Paper Menu >>
Journal Menu >>
 J. Biomedical Science and Engineering, 2009, 2, 123-127 Published Online April 2009 in SciRes. http://www.scirp.org/journal/jbise JBiSE 1 Bioinformatics analysis and characteristics of envelop glycoprotein E epitopes of dengue virus Hua Zhong1, Wei Zhao1, Liang Peng1, Shan-Feng Li1, Hong Cao1 1Department of Microbiology, School of Public Health and Tropical Medicine, Southern Medical University, Guangzhou, Guangdong 510515, China. Corre- spondence should be addressed to Hong Cao (gzhcao@fimmu.com), Tel.:+86 20 61648723. Received Sep. 4th, 2008; revised Dec. 22nd, 2008; accepted Jan. 8th, 2009 ABSTRACT The major envelope glycoprotein E of dengue (DEN) virus plays a central role in the biology of flaviviruses. It is capable of inducing a protective immune response in vivo and responsible for the viral binding to the cellular receptor. The crystal structures of glycoprotein E ectodomains have already been determined. However, it is still un- clear where the well-defined B-cell epitopes for glycoprotein E which induce the neutralizing an- tibodies locates. Thus, in order to characterize the role of glycoprotein E in the pathogenesis of dengue virus infection, we first used network servers (http://bio.dfci. harvard.edu/Tools/ & http://www. imtech. res. in) to predict and analyze the well defined B-cell and T-cell epitopes of the glycoprotein of the DEN-1 HAWAII strain. Then based on the highly conserved envelop glyco- protein amino acids, the hydrophilicity, antigenic- ity, accessibility and flexibility of envelop glyco- protein E were further predicted by using Biotic softwares (DNASTAR) and network servers (http://bio. dfci.harvard.edu/Tools/), the secondary structure was putatively obtained. In our study, the sequence at 281-295 amino acid (aa) for den- gue virus type 1 HAWAII strain and the sequence at 345-359, 383-397 for dengue virus type 2 NGC strain were predicted as the more prevalent epi- topes by using multiple parameters and different analysis softwares, respectively. Two epitopes of DEN-2 and one of DEN-1 locate on the domain Ш and domainⅡ of the protein E, respectively. Sub- sequently, further studies will be carried out to examine the antigenicity and protection of the synthetic peptides with higher scores in the av- erage antigen index (AI) and better hydrophilic properties determined by our data. Keywords: Dengue Virus, Glycoprotein E, Epi- tope, Bioinformatics 1. INTRODUCTION Dengue virus, a flavivirus belonging to the