 Advances in Microbiology, 2013, 3, 442-449 http://dx.doi.org/10.4236/aim.2013.35060 Published Online September 2013 (http://www.scirp.org/journal/aim) Methods for Detection of Viruses in Water and Wastewater Alicja Hryniszyn1, Magdalena Skonieczna2, Jarosław Wiszniowski1 1Environmental Biotechnology Department, Silesian University of Technology, Gliwice, Poland 2Biosystems Group, Institute of Automatic Control, Silesian University of Technology, Gliwice, Poland Email: rie8@polsl.pl Received July 3, 2013; revised August 3, 2013; accepted August 13, 2013 Copyright © 2013 Alicja Hryniszyn et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. ABSTRACT Viruses present in water might be harmful for human health and life. Nowadays over 100 pathogenic human virus spe- cies occur in water polluted with sewage. Chlorination, which is the most popular disinfection method is not able to remove easily viruses from treated water. Due to this, it’s necessary to detect viruses in water before treatment in order to determine disinfectant dose and to ensure the sanitary safety level of treated water. The aim of this article is to review viruses detection methods as well as the problems related to implementation of those methods in analysis of water and wastewater samples. Keywords: Viruses; Analytical Problems; Environmental Samples; Detection Methods 1. Introduction Nowadays water is used for consuming, irrigation or rec- reation. Water quality, independently of its use is very important for human health both in developed countries and developing countries. World Health Organization (WHO) guidelines involving quality of drinking water emphasizes the need of clean, void of viruses and para- sites water, which is supplied to consumers. Polish legis- lation does not take into consideration detection of viruses in drinking water and sewage which are used in irriga- tion, and recreation reservoirs. Most common human dis- eases include acute inflammation of intestines and stom- ach. Among main viruses, which are responsible for dis- eases of the digestive system are rotaviruses (20% to 30% cases), adenoviruses (5% cases), human caliciviruses (5% to 10% cases) and astroviruses (5% cases) [1]. Some of diseases caused by enteroviruses might cause death. For example, among Hepatitis E Virus infected women’s mortality rate is 20% [2]. Other diseases can lead to se- rious complications such as inflammation of the heart muscle, inflammation of meninges, inflammation of brain, respiratory disorder and acute liver failure [2-4]. Basic source of enteroviruses is feces from infected people (viruses are excreted in big amounts, account from 105 to 1011 virus particles per gram of stool). Excreta gets to sewage, solid waste or they can create with rainfall water or meltwater surface outflow. Viruses can infiltrate from surface outflow to surface water. Enteroviruses get to human organism as a result of infected crustaceans con- sumption, infected crop and contaminated drinking water consumption, polluted air inhalation (aerosoles) or bath- ing in contaminated water reservoirs. The most effective virus removal method from water or wastewater is ozonation. Application of concentration of residual ozone amount to 0.51 mg/l and contact time amount 2 minutes (in temperature 20˚C, with pH 7.2) enables Polio 1 virus inactivation [5]. Nevertheless, the most popular disinfec- tion method is water chlorination. The fact that water or sewage chlorination enables bacteria removal is not equivalent with the fact that it can eliminate viruses. For example, in order to poliovirus total inactivation in mu- nicipal wastewater, the concentration of residual chlorine should be 9.0 mg/l (contact time should be 30 minutes) but for proper sewage disinfection in case of bacteria Salmonella lower concentration of residual chlorine is required (from 1.0 to 2.0 mg/l) [6]. Therefore, in order to ensure drinking water/waste- water sanitary safety, appropriate disinfectant dose and contact time selection in treated medium, are very im- portant analytical techniques and methods, which enable detection of viruses. Detection of viruses in sewage and drinking water requests detection methods which are: sensitive, resistant for false-positive results and enabling full automation. Moreover applied methods should be C opyright © 2013 SciRes. AiM  A. HRYNISZYN ET AL. 443 fast and inexpensive. Method, which fulfils all needed re- quirements, as yet was not worked out. 2. Virus Detection Methods in Water Environment Virus detection methods are applied for many years in medical diagnosis. Some of these methods were modified in order to be utilized in analysis of environmental sam- ples. For analysis of