 Food and Nutrition Sciences, 2013, 4, 49-58 http://dx.doi.org/10.4236/fns.2013.49A2008 Published Online September 2013 (http://www.scirp.org/journal/fns) Selective Lactococcus Enumeration in Raw Milk* Laetitia Gemelas, Véronique Rigobello, Maï Huong Ly-Chatain, Yann Demarigny Isara-Lyon, Unité BioDyMIA, Lyon, France. Email: lgemelas@isara.fr Received February 13th, 2013; revised March 13th, 2013; accepted March 20th, 2013 Copyright © 2013 Laetitia Gemelas et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. ABSTRACT The Lactococcus diversity in cow and goat raw milk was investigated. To do so, a protocol had to be established for the specific enumeration of lactococci. Eight agar media and one control medium were analysed to compare their profi- ciency in evaluating the Lactococcus population in raw milk: M17 Nal, Elliker, modified Elliker, PCA + milk, modified KCA, modified Chalmers, Turner, FSDA. The M17 medium was used as reference. Eighteen pure strains were tested on these media for their selectivity towards lactococci: six Lactococcus species or subspecies, three Leuconostoc, three Enterococcus, two Lactobacillus, one Streptococcus thermophilus, one Pseudomonas fluorescens, one Escherichia coli and one Staphylococcus aureus. All these bacteria were chosen for their regular presence in raw milk. The KCA me- dium proved to be the most selective towards lactococci, on condition that 1) we discriminated the colonies using the catalase test and 2) we subtracted the Enterococcus population counted on BEA. However, it was not possible to sepa- rate the Streptococcus from the Lactococcus colonies on KCA. The “Lactococcus-like” population including these two genera was estimated at a mean level of 3.18 log(cfu)/mL and 4.14 log(cfu)/mL in cow and goat raw milk respectively. This is consistent with the data already published. Keywords: Lactococcus; Culture Media; Raw Milk; LAB; Modified KCA 1. Introduction Raw milk include different microflora traditionally grouped into three categories: positive, negative and neu- tral. Technological microflora is considered as positive. Among them, lactic acid bacteria (LAB), i.e. Lactococ- cus, Leuconostoc, homo and heterofermentative Lacto- bacilli, Pediococcus, and cheese surface bacteria—Mi- crococcus, non-pathogenic Staphylococcus, yeasts and moulds—are generally referred to. Spoiling bacteria— negative microflora—include for instance the Pseudo- monas and Escherichia genera. Neutral bacteria, among them many Archaebacteria, are considered to have no effect on milk quality [1,2]. Raw milk LAB are known to contribute positively to cheese making. Despite being at a much lower level compared with the starter bacteria (3 log(cfu)/mL vs 6 to 7 log(cfu)/mL), wild LAB are able to participate in the acidification step and in the ripening [3,4] reaching levels as high as 7 - 8 log(cfu)/g [5]. Their enzymes help to modify the physico-chemical and bio- chemical environment of the cheese which allows the aromatic balance to develop [6]. In light of these obser- vations, cheese makers are divided over the necessity to control pathogens, on the one hand, by lowering the total bacterial count and their will, on the other, to favour positive microflora [7,8]. This position which has proven to be antinomic until now can be justified by two expla- nations. The microbial flux within the farm which leads to the microbial enrichment of the milk is still unknown. A plate medium designed for the specific enumeration of positive microflora is lacking. This is particularly true for the Lactococcus population. Lactococci, Qualified Presumption of Safety (QPS) microorganisms, are a technological microflora of impor- tance. Wild lactococcal strains rapidly grow during the first steps of cheese making due to their proteolytic en- zymes. Through the production of lactic acid, they par- ticipate in curd formation and contribute positively to the taste and texture characteristics of the cheeses [9]. In spite of these interesting technological abilities, the level of lactococci in raw milk is still unknown. No specific medium in which to count them is currently available because of the complex nutritional requirements of these bacteria. Consequently, rich non-selective media are used to study them. But these agar plates allow many other bacteria to form colonies exhibiting the same morpho- *Diversity of the Lactococcus type population in cow and goat’s raw milks. Copyright © 2013 SciRes. FNS  Selective Lactococcus Enumeration in Raw Milk 50 type as that of lactococci. Among the media mentioned in the literature to culti- vate lactococci, M17 is frequently put forward as plate count agar and broth [10]. But the reducing power of lactococci linked with organic acid production is also tested on Turner agar and modified KCA [11,12]. Prote- olytic and non-proteolytic strains are separated on FSDA agar (Fast Slow Differential Agar) and PCA (Plate Count Agar) supplemented with milk (1%) [13]. Inhibitors of gram negative bacteria (i.e. sodium azide) and acidity indicator (i.e. bromocresol purple) are sometimes added to PCA or M17 agar in order to improve their efficiency in detecting LAB and especially lactococci [14]. Modi- fied Elliker agar and modified Chalmers agar are selec- tive media which bring to light the acidifying bacteria [15,16]. 