flaviviridae family, is a mosquito-borne human pathogen that causes dengue and dengue hemorrhagic fever which is currently one of the serious public health threats throughout the tropical and subtropical regions of the world [1]. Four serotypes of DEN virus have been identified (DEN-1, 2, 3 and 4), and each of these serotypes can infect humans and cause disease. This virus shares many characteristics with other flaviviruses, having a single-stranded RNA genome surrounded by an icosahedral scaffold and cov- ered by a lipid envelope. The complete virion is 50 nm in diameter and contains an 11-kb plus-sensed RNA genome that is composed of seven nonstructural (NS) protein genes and three structural protein genes, core (C, 100 amino acids), membrane (M, 75 amino acids), and envelope (E, 495 amino acids) [2,3]. The order of pro- teins encoded is 5’-CprM(M)-E-NS1-NS2A-NS2B- NS3-NS4A-NS4B-NS5-3’[4].The 495-amino-acid (aa) envelop (E) glycoprotein, one of the three structural proteins, is the principal component of the external sur- face of the virion [5], and it is responsible for a wide range of biological activities, including binding to host cell receptors, fusion to and entry into host cells, there- fore, this protein directly affects host range, cellular tro- pism, and, in part, the virulence of the virus [2,5]. Fur- thermore, the E protein also stimulates host immunity by inducing protective and neutralizing antibodies [6]. It is a main target and important antigen for vaccine devel- opment, and many attempts have been made to elucidate the structure-function relationships of the dengue virus glycoprotein E. The crystal structures of protein E ecto- domains have already been determined. However, the location of well-defined B-cell and T-cell epitopes for glycoprotein E is largely unknown. Mapping of the B-cell and T-cell epitopes should be important for im- munoinfectomic studies of dengue virus infection. Ran- dom peptide display has been applied in antigenic epi- tope determination. However, a combination of compu- tational methods (e.g., bioinformatics) and experimental approaches of conventional biology should be a holistic way to determine the rigorous B-cell and T-cell epitopes. Thus, in order to characterize the role of glycoprotein E in the pathogenesis of dengue virus infection, we used bioinformatics and molecular approaches to predict and analyze its B-cell and T-cell antigen epitopes. Parameters SciRes Copyright © 2009 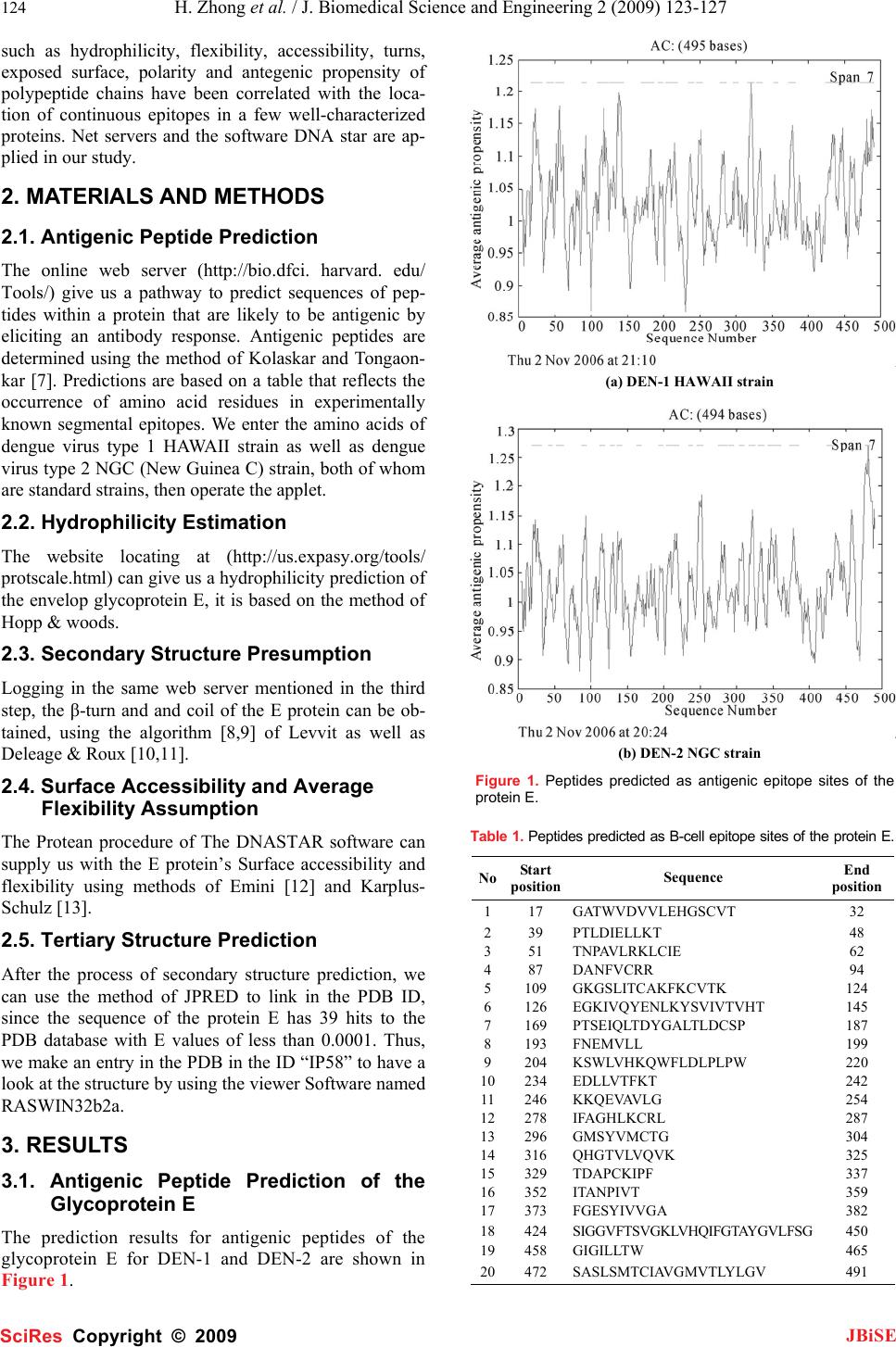 124 