion s as possible, while generating the maximum number of matchi n g re sidues. 2.3. Strategy for U4-/U6-snRNA Duplex Secondary Structure Determination We elected to start completely from the beginning in deriving our secondary structure model, in contrast to merely modifying existing models, to optimize the chances of identifying structural components not previ- ously recognized. We began by utilizing version 3.6 of the Mfold program (http://mfold.rna.albany.edu) [29] for the two genes individually from Drosophila melanogaster (fruit-fly), Homo sapiens (human), Arabidopsis thaliana (plant), Kluyveromyces lactis (yeast) and Trypanosoma brucei (flagellate). GenBank accession numbers are given in Table 1 . These structures contained a variety of potential stem-loops, and were combined to include only stem-loops held in common. The resulting U4- and U6-snRNA structures were then combined to accommo- date base-pairing between the two molecules in the two closely adjacent U6 locations previously determined by photochemical cross-linking in mammalian snRNAs [30] nd by subsequent observation of compensatory base a Copyright © 2013 SciRes. OPEN ACCESS  J. C. Vaughn et al. / Open Journal of Animal Sciences 3 (2013) 36-48 Copyright © 2013 SciRes. OPEN ACCESS 38 Table l. U4 and U6 RNA sequences utilized in this study. Organism GenBank Accession Number or Reference U6-snRNA U4-snRNA Animalia, Vertebrate Homo sapiens (human) X07425 X59361 Pan troglodytes (c h impanzee) AC146131 NW_001223167 Macaca mulatta (monkey) NW_001218112 NW_001096649 Mus musculus (mouse) X06980 AC159539 Rattus norvegicus (rat) AC120800 K00477 Canis familiaris (dog) AC188530 NW_876282 Bos taurus (cattle) NW_001492849 NW_001493540 Sus scrofa (pig) CR956385 ----- Equus caballus (horse) NW_001799704 NW_001799734 Monodelphis domestica (opossum) NW_001581906 NW_001584232 Ornithorhynchus anatinus NW_001794177 NW_001765942 (duck-billed platypus) Gallus gallus (chicken) NW_001471627 M14136 Xenopus tropicalis (frog) M31687 ----- Danio rerio (zebrafish) CU466287 NW_001514552 Animalia, Invertebrate Drosophila melanogaster (fruit-fly) X06669 D00043 Aedes aegypti (mosquito) AAGE02013372 ----- Anopheles gambiae (mosquito) NZ_AAAB02008807 ----- Culex pipiens (mosquito) AAWU01008690 AAWU01009244 Apis mellifera (honey bee) NW_001253045 ----- Nasonia vitripennis (jewel wasp) NW_001815737 AAZX01001234 Bombyx mori (silkworm moth) AADK01011346 DQ861919 Tribolium castaneum (flour beetle) AC154132 NW_001092869 Tachypleus tridentatus X53789 ----- (horseshoe crab) Ascaris lumbricoides (nematode) L22252 L22250 Caenorhabditis elegans (nematode) X07829 X07828 Schistosoma mansoni (trematode) L25920 ----- Taenia solium (tapeworm) AF529186 ----- Ly t echinus variegatus (sea urchin) ----- U37266 Strongylocentrotus purpuratus X76389 NW_001323459 (sea urchin) Fungi, Ascomycota Saccharomyces cerevisiae X12565 Siliciano et al. (1987) (budding yeast) Schizosaccharomyces pombe X14196 X15491 (fission yeast) Kluyveromyces lactis NC_006042 Guthrie & Patterson (1988) Candida albicans EU144231 EU144229 Va nde rwaltoz yma po lyspor a NZ_AAZN01000268 -----  J. C. Vaughn et al. / Open Journal of Animal Sciences 3 (2013) 36-48 39 Continued Ashbya gossypii NC_005788 ----- Fungi, Basidiomycota Erythrobasidium hasegawianum Tani & Ohshima (1991) D63682 Puccinia graminis AAWC01000866 ----- Coprinopsis cinerea AACS01000244 ----- Phanerochaete chrysosporium AADS01000210 ----- Amoebozoa, Mycetozoa Dictyostelium discoideum AY953942 AY918063 (sl ime mold ) Physarum polycephalum ----- X13840 (sl ime mold ) Amoebozoa, Con osa Entamoeba histolytica U43841 BK006131 Viridiplantae, Eudicot Arabidopsis thaliana (thale cress) X52527 X67145 Vicia faba (broad bean) Solymosy & Pollak (1993) Solymosy & Pollak (19 93 ) Pisum sativum (pea) Solymosy & Pollak (1993) X15933 Solanum lycopersicum (toma to) X51447 ----- Solanum tuberosum (potato) S83742 ----- Populus trichocar pa (Poplar) NC_008469 NC_008470 Viridiplantae, Monocot Oryza sativa (rice) NC_008405 DQ649301 Triticum aestivum (wheat) X63066 ----- Zea mays (maize) ----- Solymosy & Pollak (19 93 ) V iridiplantae, Algae Chlamydomonas reinhardtii X71486 X71485 Alveolata, Cilliophora Tetrahymena thermophila Orum et al. (1991) Orum et al. (1991) Alveolata, A picomplexa Plasmodium falciparum EF419774 EF140769 Euglenozoa Trypanosoma brucei (flagellate) X57 04 6 Solymosy & Pollak (1993) Crithidia fasciculata (flagellate) X78550 AF326336 Leishmania tarentolae (flagellate) ----- X97621 Leishmania mexicaca (flagellate) X82228 ----- Leptomonas seymouri (flagellate) X78552 AJ245951 Phytomonas sp. (flagellate) X82229 ----- mutations [12], which resulted in further simplification of potential stem-loops in the predicted duplex RNA structure. Compensatory base changes were then entered onto the Drosophila duplex structure in comp arison with the five species originally used to begin the study (above), using the alignment to assure that homologous nucleotides were being compared. We adopted the crite- rion [20] that existen ce of a helix is considered proven if there are at least two base-pair replacements. Stems as short as two base-pairs are acceptable if compensatory base changes can be demonstrated (Carl Woese, personal communication). Finally, the provisional model was compared to every species utilized in the study (Table 1), to determine the extent to which the resulting structure was univ ersal. When an otherwise proven ste m-loop was found to be absent from any taxonomic level, the timing Copyright © 2013 SciRes. OPEN ACCESS  J. C. Vaughn et al. / Open Journal of Animal Sciences 3 (2013) 36-48 40 of that loss was charted with reference to the eukaryotic phylogenetic tree [31]. 3. RESULTS AND DISCUSSION 3.1. An Improved General Secondary Structure Model for U4-/U6-snRNA The derived U4-/U6-snRNA duplex secondary struc- ture model is shown in Figure 1, and structures from representative species at different taxonomic levels in Figures 2(a)-(h). A relatively large proportion of all nu- cleotides are base-paired in our U4-/U6-snRNA model. For example, in Drosophila 58% are base-paired in U4 and 63% in U6, whereas in the Y-shaped model the cor- responding numbers are 58% and 33%. Four stem-loops (I-IV) are found to be present in the U4 structure for most species, so that our model both confirms and ex- tends the secondary structure for free U4-snRNA previ- ously proposed [32] using the phylogenetic approach with far fewer species. The existence of stem-loop IV in free U4-snRNA, proposed by the same authors, is also confirmed for all species studied by us. The overall con- formation of the structure shown in our model is very similar in every species examined, with the exception of stem-loop III in U4-snRNA which is further discussed in Section 3.2. Each stem in our model has been proven by Figure 1. General secondary structure model for Drosophila U4-/U6-snRNA duplex. The two RNAs inter- act by base-pairing within regions designated DS I and DS II. Compensatory base changes which prove the structure illustrated are boxed and were identified in the alignment with reference to the structures derived for H. sapiens, A. thaliana, K. lactis and T. brucei. The range of stem lengths found between different spe- cies in our study is shown beside each stem. Stem-loop IV in free U4-snRNA (large box) is disrupted upon binding to U6-snRNA. The putative SM-binding site (SM) is indicated. An RNA recognition motif (RRM) in chaperone RNP-4F/Prp24/p110 binds primarily to a tract within free U6-snRNA nucleotides #38-57 (13), which is indicated by a heavy vertical overlay. Copyright © 2013 SciRes. OPEN ACCESS 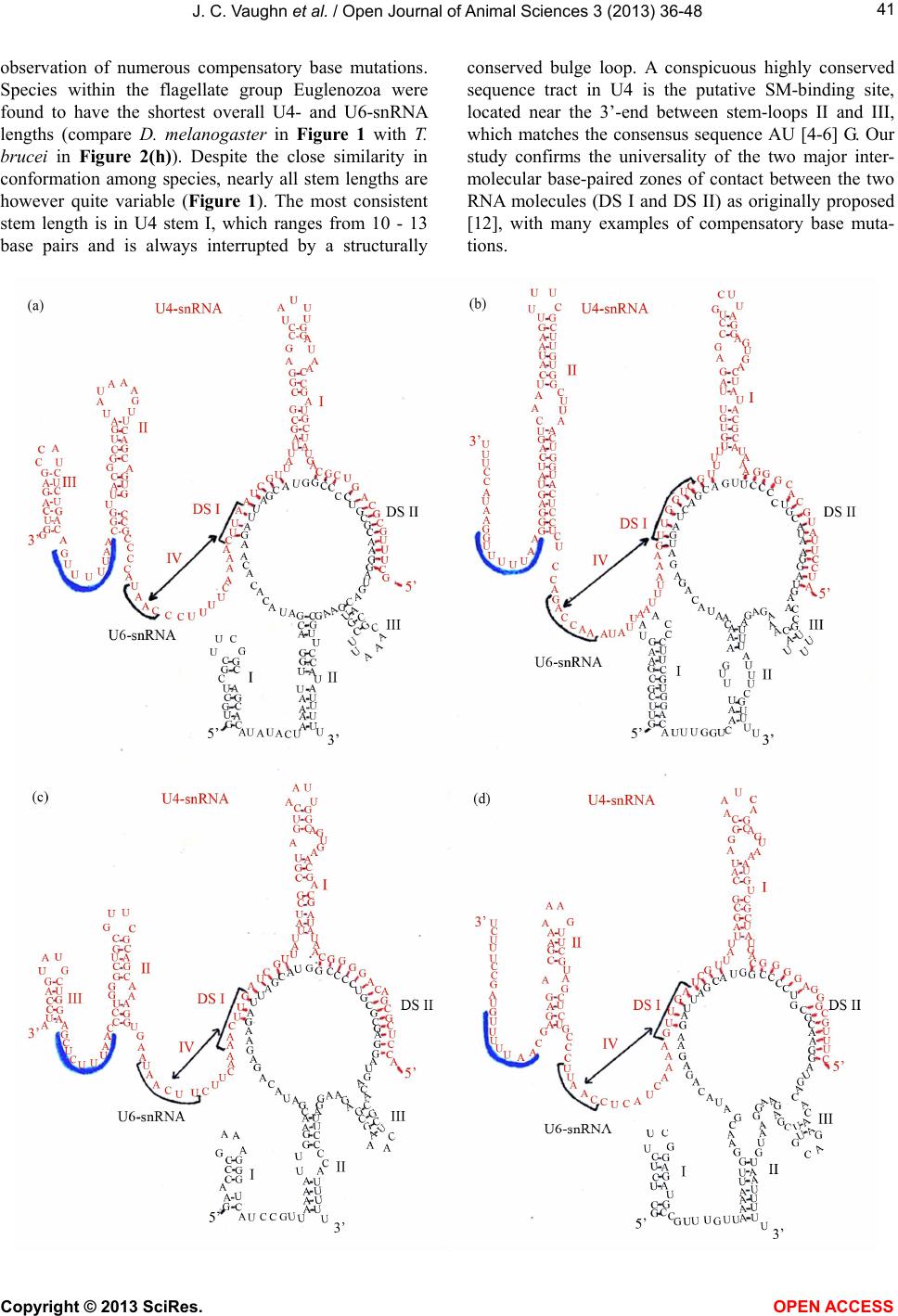 J. C. Vaughn et al. / Open Journal of Animal Sciences 3 (2013) 36-48 41 observation of numerous compensatory base mutations. Species within the flagellate group Euglenozoa were found to have the shortest overall U4- and U6-snRNA lengths (compare D. melanogaster in Figure 1 with T. brucei in Figure 2(h)). Despite the close similarity in conformation among species, nearly all stem lengths are however quite variable (Figure 1). The most consistent stem length is in U4 stem I, which ranges from 10 - 13 base pairs and is always interrupted by a structurally conserved bulge loop. A conspicuous highly conserved sequence tract in U4 is the putative SM-binding site, located near the 3’-end between stem-loops II and III, which matches the consensus sequence AU [4-6] G. Our study confirms the universality of the two major inter- molecular base-paired zones of contact between the two RNA molecules (DS I and DS II) as originally proposed [12], with many examples of compensatory base muta- tions. Copyright © 2013 SciRes. OPEN ACCESS  J. C. Vaughn et al. / Open Journal of Animal Sciences 3 (2013) 36-48 42 Figure 2. Representative U4-/U6-snRNA secondary structures from phylogenetically diverse species, folded according to our general model. (a) H. sapiens; (b) S. cerevisiae; (c) T. thermophila; (d) P. falciparum; (e) D. discoideum; (f) A. thaliana; (g) C. reinhardtii; (h) T. brucei. Labeling is as in Figure 1. The U6-snRNA nucleotide sequence is relatively highly conserved, in comparison with that for U4. Three stem-loops are also present in the U6 structure in our duplex model, which