environmental samples were adapted following methods: Polymerase Chain-Reaction (PCR) [7-12], Nucleic Acid Sequence—Based Amplification (NASBA) [13-15], microarray technique [16,17], Atomic Force Microscope (AFM) [16], fluorescent microscopy [18-23], electron microscopy [18,23-26], application of biosensors [27,28], Enzyme Linked Immunosorbent As- say (ELISA) [29-32] and flow cytometry [33-36]. Some of these methods were modified by: 1) concentration (ELISA tests, PCR and NASBA reactions, microarrays application) [7,17,29], 2) different methods combining (PCR reaction combines with plaque forming tests, ato- mic force microscopy combines with protein microarray technology) [37-39], 3) change of filter pore size (epif- luorescence microscopy) [23], 4) dilution of sample (flow cytometry) [16]. Application of Chosen Methods for Virus Detection in Water Environment As mentioned before, for viruses detection in water en- vironment combined methods can be used. For example, PCR reaction can be combined with plaque forming tests. Polymerase chain reaction is used to amplification of specific deoxyribonucleic acid (DNA) sequence. In this reaction double stranded DNA, which is called template DNA is amplified. During several years PCR technique, due to its high specificity, was adopted to detection of enteroviruses and Hepatitis A virus (HAV) in environ- ment [40-43]. However, this technique has got a lot of disadvantages. The main of them is low sensitivity. Al- though PCR reaction is very sensitive (it enables detec- tion of a single virus), it still does not fulfill standards which are determined for virus detection. Even if viruses which are present in water are concentrated, volume of environmental sample is still too large for PCR reaction. Secondly, it does not enable univocal determination if detected virus is infectious or even not if result is posi- tive. Moreover, PCR reaction enables only detection of one type of virus at one time and in water exist many types of them [7]. Viruses are capable to forming plaques on bacterial lawn or cell layers and cause their morphological alterna- tions which can be observed by light microscope [44,45]. Nevertheless, the plaque forming test is time consuming and poor in specificity [7]. PCR reaction combined with plaque forming tests was developed to simultaneous de- tection of Poliovirus, Coxsackie virus, Echovirus and Hepatitis A virus in water [7]. Water samples were in- oculated with mixture containing Poliovirus type, strain Lsc, CoxB3 or Echovirus. Contrary to PCR reaction used in medical diagnosis both samples, inoculated and uni- noculated, were concentrated by electropositive filter. Afterwards, recovery of viruses was determined and short-term culture was set. Thereafter ribonucleic acid (RNA) was extracted and purified. Viral nucleic acid was detected by multiplex-PCR and than identified by semi- nested PCR [7]. Semi-nested PCR is a kind of nested PCR. Nested PCR, consist of carrying out PCR reaction, then repetition of PCR with second pair of starters, which are located closer to center of amplified DNA fragment. When in repetition of PCR reaction only one starter which was used before is changed, then this method is called semi-nested PCR [46]. For detection of viruses in water environment also Nucleic Acid Sequence Based Amplification (NASBA) is used. NASBA technique is used for RNA detection. It abuses three enzymes: T7 RNA polymerase, reverse tran- scriptase and RNase H which enable amplification of one stranded template RNA. NASBA was implicated to de- terminate concentration of viruses in wastewater from wastewater treatment in Saint-Nicolas in Canada. Sam- ples were collected from: raw waste water, waste water after aerobic digestion with activated sludge and waste water treated by UV. The samples were concentrated be- fore analysis (precipitation of viruses from solution with the aid of 8% polyethylenoglycol 600 and centrifugation) [15]. In order to estimate influence of matrix before vi- ruses detection waste water samples were analyzed for total bacteria and fecal coliforms. Afterwards, samples were inoculated with HAV (106 PFU/ml). Then each 5 µl of sample were heated to 100˚C (lysis of viral cells) and were analyzed by NASBA. Analysis of amplification product was subjected to dot-blot hybridization [15]. Al- though significantly bacterial contamination of raw waste water (2 × 105 fecal coliforms/ml) results of analysis showed strong specific to HAV signal. This indicates that presence of bacteria do not influence on results of analy- sis. Detection limit of this method for Hepatitis A Virus is 4 × 102 PFU/ml. On the other hand, bacterial presence in the sample might negatively