H. Zhong et al. / J. Biomedical Science and Engineering 2 (2009) 123-127 SciRes Copyright © 2009 JBiSE such as hydrophilicity, flexibility, accessibility, turns, exposed surface, polarity and antegenic propensity of polypeptide chains have been correlated with the loca- tion of continuous epitopes in a few well-characterized proteins. Net servers and the software DNA star are ap- plied in our study. 2. MATERIALS AND METHODS 2.1. Antigenic Peptide Prediction The online web server (http://bio.dfci. harvard. edu/ Tools/) give us a pathway to predict sequences of pep- tides within a protein that are likely to be antigenic by eliciting an antibody response. Antigenic peptides are determined using the method of Kolaskar and Tongaon- kar [7]. Predictions are based on a table that reflects the occurrence of amino acid residues in experimentally known segmental epitopes. We enter the amino acids of dengue virus type 1 HAWAII strain as well as dengue virus type 2 NGC (New Guinea C) strain, both of whom are standard strains, then operate the applet. 2.2. Hydrophilicity Estimation The website locating at (http://us.expasy.org/tools/ protscale.html) can give us a hydrophilicity prediction of the envelop glycoprotein E, it is based on the method of Hopp & woods. 2.3. Secondary Structure Presumption Logging in the same web server mentioned in the third step, the β-turn and and coil of the E protein can be ob- tained, using the algorithm [8,9] of Levvit as well as Deleage & Roux [10,11]. 2.4. Surface Accessibility and Average Flexibility Assumption The Protean procedure of The DNASTAR software can supply us with the E protein’s Surface accessibility and flexibility using methods of Emini [12] and Karplus- Schulz [13]. 2.5. Tertiary Structure Prediction After the process of secondary structure prediction, we can use the method of JPRED to link in the PDB ID, since the sequence of the protein E has 39 hits to the PDB database with E values of less than 0.0001. Thus, we make an entry in the PDB in the ID “IP58” to have a look at the structure by using the viewer Software named RASWIN32b2a. 3. RESULTS 3.1. Antigenic Peptide Prediction of the Glycoprotein E The prediction results for antigenic peptides of the glycoprotein E for DEN-1 and DEN-2 are shown in Figure 1. (a) DEN-1 HAWAII strain (b) DEN-2 NGC strain Figure 1. Peptides predicted as antigenic epitope sites of the protein E. Table 1. Peptides predicted as B-cell epitope sites of the protein E. No Start position Sequence End position 