is in contrast to the Y-shaped model in which only stem-loop I is shown. In our model the 3’-end of U6-snRNA is incorporated into the structu re to form stem-loop II in every species examined, albeit in some cases with a central bulge loop or absence of base-pairing at the top of the stem. U6 stem-loop II is proven by observation of compensatory base mutations, and is not shown in other models. A second U6 structural feature in our model which is not shown in other models is a short stem-loop III. This stem-loop is only two base-pairs long in many species, but is proven by obser- Copyright © 2013 SciRes. OPEN ACCESS  J. C. Vaughn et al. / Open Journal of Animal Sciences 3 (2013) 36-48 43 vation of compensatory base mutations. We did however fail to observe this stem-loop in the fungus C. albicans and in E. histolyt i ca, showing that it is not universal. 3.2. The General Secondary Structure Model is Not Universal and Multiple Independent U4-snRNA Stem-Loop III Losses Have Occurred During Evolution Representative structures for a diverse selection of evolutionarily distant species show that the general model is not universal. The most striking example is in the absence of U4-snRNA stem-loop III (Figures 2(b), (d), (e)), otherwise proven by observation of numerous compensatory base changes. The absence of this stem- loop has previously been noted in secondary structures for various species of yeast and slime molds [12,25,32, 33]. It has been suggested that the absence of this stem-loop is correlated with phylogenetic depth, imply- ing that this structural feature was not present in the ear- liest eukaryotes and is newly evolved [32]. We tested this hypothesis by superimposing the presence/ absence of this stem-loop onto the eukaryotic phylogenetic tree [31]. The results show that this stem-loop is present in all spe- cies among the deeply-rooted flagellate Euglenozoa ex- amined, but that three clearly independent secondary losses have occurred during evolution (Figure 3). The most recent is within Fungi, where all Ascomycete spe- cies studied have lost the stem-loop, which is however present in the Basidiomycete E. hasegawianum. An ear- lier independent loss occurred among the Amoebozoa, where the Mycetozoa slime mold species examined have lost the stem-loop but the amoeboid Conosa E. histo- lytica has not. The earliest loss is in the Alveolata, where this feature is absent in the Apicomplexa P. falciparum but not in the Cilliophora T. thermophila. 3.3. The General Secondary Structure Model Compared to the Classical Y-Shaped Model It is informative to compare the secondary structures of free U4- and U6-snRNAs with that of the duplex which is formed upon their association during spli- ceosome assembly, in consideration of the most parsi- monious solution for their association (Figure 4). An excellent free U4-snRNA secondary structure model has previously been proposed based on the phylogenetic ap- proach [32], utilizing a taxonomic diversity of species extending only as deep as the slime mold Physarum. This structure has been experimentally supported by the re- sults of enzymatic digestion studies in rat U4-snRNA [34]. The model contains four stem-loops, of which three are incorporated directly into both our model and the Y-shaped model. Stem-loop IV is disrupted in favor of intermolecular base-pairing to form DS I, upon associa- tion with U6-snRNA. Our resu lts confirm and extend the previously proposed free U4-snRNA model, showing that the structure has been retained to its origin within the flagellate group Euglenozoa (Figure 3). There ar e no differences in this part of our model in comparison to the Y-shap ed model. Previously proposed free U6-snRNA models for hu- man [35] and yeast S. cerevisiae [36] show somewhat differing structures, which are both supported by the re- sults of chemical and enzymatic probing in these species [37]. In the simplest free U6-snRNA secondary structure model, as exemplified in Drosophila (Figure 1) and hu- man, a short stem-loop is present at the 5’-end and the entire 3’-terminus is folded into one long interrupted stem-loop (Figure 4(a)). In human the chaperone p110, an ortholog of Drosophila RNP-4F, has been shown to bind primarily to free U6-snRNA nucleotides #38-57 [13], promoting unwinding of the long stem-loop and base-pairing to two closely adjacent tracts on U4-snRNA, which we have designated as DS I and DS II, followed by chaperone release. Our model and the Y- shaped model differ primarily in how they show the U6-snRNA structure within the RNA duplex. In the latter model, only the 5’-end stem-loop is retained (Figure 