influence on results of enzymatic test ELISA. In enzyme-linked immunosorbent assay is a test where ligand binds covalently to peroxi- dase. During the test ligand binds with antibody of inter- est. Unbound ligand is washed out and bound ligand in examined by addition of substrate, which are converted into colored products, as a result of enzyme activity [47]. Very important agent, which influences on sensitivity of ELISA tests is antibody affinity to antigen of interest. However, use of high affinity antibodies could be ineffi- Copyright © 2013 SciRes. AiM  A. HRYNISZYN ET AL. 444 cient if recognizable epitopes are hidden in protein structure, or if they do not have proper to recognize its conformation [48]. Main advantage of ELISA compared with other methods is its cost-effectiveness and the fact that it is relatively simple [49]. ELISA was used for de- tection of rotavirus antigen in water samples (two sam- ples of tap water, 4 samples of drinking water and 26 samples of water for domestic use) [29]. Firstly tem- perature and pH of samples were measured. Then sam- ples were transported in containers with ice to laboratory. There residual free chlorine was neutralized by addition of sodium thiosulfate solution. The sample were adjusted to pH 3.5 (1 N HCl and 0.0015 N AlCl3) and gently mixed at room temperature. The samples were filtered with aid of negatively charged filter. Filter was washed by 0.14 NaCl, pH 3.5 in order to remove excess of Al3+ ions. Then the viruses were eluted with aid of 2.9% tryptose phosphate broth (TPB), containing 6% glycine, pH 9.0, neutralized by 4 N HCl and stored at 4˚C. There- after elute was centrifugated. To samples was added mix- ture of streptomycin and penicillin (to reduce bacterial influence), Hanks salt solution and broth. The samples were adjusted to pH 7.4 by 4 N HCl, vigorously mixed and stored at −80˚C. Prepared samples were subjected to ELISA, where 96 well plates were coated by specific polyclonal antibodies. Sampled water was put to wells and was incubated with horseradish peroxidase conju- gated to specific polyclonal antibody (1 h). The wells were washed five times with Tris-buffered solution. Then buffer containing tetramethylbenzidine (TMB) and hy- drogen peroxide were added. Content of wells was mixed and samples were incubated for 10 minutes, at room temperature. At the end sulfuric acid was added to wells and absorbance of product was measured. In the same experiment 5 liters of tap water was inoculated with bo- vine rotavirus (5.7 × 104 IFU), concentrated and sub- jected to ELISA. In experiment was also used standard addition method. In this method two analysis were set simultaneously—analysis of sample of interest and ana- lysis of the same sample with addition of definite amount of standard, which is also determined substance [29]. Promising method which may be used in the near future is device which is called laboratory-on-a chip (LOC). It is a kind of biosensor. Biosensors are devices, which can responds to certain properties of analyte and convert these responses into detectable signal, the most common kind of signal is electrical [50]. LOC technology does not require manual samples preparation and measurements using sample signal detection (optical or electrochemical) do not require special laboratory environmental (outdoor analysis are enable). Moreover, lab-on-chip enables bio- chemical reactions (immunological reactions, enzymatic reactions and DNA determination). Chip is constructed with elements as: wells, channels, electrodes and filters [51]. Wells are storages of buffer, samples and waste. On it samples are directly proceed, PCR reaction is set and DNA of interest is detected. Wells and reaction chambers are connected by micro-channels. In turn, micropumps, microvalves and heaters are used to preparation of sam- ple and/or separation of reagents, agitation, incubation and separation of different biochemical reactions and physical processes [27]. Lab-on-chip technology is fast and sensitive to detect microbiological pathogens in en- vironmental samples. There are several approaches to use it in environmental microbiology. The simplest approach is placing on miniature chip a termocycler. Chip com- bines cell capturing with lyses, amplification of biologi- cal material (PCR), detection of pathogens in real time and analysis of data [27]. Comparison of described me- thods is presented in Table 1. Implementation of virus detection methods, for virus detection in water environment aside from economical agents, can come to grip with several impediments which include: considerable dilution of sample, influence of environmental matrix on analysis results and mutagenic