117 GATWVDVVLEHGSCVT 32 239 PTLDIELLKT 48 351 TNPAVLRKLCIE 62 487 DANFVCRR 94 5109 GKGSLITCAKFKCVTK 124 6126 EGKIVQYENLKYSVIVTVHT 145 7169 PTSEIQLTDYGALTLDCSP 187 8193 FNEMVLL 199 9204 KSWLVHKQWFLDLPLPW 220 10234 EDLLVTFKT 242 11246 KKQEVAVLG 254 12278 IFAGHLKCRL 287 13296 GMSYVMCTG 304 14316 QHGTVLVQVK 325 15329 TDAPCKIPF 337 16352 ITANPIVT 359 17373 FGESYIVVGA 382 18424 SIGGVFTSVGKLVHQIFGTAYGVLFSG 450 19458 GIGILLTW 465 20472 SASLSMTCIAVGMVTLYLGV 491  H. Zhong et al. / J. Biomedical Science and Engineering 2 (2009) 123-127 125 SciRes Copyright © 2009 JBiSE The B-cell (Table 1) and T-cell epitopes (Table 2) of the glycoprotein of DEN-1 HAWAII standard strain were predicted by the means of the position of amino acids with the online server respectively. 3.2. Hydrophilicity Prediction of the Glyco- protein E The prediction results for hydrophilicity of the protein E of DEN-1 and DEN-2 are diagramed in Figure 2. Table 2. Peptides predicted as T-cell epitope sites of the protein E. HLA Sites Peptides Position HLA-A2 206-215 117-126 483-492 HLA-A11 299-308 238-247 50-59 HLA-A24 298-307 439-446 325-334 HLA-B51 216-225 420-446 206-215 HLA-B60 48-57 313-322 256-265 HLA-B62 414-423 291-300 124-133 3.3. Secondary Structure Prediction The prediction of protein E’s secondary structure is shown in Figure 3. 3.4. Surface Probability of the Glycoprotein E The surface probability assumption of the protein E of DEN-1 and DEN-2 strains is diagramed in Figure 4. 3.5. Flexibility Presumption of the Glyco- protein E The Flexibility presumption of the glycoprotein E of DEN-1 and DEN-2 strains is shown in Figure 5. 3.6. Putative Tertiary Structure of the Gly- coprotein E The putative tertiary structure of the glycoprotein E is shown in Figure 6 (protein data bank). The ID is IP58. 4. DISCUSSION It is well known that although urgently needed for den- gue virus infection, specific drugs for treatment and ef- fective vaccination for prevention are currently unavail- able. This is the main reason why we have focused on the so-called Antibody-dependent enhancement (ADE). Studies suggest that during a secondary infection with a different serotype, the presence of cross-reactive, non-neutralizing antibodies enhances the efficiency with which dengue virus infects susceptible cells. A molecu- lar understanding of the events that lead to antibody neutralization, enhancement, or escape will be critical to the improvement of vaccines It is therefore important to determine which surface features on