4(c)), and no base-pairing occurs else- where except within regions DS I and DS II, so that the 3’-end is unpaired. In our model, the base of old free U6-snRNA is retained in stem-loop II, which brings the 3’-end into a duplex structure (Figure 4(b)). One set of observations in support of this structure is seen in the compensatory mutations present in this stem (Figure 1). The results of previously reported chemical and enzy- matic probing of the U4-/U6-snRNA duplex further support the model which we have proposed and not the Y-shaped model. In human, chemical reagent modifica- tions were not observed within nucleotides #27-38 or #94-106, which comprise the helix in stem-loop II in our model but which are shown in long unpaired 5’- and 3’-tracts in the Y-shaped model. These observations are indicative of a double-stranded structure here, and this interpretation is confirmed by the observation of RNase V1 cleavage 3’ to positions 33 and also 35 in the human U4-/U6-snRNA duplex [37]. This is an enzyme which cleaves specifically double-stranded RNA regions. These results have also been reported by these authors upon probing the base of free U6-snRNA stem-loop II. It has been proposed that a potential third base-paired region of contact may exist between U4- and U6-snRNA [24]. In this view, the top of our U6-snRNA stem-loop II is base-paired with a complement located within the long single-stranded U4 connective between DS I and U4 stem-loop II. We are skeptical of this proposed third zone Copyright © 2013 SciRes. OPEN ACCESS 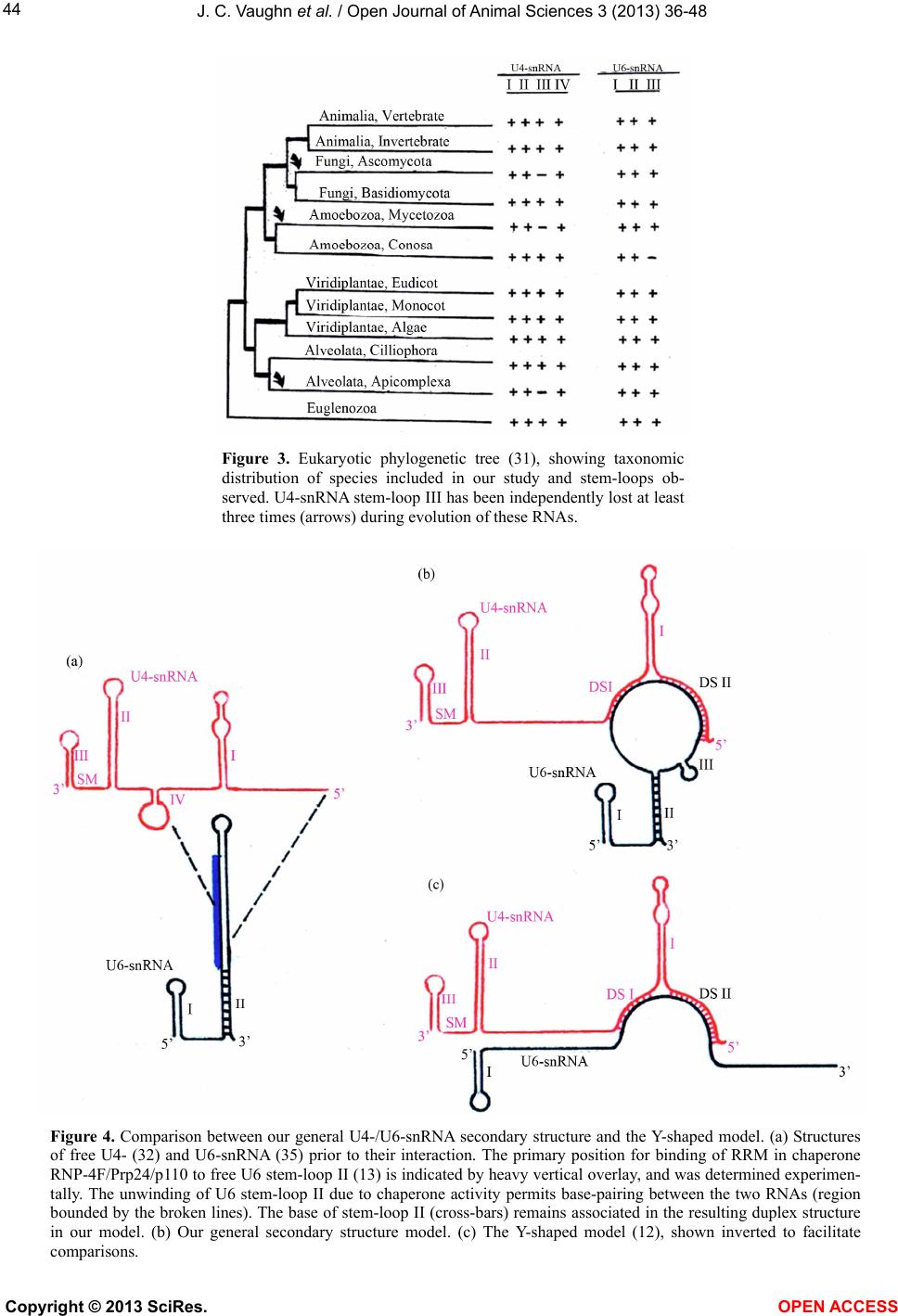 J. C. Vaughn et al. / Open Journal of Animal Sciences 3 (2013) 36-48 Copyright © 2013 SciRes. 44 Figure 3. Eukaryotic phylogenetic tree (31), showing taxonomic distribution of species included in our study and stem-loops ob- served. U4-snRNA stem-loop III has been independently lost at least three times (arrows) during evolution of these RNAs. Figure 4. Comparison between our general U4-/U6-snRNA secondary structure and the Y-shaped model. (a) Structures of free U4- (32) and U6-snRNA (35) prior to their interaction. The primary position for binding of RRM in chaperone RNP-4F/Prp24/p110 to free U6 stem-loop II (13) is indicated by heavy vertical overlay, and was determined experimen- tally. The unwinding of U6 stem-loop II due to chaperone activity permits base-pairing between the two RNAs (region bounded by the broken lines). The base of stem-loop II (cross-bars) remains associated in the resulting duplex structure in our model. (b) Our general secondary structure model. (c) The Y-shaped model (12), shown inverted to facilitate comparisons. OPEN ACCESS  J. C. Vaughn et al. / Open Journal of Animal Sciences 3 (2013) 36-48 45 of RNA/RNA interaction, since different nucleotides in the alignment must be utilized to create this structure. For example, in S. cerevisiae and K. lactis the U4 region of contact is very different from that for other species. Within free U6-snRNA, nucleotides comprising stem- loop III are contained within the long stem-loop (Figure 4(a)). The existence of stem-loop III in the duplex struc- ture is proven by observation of compensatory base mu- tations, but the stem length is reduced to only two base- pairs in many species. Chemical and enzymatic probing of the human U4-/U6-snRNA duplex [37] did not pro- vide any further clarification for existence of this stem- loop, since most of this region was contained in the site of the primer utilized. Cryo-electron microscopy of iso- lated U4-/U6-snRNA has been reported to show two major structural domains linked by a thin connective [38], in good agreement with our general secondary structure model. 3.4. Phylogenetic Depth of the Genes Encoding U4- and U6-snRNAs The secondary structure of the U4-/U6-snRNA duplex in our model is found to be identical, with the exception of multiple independent losses of U4 stem-loop III dis- cussed above, down to and including the deeply-rooted flagellate group Euglenozoa. However, extensive BLAST searches against both the Giardia [39] and Tricho monas [40] genome sequences failed to detect any U4- or U6- snRNA orthologs, using the corresponding T. brucei se- quences as bait. The diplomonads and parabasalids are generally considered to be descendants of the earliest extant eukaryotes [31], leading us to consider the impli- cations of this obser vat i on . Success in BLAST searches is dependent on the de- gree of nucleotide conservation between bait and prey sequences, in addition to the completeness and accuracy of the genomic sequence database itself. The nucleotide sequences of genes encoding U6-snRNAs are