variability of viruses. In water contaminated with faeces viruses are present in relatively small amounts (20 vi- ruses in 100 ml of ground water) [55]. Therefore often concentration of samples before exact determination of viruses content is needed. To this end there were worked out series of methods which include: concentration by absorption—elution technique, with the aid of negatively charged filter (this is a method which is used in enzy- matic test ELISA) [29], application of positively charged filter (in case of methods based on molecular biology) [7], precipitation of viruses from solution with the aid of 8% polyethylenoglycol 600 and centrifugation (in case of Nucleic Acid Sequence—Based Amplification—NASBA) [15]. In analyzed environmental samples (water) apart from viruses, there are also other substances (suspensions, humic acids, detritus or other microorganisms e.g. bacte- ria). Humic acids are important factors which inhibits PCR reaction. Although the PCR reaction is very sensi- tive (it enables detection of single virus), by the reason of inhibitors presence its sensitivity does not fulfill re- quirements which are define for detection of viruses which are present in water [37]. Suspension and detritus presence impedes counting of viruses with the aid of methods based on direct observation, such as transmis- sion electron microscopy [23,56]. On the other hand bacterial presence in the sample might negatively influ- ence on results of enzymatic test ELISA. In this case, in order to reduce bacterial influence on results of determi- nation inhibitors of growth and cell division should be added during water samples preparation [29]. Another impediment during virus detection in envi- ronmental samples is mutagenic variability. By reason of igh mutation rate (for instance, mutation rate of rotavi- h Copyright © 2013 SciRes. AiM 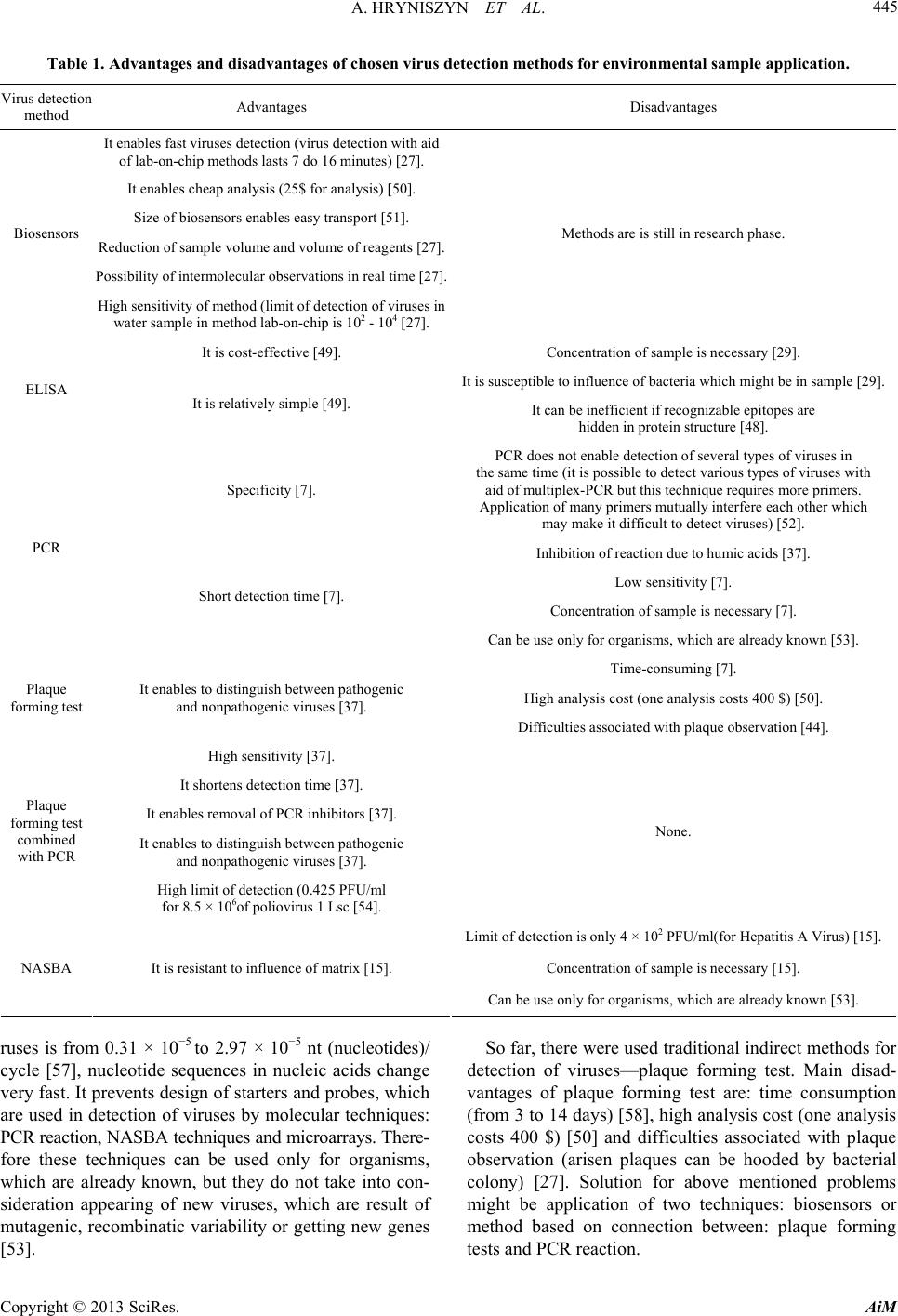 A. HRYNISZYN ET AL. Copyright © 2013 SciRes. AiM 445 Table 1. Advantages and disadvantages of chosen virus detection methods for environmental sample application. Virus detection method Advantages Disadvantages It enables fast viruses detection (virus detection with aid of lab-on-chip methods lasts 7 do 16 minutes) [27]. It enables cheap analysis (25$ for analysis) [50]. Size of biosensors enables easy transport [51]. Reduction of sample volume and volume of reagents [27]. Possibility of intermolecular observations in real time [27]. Biosensors High sensitivity of method (limit of detection of viruses in water sample in method lab-on-chip is 102 - 104 [27]. Methods are is still in research phase. It is cost-effective [49]. Concentration of sample is necessary [29]. It is susceptible to influence of bacteria which might be in sample [29]. ELISA It is relatively simple [49]. It can be inefficient if recognizable epitopes are hidden in protein structure [48]. Specificity [7]. PCR does not enable detection of several types of viruses in the same time (it is possible to detect various types of viruses with aid of multiplex-PCR but this technique requires more primers. Application of many primers mutually interfere each other which may make it difficult to detect viruses) [52]. Inhibition of reaction due to humic acids [37]. Low sensitivity [7]. Concentration of sample is necessary [7]. PCR Short detection time [7]. Can be use only for organisms, which are already known [53]. Time-consuming [7]. High analysis cost (one analysis costs 400 $) [50]. Plaque forming test It enables to distinguish between pathogenic and nonpathogenic viruses [37]. Difficulties associated with plaque observation [44]. High sensitivity [37]. It shortens detection time [37]. It enables removal of PCR inhibitors [37]. It enables to distinguish between pathogenic and nonpathogenic viruses [37]. Plaque forming test combined with PCR High limit of detection (0.425 PFU/ml for 8.5 × 106of poliovirus 1 Lsc [54]. None. Limit of detection is only 4 × 102 PFU/ml(for Hepatitis A Virus) [15]. Concentration of sample is necessary [15]. NASBA It is resistant to influence of matrix [15]. Can be use only for organisms, which are already known [53]. ruses is from 0.31 × 10−5 to 2.97 × 10−5 nt (nucleotides)/ cycle [57], nucleotide sequences in nucleic acids change very fast. It prevents design of starters and probes, which are used in detection of viruses by molecular techniques: PCR reaction, NASBA techniques and microarrays. There- fore these techniques can be used only for organisms, which are already known, but they do not take into con- sideration appearing of new viruses, which are result of mutagenic, recombinatic variability or getting new genes [53]. So far, there were used traditional indirect methods for detection of viruses—plaque forming test. Main disad- vantages of plaque forming test are: time consumption (from 3 to 14 days) [58], high analysis cost (one analysis costs 400 $) [50] and difficulties associated with plaque observation (arisen plaques can be hooded by bacterial colony) [27]. Solution for above mentioned problems might be application of two techniques: biosensors or method based on connection between: plaque forming tests and PCR reaction.  