2. Materials and Methods 2.1. Reference Strains, Culture Maintenance and Reference Media Eighteen known strains were used. Lactococcus garvieae CIP 102507, Lc plantarum CIP 102506, Enterococcus faecalis CIP103631, Ec faecium CIP106742, Leucono- stoc mesenteroides subsp. cremoris CIP103009 (referred to hereafter as Ln cremoris), Ln mesenteroides subsp. dextranicum CIP102423 (Ln dextranicum), Ln mesenter- oides subsp. mesenteroides CIP54178 (Ln mesenter- oides), Staphylococcus aureus CIP103429, Escherichia coli CIP7624 were purchased from the “Collection de l’Institut Pasteur”. Ec durans, Lc lactis subsp. lactis (Lc lactis), Lc lactis subsp. cremoris (Lc cremoris), Lc lactis subsp. hordniae (Lc hordniae), Lc lactis subsp. lactis biovar. diacetylactis (Lc diacetylactis), three Lc planta- rum strains, Streptococcus thermophilus, Lactobacillus paracasei, Lb plantarum and Pseudomonas fluorescens originated from our laboratory collection. These strains had been previously isolated from raw milk samples and carefully characterized and identified [17]. All the strains were kept frozen at −80˚C in a mixture of the culture medium and glycerol 30% (Sigma-Aldrich). After thawing, the strains were cultured in their spe- cific maintenance broth, that is: Lactococcus, Enteroco ccus and St thermophilus: M17 broth (Biokar) for 24 h at 30˚C, 30˚C and 44˚C re- spectively; Leuconostoc and Lactobacillus: MRS broth (Biokar) for 24 h at 30˚C and 37˚C respectively; Ps fluorescens, S aureus and E. coli: BHI broth (Bio- kar) for 24 h at 30˚C, 37˚C and 37˚C respectively. Purity tests were carried out in aerobic conditions at the same temperatures and on the same media, but in Petri dishes. Incubation lasted 24 h, except for Leucono- stoc strains which were incubated for 72 h and for Lac- tobacillus strains, which were incubated for 48 h in an- aerobic conditions. 2.2. Agar Media and Culture Conditions Nine agar culture media were tested. Two elective media were dedicated to the enumeration of the mesophilic aerobic lactic acid bacteria, PCA (Biokar) with 1 g/L skim milk powder (Oxoid) and Elliker (Biokar). Three selective media were used: basic M17 agar (Biokar) was amended with 0.04 g/L nalidixic acid (Sigma-Aldrich); modified Elliker agar contained 1 g/L thallium acetate (Merk Group) and 25 mg/L bromocresol purple (Chimie- Plus) to underline acidifying strains; Chalmers media in- cluded polymixin-β-sulfate (100 iu·mL−1, Sigma-Aldrich) and a redox indicator (TTC, triphenyl tetrazolium chlo- ride, Sigma-Aldrich). Three media were chosen to high- light specific biochemical features: the proteolytic active- ity was checked on FSDA and the reducing power of strains on Turner and KCA agar. These three media and the Chalmers medium were all prepared in the lab. Basic M17 agar was employed as control. The characteristics of each medium are displayed on Table 1. The nine me- dia were incubated for 48 h at 30˚C aerobically. These conditions are supposed to be optimal for the growth of the Lactococcus population. 2.3. Analytical Design The nine media quoted above were checked for their ability to favour the growth of the different Lactococcus species and for their selectivity when in the presence of other bacterial populations in pure and mixed cultures. On the basis of these two experiments, the most suitable medium was then kept for further use on raw milk. Some presumed lactococcal isolates were collected for phenol- typic and genomic identification. 2.3.1. Microbi ological An al y s i s 1) Tests on Reference Strains The eighteen strains were cultured in optimal condi- tions. As soon as optical density corresponded to the growth phase, a sample was removed for enumeration on the nine media listed above. The results obtained were compared with one another and with the control medium (M17). Each experiment was duplicated. 