among the most highly conserved of any eukaryotic genes. For ex- ample, the human and Drosophila U6-snRNA sequences are 94% identical. The U6-snRNA sequence within and immediately flanking the region of base-pairing with U4-snRNA is exception ally well conserved. For example, comparison between Drosophila and flagellate T. brucei U6 nucleotides #40-75 shows 86% identity. This degree of conservation is far greater than that observed for U4, making identification of its most ancient orthologs more difficult. It was therefore surprising that no U6-snRNA genes turned up during BLAST searches against both the diplomonad and p arabasalid g e nomes. The Giardia and Trichomonas genome annotations are well along, and we therefore asked if ANY of the U-se- ries snRNA gene sequences have been annotated in these species. Surprisingly, NONE of these genes have been found despite an ~7X coverage during sequencing. In addition, none of the genes encoding proteins which are part of the U4- and U6-snRNPs in other eukaryotes have been found (Steven Sullivan, personal communication). Annotation of the Giardia genome has also failed to de- tect any genes encoding U4- or U6-snRNA (Hilary Mor- rison, personal communication). What are the implica- tions of these observations? The spliceosome is widely viewed as having evolved from self-splicing group II introns like those in organellar protein-encoding genes as well as in many bacteria [reviewed in 41,42], which do not utilize the U-series of snRNAs. Interestingly, it has been proposed that Giardia and Trichomonas nuclear introns may represent evolutionary intermediates, show- ing characteristics of both group II and spliceosomal introns [43]. If so, then our study suggests that genes encoding U4- and U6-snRNAs, and the resultant duplex RNA which forms between them with a virtually identi- cal secondary structure among all eukaryotes, may have evolved within the flagellate group Euglenozoa. 3.5. A Potential Secondary RNP-4F Chaperone Recognition Site in the 5’-UTR of Drosophila rnp-4f Pre-mRNA May Play a Key Role in Controlling Its Own Expression We have previously described a long evolutionarily- conserved potential stem-loop which arises by base- pairing between all of the rnp-4f pre-mRNA intron 0 and part of adjacent exon 2 in D. melanogaster [6,8]. We have recently shown using RNA electrophoretic mobility shift assay that retention of intron 0 within the rnp-4f 5’-UTR is correlated with binding of a dADAR protein isoform, and that an unidentified second protein sus- pected to be RNP-4F also binds to this stem-loop [9]. Subsequent work employing RNAi technology showed that this dADAR protein is the truncated isoform [11]. We have proposed a negative feedback model for regu- lating expression of rnp-4f mRNA under conditions of RNP-4F excess within the developing fly central nervous system [6]. If this hypothesis is correct, then the con- served long stem-loop would be expected to contain a nucleotide recognition sequence to which RNP-4F could potentially bind, in competition with its preferred bind- ing to a conserved sequence tract within the long stem- loop of free U6-snRNA [13]. In Drosophila U6-snRNA the conserved sequence contains nucleotides between positions #38-5 7, although an even shorter sequen ce may suffice for chaperone binding, but th is possibility has not yet been tested. Examination of the Drosophila con- served rnp-4f 177-nt stem-loop nucleotide sequence/ structure shows that a 12-nt tract closely resembling the preferred U6-snRNA binding site is indeed present (Fig- re 5(b), (c)). An additional similarity between the u Copyright © 2013 SciRes. OPEN ACCESS 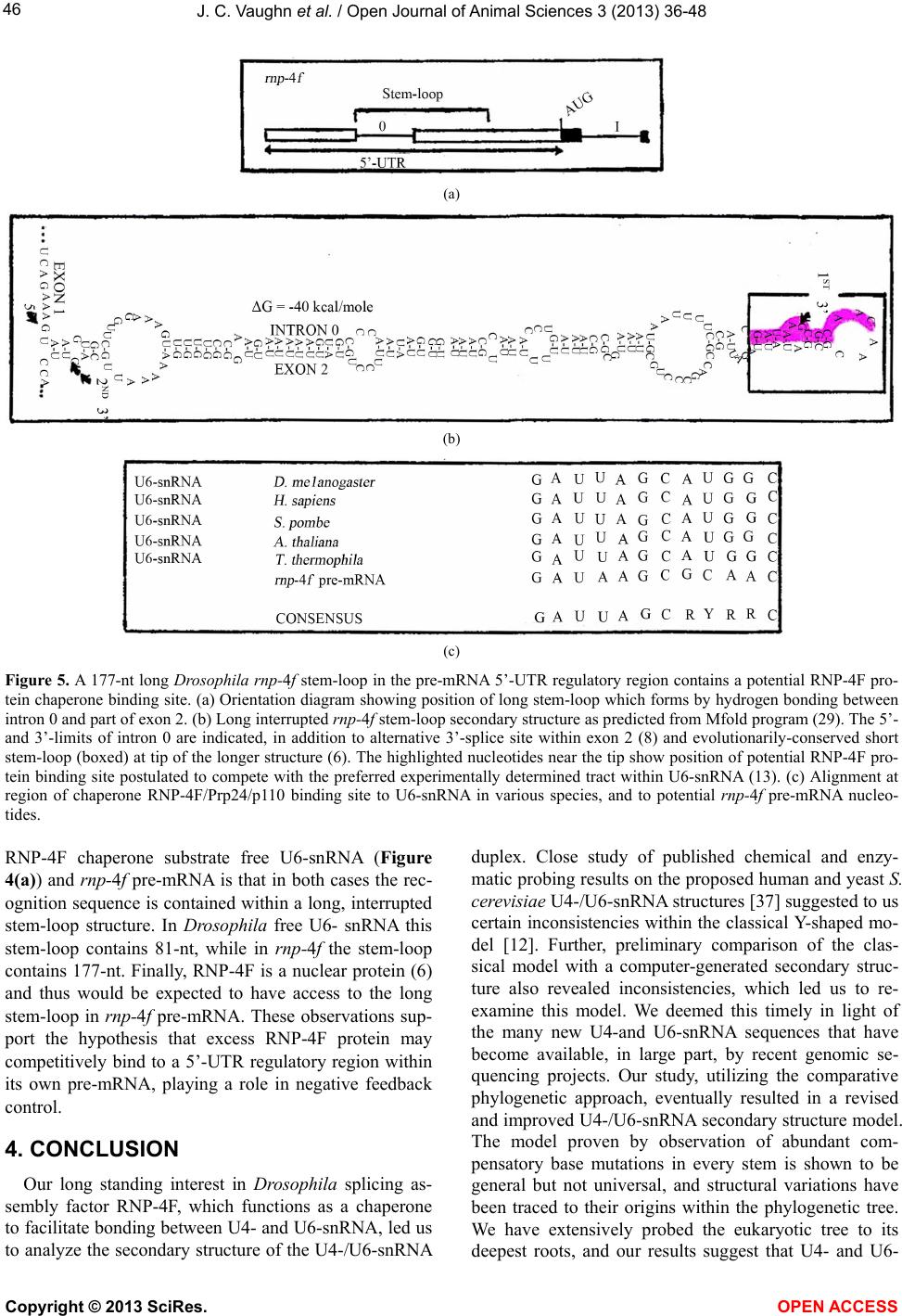 J. C. Vaughn et al. / Open Journal of Animal Sciences 3 (2013) 36-48 46 (a) (b) (c) Figure 5. A 177-nt long Drosophila rnp-4f stem-loop in the pre-mRNA 5’-UTR regulatory region contains a potential RNP-4F pro- tein chaperone binding site. (a) Orientation diagram showing position of long stem-loop which forms by hydrogen bonding between intron 0 and part of exon 2. (b) Long interrupted rnp-4f stem-loop secondary structure as predicted from Mfold program (29). The 5’- and 3’-limits of intron 0 are indicated, in addition to alternative 3’-splice site within exon 2 (8) and evolutionarily-conserved short stem-loop (boxed) at tip of the longer structure (6). The highlighted nucleotides near the tip show position of potential RNP-4F pro- tein binding site postulated to compete with the preferred experimentally determined tract within U6-snRNA (13). (c) Alignment at region of chaperone RNP-4F/Prp24/p110 binding site to U6-snRNA in various species, and to potential rnp-4f pre-mRNA nucleo- tides. RNP-4F chaperone substrate free U6-snRNA (Figure 4(a)) and rnp-4f pre-mRNA is that in both cases the rec- ognition sequence is con tained within a long, interrupted stem-loop structure. In Drosophila free U6- snRNA this stem-loop contains 81-nt, while in rnp-4f the stem-loop contains 177-nt. Finally, RNP-4F is a nuclear protein (6) and thus would be expected to have access to the long stem-loop in rnp-4f pre-mRNA. These observations sup- port the hypothesis that excess RNP-4F protein may competitively bind to a 5’-UTR regulatory region within its own pre-mRNA, playing a role in negative feedback control. 