the dengue virion are responsible for inducing protective or enhancing immune response in the different serotypes. Thus, the structural and functional organizations of the dengue virus proteins are of central interests for the understand- ing of the biology of dengue virus and the mechanisms of virus-cell interactions. The dengue E ectodomain consists of structurally dis- tinct domains: Ⅰ, Ⅱ and Ⅲ [14]. The domain Ⅲ ap- pears to play an important part in host cell receptor binding for viral entry and in inducing protective immu- nity. The rigorous B-cell and T-cell epitopes were not identified yet. In our study, we focused on the character- izing the B-cell and T-cell epitopes of dengue virus en- velop E glycoprotein by deploying the bioinformatics approaches, the sequence at 281-295 amino acid (aa) for dengue virus type 1 HAWAII strain and the sequence at 345-359, 383-397 for dengue virus type 2 NGC strain were predicted as the more prevalent epitopes by using multiple parameters and different analysis softwares, respectively. The sequences selected not only have higher scores in the average antigen index (AI), which could predict the antigen epitope of envelop glycopro- tein E, but also showed better hydrophilic properties. Two epitopes of DEN-2 and one of DEN-1 locate on (a) DEN-1 HAWAII strain (b) DEN-2 NGC strain Figure 2. The hydrophilicity of the protein E. 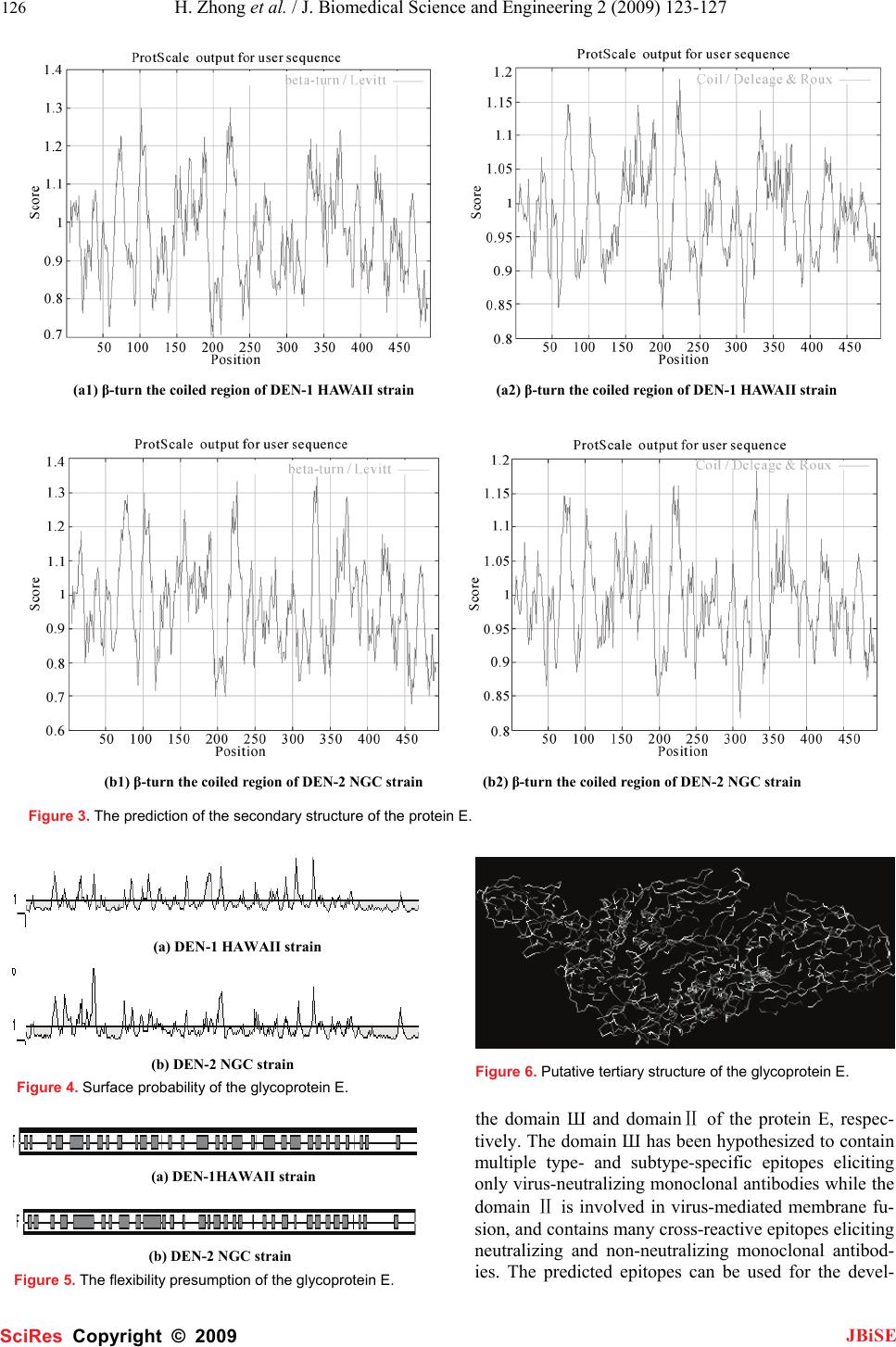 126 H. Zhong et al. / J. Biomedical Science and Engineering 2 (2009) 123-127 SciRes Copyright © 2009 JBiSE (a1) β-turn the coiled region of DEN-1 HAWAII strain (a2) β-turn the coiled region of DEN-1 HAWAII strain (b1) β-turn the coiled region of DEN-2 NGC strain (b2) β-turn the coiled region of DEN-2 NGC strain Figure 3. The prediction of the secondary structure of the protein E. (a) DEN-1 HAWAII strain (b) DEN-2 NGC strain Figure 4. Surface probability of the glycoprotein E. (a) DEN-1HAWAII strain (b) DEN-2 NGC strain Figure 5. The flexibility presumption of the glycoprotein E. Figure 6. Putative tertiary structure of the glycoprotein E. the domain Ш and domainⅡ of the protein E, respec- tively. The domain Ш has been hypothesized to contain multiple type- and subtype-specific epitopes eliciting only virus-neutralizing monoclonal antibodies while the domain Ⅱ is involved in virus-mediated membrane fu- sion, and contains many cross-reactive epitopes eliciting neutralizing and non-neutralizing monoclonal antibod- ies. The predicted epitopes can be used for the devel-  