A. HRYNISZYN ET AL. 446 Application of biosensors enables detection of viruses during 7 to 16 minutes [27]. In addition this technique is very sensitive (limit of detection of this method is 102 to 104 cells/ml) and enables observation of intermolecular interactions in real time [27,59]. Moreover analysis of environmental samples with the aid of biosensors are relatively cheap (25 $ for water sample per analysis) [50]. However among advantages of second method, men- tioned above, is high sensitivity (for example in semi- nested PCR sensitivity for Poliovirus is 2.4 PFU/ml), possibility of distinguishing between pathogenic and non-pathogenic viruses [37], and ability to remove in- hibitors of PCR reaction (e.g. humic acids), which are present in primary matrix of sample. Both of mentioned methods can shorten time of virus detection in water samples. Plaque froming tests connected with PCR reaction en- able detection of viruses present in water in time of 2 to 4 days [37]. In this method detection time is shorter than detection time in traditional plaque forming test, within shorter incubation period. Techniques based on biosen- sors in terms of time saving are much better [27]. 3. Summary and Discussion Considering that, monitoring of viruses present in treated and untreated water is necessary (guidelines of World Health Organization (WHO), associated with quality of drinking water), analysis of water samples is needful. After review, is considered that traditional methods of viruses detection, such as plaque forming tests, mainly due to individual cost of analysis, are not popular in monitoring of water environment [50]. Also immunolo- gical tests and techniques, based on direct observation of viruses, become less applicable than techniques based on molecular biology. Techniques based on direct observa- tion of viruses did not enable detection of viruses, which are present in environment in concentrations above 106/ml (samples are concentrated, what results in high concentration of contaminants on microscope measured grid surface). An example is transmission electron mi- croscopy [23,56]. In addition, phenomenon of autofluo- rescence, which occurs in epifluorescence microscopy, does not make method based on direct observation of viruses attractive. However, enzymatic tests, such ELISA, are fallible, if recognizable epitopes are hidden in protein structure, or if they do not have proper to recognize its conformation. Nowadays, techniques based on molecular biology are modified by connecting plaque forming tests with PCR, or are replace by biosensors. This eventuate from high mutagenic variability, which makes starters and probes designing difficult and complexity of matrix, in which may occur substances inhibiting PCR reactions and NASBA. 4. Conclusions Discussed techniques, for viruses detection, are used on a large scale in medical diagnosis, but are less popular in environmental analysis of water and sewage samples. The reason of this fact is that during detection of viruses it occurs numerous impediments, which include: high dilution of samples, significant matrix influence on re- sults/reliability of analysis and mutagenic variability of viruses. After review, it can be claimed that most appli- cable techniques can be: connection of cell cultures (pla- que forming tests) with molecular biology techniques (PCR) and biosensors. However the most optimal solu- tion seems to be biosensors application due to time sav- ing and unit prize of analysis. Nowadays they are appli- cative on small-scale as systems which are used to run PCR reaction on material from environmental samples, because they are still in research phases [27]. Both in Poland and highly developed countries’ law, the main emphasis is putting on health safety of drinking water in terms of content of pathogenic bacteria and parasites. Formal constraints involving monitoring of viruses do not exist. We should pay attention to the fact that applied disinfectant is not always able to deactivate enteroviruses present in treated water. Researches have shown that in the United States of America every year the treatment of diseases caused by enteroviruses costs 5 to 6 milliard dollars [50]. Implementation of methods which