4. CONCLUSION Our long standing interest in Drosophila splicing as- sembly factor RNP-4F, which functions as a chaperone to facilitate bonding between U4- and U6-snRNA, led us to analyze the secondary structure of the U4-/U6-snRNA duplex. Close study of published chemical and enzy- matic probing results on th e proposed human and yeast S. cerevisiae U4-/U6-snRNA structures [37] suggested to us certain inconsistencies within the classical Y-shaped mo- del [12]. Further, preliminary comparison of the clas- sical model with a computer-generated secondary struc- ture also revealed inconsistencies, which led us to re- examine this model. We deemed this timely in light of the many new U4-and U6-snRNA sequences that have become available, in large part, by recent genomic se- quencing projects. Our study, utilizing the comparative phylogenetic approach, eventually resulted in a revised and improved U4-/U6-snRNA secondary structure model. The model proven by observation of abundant com- pensatory base mutations in every stem is shown to be general but not universal, and structural variations have been traced to their origins within the phylogenetic tree. We have extensively probed the eukaryotic tree to its deepest roots, and our results suggest that U4- and U6- Copyright © 2013 SciRes. OPEN ACCESS  J. C. Vaughn et al. / Open Journal of Animal Sciences 3 (2013) 36-48 47 snRNAs apparently evolved after the emergence of lines leading to the diplomonad Giardia and the parabasalid Trichomonas, but once established they have maintained a remarkably well conserved U4-/U6-snRNA secondary structure extending to, and including, the flagellates among the Euglenozoa. An unexpected result of this study was discovery of a potential competitive binding site for Drosophila splicing assembly factor RNP-4F to a 5’-UTR regulatory region within its own pre-mRNA, which may play a role in negative feedback control [6]. This negative feedback expression control model awaits experimental testing. 5. ACKNOWLEDGEMENTS We wish to thank the late Carl Woese for his helpful discussions of RNA secondary structure determinations over more than two decades, to whom this study is dedicated. We also thank Jane Carlton and Steven Sullivan for helpful discussion bearing on Giardia and Trichomonas splicing machinery components. Peter Dobler is to be thanked for his help in constructing the secondary structure figures in the manuscript. This work was primarily supported by National Institutes of Health (NIH) Grant 1-R15-GM070802-01 and 1-R15-GM093895-01 to J. Vaughn. REFERENCES [1] Banfi, S., Borsani, G., Rossi, E., Bernard, L., Guffanti, A., Rubboli, F., Marchitiello, A., Giglio, S., Coluccia, E. and Zollo, M. (1996). Identification and mapping of human cDNAs homologous to Drosophila mutant genes through EST database searching. Nature Genetics, 13, 167-174. http://dx.doi.org/10.1038/ng0696-167 [2] Fortini, M.E., Skupski, M.P., Boguski, M.S. and Hariha- ran, I.K. (2000) A survey of human disease gene coun- terparts in the Drosophila genome. The Journal of Cell Biology, 150, F23-F30. http://dx.doi.org/10.1083/jcb.150.2.F23 [3] Petschek, J.P., Scheckelhoff, M.R., Mermer, M.J. and Vaughn, J.C. (1997) RNA editing and alternative splicing generate mRNA transcript diversity from the Drosophila 4f-rnp locus. Gene, 204, 267-276. http://dx.doi.org/10.1016/S0378-1119(97)00465-4 [4] Feiber, A.L., Rangarajan, J. and Vaughn, J.C. (2002) The evolution of single-copy Drosophila nuclear 4f-rnp genes, spliceosomal intron losses create polymorphic alleles. Journal of Molecular Evolution, 55, 401-413. http://dx.doi.org/10.1007/s00239-002-2336-y [5] Peters, N.T., Rohrbach, J.A., Zalewski, B.A., Byrkett, C.M. and Vaughn, J.C. (2003) RNA editing and regula- tion of Drosophila 4f-rnp expression by sas-10 antisense readthrough mRNA transcripts. RNA, 9, 698-710. http://dx.doi.org/10.1261/rna.2120703 [6] Fetherson, R.A., Strock, S.B., White, K.N. and Vaughn, J.C. (2006) Alternative