H. Zhong et al. / J. Biomedical Science and Engineering 2 (2009) 123-127 127 SciRes Copyright © 2009 JBiSE opment of vaccine and the dissection of the ADE effect. The further experimental studies will be performed to determine the immunogenicity and protection effect of peptides with higher scores in the average antigen index (AI) and better hydrophilic properties, and to identify vaccine candidates. ACKNOWLEDGEMENTS This work was supported by Guangzhou Key Technology R&D Pro- gram (No.2008Z1-E401 to H. Cao). REFERENCES [1] T. Monath.(1994) Dengue: the risk to developed and developing countries. Proc Natl Acad Sci USA 91, 2395-2400. [2] R. J. Kuhn, W. Zhang, M. G. Rossmann. (2002) Structure of dengue virus: implications for flavivirus organization, matu- -ration, and fusion. Cell 108, 717-725. [3] E. A. Henchal, J. R. Putnak. (1990) The dengue viruses. Clin Microbiol Rev 3, 376-396. [4] C. M. Rice, B. N. Fields, D. M. Knipe, P. M. Howley. (1996)Flaviviridae: the viruses and their replication. Virology 3, 931-959. [5] J. T. Roehrig. Immunochemistry of the dengue viruses, 199-219 In: D. J. Gubler and G. Kuno (eds), Dengue and dengue hemorrhagic fever. CAB International, New York, N. Y. 1997. [6] G. J. Chang, Molecular biology of dengue viruses, 175-198 In : D. J. Gubler, G. Kuno (eds.), Dengue and Dengue Hemorrhagic Fever. CAB International, London, 1997. [7] A. S. Kolaskar, P. C. Tongaonkar. (1990) A semi-empirical method for prediction of antigenic determinants on protein anti- gens, FEBS Lett 276, 172-174. [8] M. Levitt, J. Greer. (1977) Automatic identification of secondary structure in globular proteins. J Mol Biol 114, 181-239. [9] J Cheng. (2008) A multi-template combination algorithm for protein comparative modeling. BMC Structural Biology 8, 18-36. [10] G. Deleage, B. Roux. (1987) An algorithm for protein secondary structure prediction based on class prediction. Protein Eng 1, 289-294. [11] J. Martin, J. F. Gibrat, F. Rodolphe. (2006) Analysis of an opti- mal hidden Markov model for secondary structure prediction. BMC Structural Biology 6, 25-45. [12] E. A. Emini, J. V. Hughes, D. S. Peflow. (1985) Induction of hepa-titis A vires-neutralizing antibody by a vires-specific syn- thetic peptide. J Virol 55,836-839. [13] P. A. Karplus, G. Schultz. (1985) Prediction of chain flexibility in proteins. Naturwissenschaften 72, 212-213. [14] Y. Modis, S. Ogata, D. Clements. (2003) A ligand-binding pockets in the dengue virus envelop glycoprotein. Science 100, 6986-6991. |