enables monitoring of viruses present in water and food beyond fairly economical issues (charges for treat- ment) has arguably its social overtone. To date, no guidelines/standard methods, which enable monitoring of viruses presence in environmental samples have been worked out. 5. Acknowledgements This work was supported by grant BKM/514/RAU-1/ 2013 t. 26 from Silesian University of Technology in Gliwice, Poland. REFERENCES [1] U. Desselberger and J. Gray, “Viral Gastroenteritis: Causes, Pathophysiology, Immunology, Treatment and Epidemi- ology,” In: U. Desselberger and J. Gray, Eds., Viral Gas- troenteritis, Elsevier Science B.V., AE Amsterdam, 2003, pp. 1-8. [2] G. Virella, “Mikrobiologia i Choroby Zakaźne,” 3rd Edi- tion, Wydawnictwo Urban & Partner, Wrocław, 2000. [3] A. Bosh, “Human Enteric Viruses in the Water Environ- ment: A Mini Review,” International Microbiology, Vol. 1, No. 3, 1998, pp. 191-196. [4] W. Irving, D. Ala’Aldeen and T. Boswell, “Mikrobiolo- gia Medyczna. Krótkie Wykłady,” Wydawnictwo Nau- kowe PWN, Warszawa, 2008. Copyright © 2013 SciRes. AiM  A. HRYNISZYN ET AL. 447 [5] D. Roy, P. K. Wong, R. S. Engelbrecht and E. S. Chian, “Mechanism of Enteroviral Inactivation by Ozone,” Ap- plied Environmental Microbiology, Vol. 41, No. 3, 1981, pp. 718-723. [6] Z. Dymaczewski, J. A. Oleszkiewicz and M. M. Sozański, “Poradnik Eksploatatora Oczyszczalni Ścieków,” 2nd Edition, Wydawnictwo PZITS, Poznań, 1997. [7] W. Li, X. Wang, C. Q. Yuan, J. L. Zheng, M. Jin, N. Song, et al., “Detection of Enteroviruses and Hepatitis a Virus in Water by Consensus Primer Multiplex RT- PCR,” Word Journal of Gastroenterology, Vol. 8, No. 4, 2002, pp. 699-702. [8] S. Pina, M. Pulig, F. Lucena, J. Jofre and R. Girones, “Viral Pollution in the Environment and Shellfish: Hu- man Adenovirus Detection by PCR as an Index of Human Viruses,” Applied and Environmental Microbiology, Vol. 64, No. 9, 1998, pp. 3376-3382. [9] S. H. Lee, C. Lee, K. W. Lee, H. B. Cho and S.-J. Kim, “The Simultaneous Detection of Both Enteroviruses and Adenoviruses in Environmental Water Samples Including Tap Water with an Integrated Cell Culture-Multiplex- Nested-PCR Procedure,” Journal of Applied Microbiol- ogy, Vol. 98, No. 5, 2005, pp. 1020-1029. doi:10.1111/j.1365-2672.2004.02496.x [10] H. B. Cho, S. H. Lee, J. C. Cho and S. J. Kim, “Detection of Adenoviruses and Enteroviruses in Tap Water and River Water by Reverse Transcription Multiplex PCR,” Canadian Journal of Microbiology, Vol. 46, No. 5, 2000, pp. 417-424. doi:10.1139/w00-014 [11] G. S. Fout, B. C. Martinson, M. W. N. Moyer and D. R. Dahling, “A Multiplex Reverse Transcription-PCR Me- thod for Detection of Human Enteric Viruses in Ground- water,” Applied and Environmental Microbiology, Vol. 69, No. 6, 2003, pp. 3158-3164. doi:10.1128/AEM.69.6.3158-3164.2003 [12] E. Schvoerer, F. Bonnet, V. Dubois, G. Cazaux, R. Ser- ceau, H. J. A. Fleury, et al., “PCR Detection of Human Enteric Viruses in Bathing Areas, Waste Waters and Hu- man Stools in Southwestern France,” Research in Micro- biology, Vol. 151, No. 8, 2000, pp. 693-701. doi:10.1016/S0923-2508(00)90132-3 [13] S. A. Rutjes, H. H. J. L. van den Berg, W. J. Lodder and A. M. de Roda Husman, “Real-Time Detection of Noro- viruses in Surface Water by Use of a Broadly Reactive Nucleic Acid Sequence-Based Amplification Assay,” Ap- plied and Environmental Microbiology, Vol. 72, No. 8, 2006, pp. 5349-5358. doi:10.1128/AEM.00751-06 [14] J. Jean, B. Blais, A. Darvau and I. Fliss, “Simultaneous Detection and Identification of Hepatitis A Virus and Ro- tavirus by Multiplex Nucleic Acid Sequence-Based Am- plification (NASBA) and Microtiter Plate Hybridization System,” Journal of Virological Methods, Vol. 105, No. 1, 2002, pp. 123-132. doi:10.1016/S0166-0934(02)00096-4 [15] J. Jean, B. Blais and I. Fliss, “Detection of Hepatitis A Virus by the Nucleic Acid Sequence-Based Amplification Technique and Comparison with Reverse Transcription- PCR,” Applied and Environmental Microbiology, Vol. 67, No. 12, 2001, pp. 5593-5600. doi:10.1128/AEM.67.12.5593-5600.2001 [16] S. R. Nettikadan, J. C. Johnson, C. Mosher and E. Hen- derson, “Virus Particle Detection by Solid Phase Immu- nocapture and Atomic Force Microscopy,” Biochemical and Biophysical Research Communications, Vol. 311, No. 2, 2003, pp. 540-545. doi:10.1016/j.bbrc.2003.10.022 [17] F. S. Alhamlan, M. M. Ederer, T. L. Green, C. K. Brink- man, E. R. Coats and R. L. Crawford, “A Novel Screen- ing Tool Using Microarray and PCR to Detect Pathogens in Agricultural Impacted Waters,” International Journal of Environmental Protection, Vol. 3, No. 3, 2013, pp. 17- 32. [18] S. H. Hara, K. Terauchi and I. Koike, “Abundance of Vi- ruses in