pre-mRNA splicing in Drosophila spliceosomal assembly factor RNP-4F during develop- ment. Gene, 371, 234-245. http://dx.doi.org/10.1016/j.gene.2005.12.025 [7] Chen, J., Concel, V.J., Bhatla, S., Rajeshwaran, R., Smith, D.L.H., Varadarajan, M., Backscheider, K.L., Bockrath, R.A., Petschek, J.P. and Vaughn, J.C. (2007) Alternative splicing of an rnp-4f mRNA isoform retaining an evolu- tionarily-conserved 5’-UTR intronic element is develop- mentally regulated and shown via RNAi to be essential for normal central nervous system development in Dro- sophila melanogaster. Gene, 399, 91-104. http://dx.doi.org/10.1016/j.gene.2007.04.038 [8] Chen, J., Lakshmi, G.G., Hays, D.L., McDowell, K.M., Ma, E. and Vaughn, J.C. (2009) Spatial and temporal ex- pression of dADAR mRNA and protein isoforms during embryogenesis in Drosophila melanogaster. Differentia- tion, 78, 312-320. http://dx.doi.org/10.1016/j.diff.2009.08.003 [9] Lakshmi, G.G., Ghosh, S., Jones, G.P., Parikh, R., Rawlins, B.A. and Vaughn, J.C. (2012) An RNA electrophoretic mobility shift and mutational analysis of rnp-4f 5’-UTR intron splicing regulatory proteins in Drosophila reveals a novel new role for a dADAR protein isoform. Gene, 511, 161-168. http://dx.doi.org/10.1016/j.gene.2012.09.088 [10] Chen, J., Yang, J.T., Doctor, D.L., Rawlins, B.A., Shields, B.C. and Vaughn, J.C. (2013) 5’-UTR mediated transla- tional control of splicing assembly factor RNP-4F ex- pression during development of the Drosophila central nervous system. Gene, 528,154-162. [11] Ghosh, S., Wang, Y., Cook, J.A., Chhiba, L. and Vaughn, J.C. (2013) A molecular, phylogenetic and functional study of the dADAR mRNA truncated isoform during Drosophila embryonic development. Open Journal of Animal Sciences, (In press). [12] Brow, D.A. and Guthrie, C. (1988) Spliceosomal RNA U6 is remarkab ly conserved from yeast to ma mmals. Na- ture, 334, 213-218. http://dx.doi.org/10.1038/334213a0 [13] Bell, M., Schreiner, S., Damianov, A., Reddy, R. and Bindereif, A. (2002) p110, a novel human U6 snRNP protein and U4/U6 snRNP recycling factor. The EMBO Journal, 21, 2724-2735. http://dx.doi.org/10.1093/emboj/21.11.2724 [14] Rader, S.D. and Guthrie, C. (2002) A conserved Lsm- interaction motif in Prp24 required for efficient U4/U6 di-snRNP formation. RNA, 8, 1378-1392. http://dx.doi.org/10.1017/S1355838202020010 [15] Karaduman, R., Fabrizio, P., Hartmuth, K., Urlaub, H. and Luhrmann, R. (2006) RNA structure and RNA-pro- tein interactions in purified yeast U6 snRNPs. Journal of Molecular Bi ology, 356, 1248-1262. http://dx.doi.org/10.1016/j.jmb.2005.12.013 [16] Bae, E., Reiter, N.J., Bingman, C.A., Kwan, S.S., Lee, D., Phillips, G.N., Butcher, S.E. and Brow, D.A. (2007) Structure and interactions of the first three RNA recogni- tion motifs of splicing factor Prp24. Journal of Molecular Biology, 367, 1447-1458. http://dx.doi.org/10.1016/j.jmb.2007.01.078 [17] Moore, M.J., Query, C.C. and Sharp, P.A. (1993) Splicing of precursors to mRNA by the spliceosome. In: Gesteland, R.F. and Atkins, J.F., Ed., The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 303-357. Copyright © 2013 SciRes. OPEN ACCESS  J. C. Vaughn et al. / Open Journal of Animal Sciences 3 (2013) 36-48 Copyright © 2013 SciRes. OPEN ACCESS 48 [18] Will, C.L. and Luhrmann, R. (2006) Spliceosome struc- ture and function. In: Gesteland, R.F., Cech, T.R. and At- kins, J.F., Ed., The RNA World. Cold Spring Harbor Labo- ratory Press, Cold Spring Harbor, New York, 369-400. [19] Jurica, M.S. and Moore, M.J. (2003) Pre-mRNA splicing, awash in a sea of proteins. Molecular Cell, 12, 5-14. http://dx.doi.org/10.1016/S1097-2765(03)00270-3 [20] Noller, H.F., Kop, J.A., Wheaton, V., Brosius, J., Gutell, R.R., Kopylov, A.M., Dohme, F., Herr, W., Stahl, D.A., Gupta, R. and Woese, C.R. (1981) Secondary structure model for 23S ribosomal RNA. Nucleic Acids Research, 9, 6167-6189. http://dx.doi.org/10.1093/nar/9.22.6167 [21] Vaughn, J.C., Sperbeck, S.J., Ramsey, W.J. and Lawrence, C.B. (1984) A universal model for the secondary structure of 5.8S ribosomal RNA molecules, their contact sites with 28S ribosomal RNAs, and their prokaryotic equiva- lent. Nucleic Acids Research, 12, 7479-7502. http://dx.doi.org/10.1093/nar/12.19.7479 [22] Orum, H., Nielsen, H. and Engberg, J. (1991) Spli- ceosomal small nuclear RNAs of Tetrahymena thermo- phila and some possible snRNA-snRNA base-pairing in- teractions. Journal of Molecular Biology, 222, 219-232. http://dx.doi.org/10.1016/0022-2836(91)90208-N [23] Hofmann, C.J.B., Marshallsay, C., Waibel, F. and Fili- powicz, W. (1992) Characterization of the genes encoding U4 small nuclear