Marine Waters: Assessment by Epifluorescence and Transmission Electron Microscopy,” Applied and En- vironmental Microbiology, Vol. 57, No. 9, 1991, pp. 2731-2734. [19] K. P. Henneses and C. A. Suttle, “Direct Counts of Vi- ruses in Natural Waters and Laboratory Cultures by Epif- luorescence Microscopy,” Limnology and Oceanography, Vol. 40, No. 6, 1995, pp. 1050-1055. doi:10.4319/lo.1995.40.6.1050 [20] R. T. Noble and J. A. Fuhrman, “Use of SYBR Green I for Rapid Epifluorescence Counts of Marine Viruses and Bacteria,” Aquatic Microbial Ecology, Vol. 14, No. 2, 1998, pp. 113-118. doi:10.3354/ame014113 [21] A. Shibata, Y. Goto, H. Saito, T. Kikuchi, T. Toda and S. Taguchi, “Comparison of SYBR Green I and SYBR Gold Stains for Enumerating Bacteria and Viruses by Epifluo- rescence Microscopy,” Aquatic Microbial Ecology, Vol. 43, No. 3, 2006, pp. 221-231. [22] K. Wen, A. C. Ortmann and C. A. Suttle, “Accurate Es- timation of Viral Abundance by Epifluorescence Micros- copy,” Applied and Environmental Microbiology, Vol. 70, No. 7, 2004, pp. 3862-3867. doi:10.1128/AEM.70.7.3862-3867.2004 [23] M. G. Weinbauer and C. A. Suttle, “Comparison of Epif- luorescence and Transmission Electron Microscopy for Counting Viruses in Natural Marine Waters,” Aquatic Microbial Ecology, Vol. 13, No. 3, 1997, pp. 225-232. doi:10.3354/ame013225 [24] Y. Bettarel, T. Sime-Ngando, C. Amblard and H. Laveran, “A Comparison of Methods for Counting Viruses in Aquatic Systems,” Applied and Environmental Microbi- ology, Vol. 66, No. 6, 2000, pp. 2283-2289. doi:10.1128/AEM.66.6.2283-2289.2000 [25] M. C. Alonso, J. Rodríguez and J. J. Borrego, “Enumera- tion and Isolation of Viral Particles from Oligotrophic Marine Environments by Tangential Flow Filtration,” In- ternational Microbiology, Vol. 2, No. 4, 1999, pp. 227- 232. [26] K. E. Wommack, R. T. Hill, M. Kessel, E. Russek-Cohen and R. R. Colwell, “Distribution of Viruses in the Chesa- peake Bay,” Applied and Environmental Microbiology, Vol. 58, No. 9, 1992, pp. 2965-2970. [27] W. T. Liu and L. Zhu, “Environmental Microbiology-on- Chip and Its Future Impacts,” Trends in Biotechnology, Vol. 23, No. 4, 2005, pp. 174-178. doi:10.1016/j.tibtech.2005.02.004 [28] O. V. Rengevych, Y. M. Shirshov, Y. V. Ushenin and G. Copyright © 2013 SciRes. AiM  A. HRYNISZYN ET AL. 448 V. Bektov, “Separate Determination of Thickness and Optical Parameters by Surface Plasmon Resonance: Ac- curacy Consideration,” Semiconductor Physics, Quantum Electronics & Optoelectronics, Vol. 2, No. 2, 1999, pp. 28-35. [29] L. Kittigul, B. Raengsakulrach,S. Siritanikorn, R. Kanyok, P. Diraphat, V. Thirawutch, et al., “Detection of Poliovi- rus, Hepatitis A Virus and Rotavirus from Sewage and Water Samples,” The Southeast Asian Journal Health of Tropical Medicine and Public Health, Vol. 31, No. 1, 2000, pp. 41-45. [30] A. M. Nasser and T. G. Metcalf, “An A-ELISA to Detect Hepatitis A Virus in Estuarine Samples,” Applied and En- vironmental Microbiology, Vol. 53, No. 5, 1987, pp. 1192-1195. [31] N. A. El-Esnaw, “Examination for Hepatitis E Virus in Wastewater Treatment Plants and Workers by Nested RT-PCR and ELISA,” The Journal of the Egyptian Public Health Association, Vol. 75, No. 1-2, 2000, pp. 219-231. [32] M. K. Nishida, V. L. A. Ruiz and F. Gregori, “Detection of Rotavirus from Pig Livestock Wastewater of São Paulo State, Brazil,” Ars Veterinaria, Vol. 25, No. 3, 2009, pp. 136-141. [33] D Marie, C. P. D. Brussard, R. Thyrhaug and G. Bratbak, “Enumeration of Marine Viruses in Culture and Natural Samples by Flow Cytometry,” Applied and Environ- mental Microbiology, Vol. 65, No. 1, 1999, pp. 45-52. [34] F. X. Abad, R. M. Pinto and A. Bosch, “Flow Cytometry Detection of Infectious Rotaviruses in Environmental and Clinical Samples,” Applied and Environmental Micro- biology, Vol. 64, No. 7, 1998, pp. 2392-2396. [35] C. P. D. Brussaard, “Optimization of Procedures for Counting Viruses by Flow Cytometry,” Applied and En- vironmental Microbiology, Vol. 70, No. 3, 2004, pp. 1506-1513. doi:10.1128/AEM.70.3.1506-1513.2004 [36] F. Chen, J. R. Lu, B. J. Binder, Y. C. Liu and R. E. Hod- son, “Application of Digital Image Analysis and Flow Cytometry to Enumerate Marine Viruses Stained with SYBR Gold,” Applied and Environmental Microbiology, Vol. 67, No. 2, 2001, pp. 539-545. doi:10.1128/AEM.67.2.539-545.2001 [37] T. Straub, J. L. Pepper and C. P. Gerba, “Comparison of PCR and Cell Culture for Detection of Enteroviruses in Sludge of Their Transport,” Applied and Environmental Microbiology, Vol. 61, No. 5, 1995, pp. 2066-2068. [38] B. B. Haab, M. J. Dunham and P. O. Brown, “Protein Microarray for Highly Parallel Detection and Quantifica- tion of Specific Proteins and Antibodies in Complex So- lutions,” Genome Biology, Vol. 2, No. 2, 2000, pp. 1-13. [39] H. Zhu and M. Snyder, “Protein Arrays and Microarrays,” Current Opinion in Chemical Biology, Vol. 5, No. 1, 2001, pp. 40-45. doi:10.1016/S1367-5931(00)00170-8 [40] D. Egger, L. Pasamontes, M. Ostermayer and K. Bienz, “Reverse Transcription Multiplex PCR for Differentiation between Polio and Enteroviruses from Clinical and En- vironmental Samples,” Journal of Clinical Microbiology, Vol. 33, No. 6, 1995, pp. 1442-1447. [41] K. A. Reynolds, C. P. Gerba and J. L. Pepper, “Detection of Infectious Enteroviruses by an Integrated Cell Culture- PCR Procedure,” Applied and Environmental Microbiol- ogy, Vol. 62, No. 4, 1996, pp. 1424-1427. [42] F. L. Guyader, E. Dubois, D. Menard and M. Pommepuy, “Detection of Hepatitis A Virus, Rotavirus and Enterovi- rus in Naturally Contaminated Shellfish and Sediment by Reverse Transcription-Seminested PCR,” Applied and Environmental Microbiology, Vol. 60, No. 10, 1994, pp. 3665-3671. [43] C. Gantzer, S. Senouci, A. Maul, Y. Levi and L. Schwarz- bold, “Enteroviruses Genomes in Wastewater: Concentra- tion on Glass Wool and Glass Powder and Detection by RT-PCR,” Journal of Virological Methods, Vol. 65, No. 2, 1997, pp. 265-271. doi:10.1016/S0166-0934(97)02193-9 [44] J. M. Łoś, P. Golec, G. Węgrzyn, A. Węgrzyn and M. Łoś, “Simple Method for Plating Escherichia coli Bacte- riophage Forming Very Small Plaques or No Plaques un- der Standard Conditions,” Applied and Environmental Microbiology, Vol. 74, No. 16, 2008, pp. 5113-5120. doi:10.1128/AEM.00306-08 [45] J. C. Espinoza and J. Kuznar, “Rapid Simultanous Detec- tion and Quantitation of Infectious Pancreatic Necrosis Virus (IPNV),” Journal of Virological Methods, Vol. 105, No. 1, 2002, pp. 81-85. doi:10.1016/S0166-0934(02)00083-6 [46] A. Raszka, A. Ziembińska and A. Wiechetek, “Metody i Techniki Biologii Molekularnej w Biotechnologii Środo- wiskowej,” Technical Transactions. Environmental Engi- neering, Vol. 2, No. 2, 2009, pp. 101-114. [47] P. M. Lydyard, A. Whelan and M. W. Fanger, “Immuno- logia. Krókie Wykłady,” 2nd Edition, Wydawnictwo Nau- kowe PWN, Warszawa, 2006. [48] J. M. Brown, D.M. Coates and R. J. Phillpots, “Evalua- tion of Monoclonal Antibodies for Generic Detection of Flaviviruses by ELISA,” Journal of Virological Methods, Vol. 62, No. 1, 1996, pp. 143-151. doi:10.1016/S0166-0934(96)02095-2 [49] R. D. Saville, N. T. Constantine, F. R. Cleghorn, N. Jack, C. Bartholomew, J. Edwards, et al., “Fourth-Generation Enzyme-Linked Immunosorbent Assay for Simultanous Detection of Human Immunodeficiency Virus Antigen and Antibody,” Journal of Clinical Microbiology, Vol. 39, 2001, pp. 2518-2524. doi:10.1128/JCM.39.7.2518-2524.2001 [50] R. Yadav and S. Dwivedi, “Trends and Perspectives of Biosensors for Food and Environmental Virology,” Food and Environmental Virology, Vol. 2, No. 2, 2010, pp. 53-63. doi:10.1007/s12560-010-9034-5 [51] D. R. Reyes, D. Jossifidis and P. A. Auroux, “Micro Total Analysis Systems. 1. Introduction, Theory and Technol- ogy,” Analytical Chemistry, Vol. 74, No. 12, 2002, pp. 2623-2636. doi:10.1021/ac0202435 [52] Y. L. Tsai, B. Tran, L. R. Sangermano and C. J. Palmer, “Detection of Poliovirus, Hepatitis A Virus, and Rotavi- rus by Triplex Reverse Transcriptase PCR,” Applied and Environmental Microbiology, Vol. 60, No. 7, 1994, pp. 2400-2700. [53] K. R. Everett, J. Rees-George, I. P. S. Puspharajah, B. J. Jansen and Z. Luo, “Advantages and Disadvantages of Microarrays to Study Microbial Population Dynamics,” Copyright © 2013 SciRes. AiM  A. HRYNISZYN ET AL. Copyright © 2013 SciRes. AiM 449 New Zealand Plant Protection Society Journals, Vol. 63, 2010, pp. 1-6. [54] H. B. Balkin and A. B. Margolin, “Detection of Poliovi- rus by ICC/qPCR in Concentrated Water Samples Has Greater Sensitivity and Is Less Costly Using BGM Cells in Suspension as Compared to Monolayers,” Virology Journal, Vol. 7, No. 282, 2010, pp. 1-4. [55] Y. Marzouk and S. M. Goyal, “Prevalence of Enterovi- ruses in Ground Water of Israel,” Ground Water, Vol. 17, No. 5, 1979, pp. 487-491. doi:10.1111/j.1745-6584.1979.tb03345.x [56] K. P. Hennes and C. A. Suttle, “Direct Counts of Viruses in Natural Waters and Laboratory Cultures by Epifluo- rescence Microscopy,” Limnology and Oceanography, Vol. 40, No. 6, 1995, pp. 1050-1055. doi:10.4319/lo.1995.40.6.1050 [57] A. Piekarowicz, “Podstawy Wirusologii Molekularnej,” Wydawnictwo Naukowe PWN, Warszawa, 2004. [58] S. J. Martin, “The Biochemistry of Viruses,” Cambridge University Press, Cambridge, 1978. [59] M. Keusgen, “Biosensors: New Approaches in Drug Dis- covery,” Naturwissenschaften, Vol. 89, No. 10, 2002, pp. 433-444. doi:10.1007/s00114-002-0358-3
|