RNAs in Arabidopsis thaliana. Mo- lecular Biology Reports, 17, 21-28. http://dx.doi.org/10.1007/BF01006396 [24] Jakab, G., Mougin, A., Kis, M., Pollak, T., Antal, M., Branlant, C. and Solymosy, F. (1997) Chlamydomonas U2, U4 and U6 snRNAs. An evolutionary conserved pu- tative third interaction between U4 and U6 snRNAs which has a counterpart in the U4atac-U6atac snRNA du- plex. Biochimie, 79, 387-395. http://dx.doi.org/10.1016/S0300-9084(97)86148-2 [25] Hinas, A., Larsson, P., Avesson, L., Kirsebom, L.A., Vir- tanen, A. and Soderbom, F. (2006) Identification of the major spliceosomal RNAs in Dictyostelium discoideum reveals developmentally regulated U2 variants and poly- adenylated snRNAs. Eukaryot. Cell , 5, 924-934. http://dx.doi.org/10.1128/EC.00065-06 [26] Davis, C.A., Brown, M.P.S. and Singh, U. (2007) Func- tional characterization of spliceosomal introns and identi- fication of U2, U4, and U5 snRNAs in the deep-branch- ing eukaryote Entamoeba histolytica. Eukaryotic Cell, 6, 940-948. http://dx.doi.org/10.1128/EC.00059-07 [27] Gu, J., Chen, Y. and Reddy, R. (1998) Small RNA data- base. Nucl. Acids Res. 26, 160-162. http://dx.doi.org/10.1093/nar/26.1.160 [28] Stark, A., Lin, M.F., Kheradpour, P., et al. (2007) Discov- ery of functional elements in 12 Drosophila genomes us- ing evolutionary signatures. Nature, 450, 219-232. http://dx.doi.org/10.1038/nature06340 [29] Zuker, M. (2003) Mfold web server for nucleic acid fold- ing and hybridization prediction. Nucleic Acids Research, 31, 3406-3415. http://dx.doi.org/10.1093/nar/gkg595 [30] Rinke, J., Appel, B., Digweed, M. and Luhrmann, R. (1985) Localization of a base paired interaction between small nuclear RNAs U4 and U6 in intact U4/U6 ribonu- cleoprotein particles by psoralem cross-linking. Journal of Molecular Biology, 185, 721-731. http://dx.doi.org/10.1016/0022-2836(85)90057-9 [31] Steenkamp, E.T., Wright, J. and Baldauf, S.L. (2006). The protistan origins of animals and fungi. Molecular Biology and Evolution, 23, 93-106. http://dx.doi.org/10.1093/molbev/msj011 [32] Myslinski, E. and Branlant, C. (1991) A phylogenetic study of U4 snRNA reveals the existence of an evolu- tionarily conserved secondary structure corresponding to “free” U4 snRNA. Biochimie, 73, 17-28. http://dx.doi.org/10.1016/0300-9084(91)90069-D [33] Mitrovich, Q.M. and Guthrie, C. (2007) Evolution of small nuclear RNAs in S. cerevisiae, C. albicans, and other hemiascomycetous yeasts. RNA, 13, 2066-2080. http://dx.doi.org/10.1261/rna.766607 [34] Krol, A., Branlant, C., Lazar, E., Gallinaro, H. and Jacob, M. (1988) Primary and secondary structures of chicken, rat and man nuclear U4 RNAs. Homologies with U1 and U5 RNAs. Nucleic Acids Resear c h, 9, 2699-2716. http://dx.doi.org/10.1093/nar/9.12.2699 [35] Epstein, P., Reddy, R., Henning, D. and Busch, H. (1980). The nucleotide sequence of nuclear U6 (4.7S) RNA. Journal of Molecular Biology, 255, 8901-8906. [36] Fortner, D.M., Troy, R.G. and Brow, D.A. (1994) A stem/loop in U6 RNA defines a conformational switch required for pre-mRNA splicing. Genes & Development, 8, 221-233. http://dx.doi.org/10.1101/gad.8.2.221 [37] Mougin, A., Gottschalk, A., Fabrizio, P., Luhrmann, R. and Branlant, C. (2002) Direct probing of RNA structure and RNA-protein interactions in purified HeLa cell’s and yeast spliceosomal U4/U6.U5 tri-snRNP particles. Jour- nal of Molecular Biology, 317, 631-649. http://dx.doi.org/10.1006/jmbi.2002.5451 [38] Stark, H. and Luhrmann, R. (2006) Cryo-electron mi- croscopy of spliceosomal components. Annual Review of Biophysics and Biomolecular Structure, 35, 435-457. http://dx.doi.org/10.1146/annurev.biophys.35.040405.101 953 [39] McArthur, A.G., Morrison, H.G., Nixon, J.E.J., et al. (2000) The Giardia genome project database. FEMS Mi- crobiology Letters, 189, 271-273. http: //d x.doi.org/10. 1111/j.1574-6968.2000.tb09242.x [40] Carlton, J.M., Hirt, R.P., Silva, J.C., et al. (2007) Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science, 315, 207-212. http://dx.doi.org/10.1126/science.1132894 [41] Bonen, L. and Vogel, J. (2001) The ins and outs of group II introns. Trends in Genetics, 17, 322-331. http://dx.doi.org/10.1016/S0168-9525(01)02324-1 [42] Fedorova, O. and Zingler, N. (2007) Group II introns, structure, folding and splicing mechanism. The Journal of Biological Chemistry, 388, 665-678. http://dx.doi.org/10.1515/BC.2007.090 [43] Vanacova, S., Yan, W., Carlton, J.M. and Johnson, P.J. (2005) Spliceosomal introns in the deep-branching eu- karyote Trichomonas vaginalis. Proceedings of the Na- tional Academy of Sciences, 102, 4430-4435. http://dx.doi.org/10.1073/pnas.0407500102
|