2) Test on Re-Seeded Model Milk (RMM) Re-seeded model milks (RMM) were performed as de- scribed by Dalmasso et al. [17]: pasteurized milk was seeded with the eighteen strains quoted above so as to mimic the habitual levels found in raw milk (Table 2). The RMM allowed us to test the proficiency of the media in evaluating the Lactococcus population in a mix of bacteria. 3) Milk Sampling Milk samples were taken from the milk tank just after Copyright © 2013 SciRes. FNS  Selective Lactococcus Enumeration in Raw Milk Copyright © 2013 SciRes. FNS 51 Table 1. Characteristics and composition of nine media dedicated to the enumeration of the Lactococcus population. Constituents (g·L−1, except steril whey) M17a [10] M17 Nala PCAb + milk 1% [14] Ellikerb [18] Modified Ellikerb [15] FSDAb [13] Modified Chalmersb [16] Modified KCAb [12] Turnerb [11] Tryptone 2.5 2.5 5 20 20 - - 17 5 Polypeptone 2.5 2.5 - - - - 3 - - Soytone 5 5 - - - - - - Yeast extract 2.5 2.5 2.5 5 5 - 3 4.25 5 Meat extract 5 5 - - - - 3 - - Gelatine - - - 2.5 2.5 - - 2.1 - Milkc - - 10 - - 100 - - - Glucose - - 1 5 5 - 20 4.25 0.5 Lactose 5 5 - 5 5 - 20 4.25 - Saccharose - - - 5 5 - - - - Ascorbic acid 0.5 0.5 - 0.5 0.5 - - - - NaCl - - - 4 4 - - 3.4 - MgSO4 0.25 0.25 - - - - - - - β-glycerophosphate disodium 19 19 - - - 19 - - - K2HPO4 - - - - - - - - 2 Calcium lactate - - - - - - - 6.8 - Sodium citrate - - - - - - - 1.7 - Calcium citrated - - - - - - - 5 - Whey (v/v)e - - - - - - - 0.1 - L-arginin - - - - - - - - 2 Bromocresol purple - - - - 0.025 - - - - TTCf - - - - - - - 0.1 0.05 Litmus - - - - - 1 - - - Neutral red - - - - - - 0.005 - - CaCO3 - - - - - - 20 - - Thallium acetate - - - 1 - - - - Nalidixic acid - 0.04 - - - - - - - Sodium acetate - - - 1.5 1.5 - - - - Polymixin-β-sulfatef - - - - - - 100 i.u./mL - - Agar 15 15 12 15 15 10 15 12.8 15 Buffered pH 7.1 - 7.2 7.1 - 7.2 - - - - 6 - - M17, PCA, Elliker are ready to use (Biokar), the other media are laboratory-made. aMedia sterilised at 115˚C, 20 min; bMedia sterilised at 121˚C, 15 min; cRe- formed skim milk powder separately sterilised at 110˚C, 10 min; dCalcium citrate separately sterilised at 121˚C, 15 min; eWhey separately sterilised at 110˚C, 10 min; fTTC (triphényl tetrazolium chloride) and Polymixin-β-sulfate sterilised by filtration (0.45 µm). milking. Eight milk samples were collected on eight farms from the Rhône-Alpes Region (France): Cf1, Cf2, Cf3, Cf4, Cf5, Cf6, Gf1 and Gf2 (“C”, “G” and “f” refer respectively to cow, goat and fresh milk); three raw cow milk samples came from three farms in the Massif Cen- tral Region: Cf7, Cf8 and Cf9. Six raw cow milk and two  Selective Lactococcus Enumeration in Raw Milk 52 Table 2. Theoretical bacterial concentrations in the re-seeded milk samples. Data expressed in log(cfu)/mL. Lc lactis Lc cremoris Lc hordniae Lc plantarum Lc garvieae Lc diacetylactis Ln mesenteroides Ln dextranicum Ln cremoris Ec faecalis Ec faecalis Ec durans Lb plantarum Lb paracasei St thermophilus E coli Ps fluorescens S aureus Total population Level 2.3 2.3 1.4 1.4 1.4 1.4 2.2 2.2 2.2 1.5 1.5 1.5 1.7 1.7 2.7 2.7 2.0 2.3 3.4 raw goat milk samples were collected from eight farms in the Franche-Comté Region: Ct1, Ct2, Ct3, Ct4, Ct5, Ct6, Gf3 and Gf4 (“t” means that the milk was thawed before use). Thawed samples were kept at −80˚C until analysis (within one week). Fresh samples were stored at 0/+4˚C and analysed within 12 hours after milk sampling. The colonies from the six samples Cf1, Cf2, Cf3, Gf1, Ct1 and Ct2 were first counted. Some catalase negative colonies thought to belong to the Lactococcus genus were then purified on M17 agar in view of phenotypic and genotypic characterisation. Each isolate was stored at −80˚C in a mix of medium and 30% glycerol. The other samples were just checked for the enumeration of the Lactococcus population. 2.3.2. Phenot y pi c Ch aracteriz ati on Four tests were chosen for their aptness in identifying the Lactococcus genus. The absence of the catalase was at- tested by pouring a drop of H2O2 directly on each colony (no gas production). Microscopic observation after Gram stain indicated the shape (cocci), the arrangement (small chains) and the position of the bacteria (Gram+). The type of lactic acid isomer produced was determined using the D-/L-lactate enzymatic test (ENZYTEC, R-Biopharm). This test was performed on the supernatant of a 24 h culture of each bacterium. Lactococcus, Streptococcus and Ente roco ccus strains produce L-lactate whereas Leu- conostoc strains produce D-lactate, and heterofermenta- tive Lactobacillus bacteria produce D-, L- or a mix of D- and L-lactate. BEA (Biokar) was used to separate pre- sumed lactococci from enterococci. Lactococci are gen- erally unable to develop on BEA due to bile salt inhi- bition. BEA is dedicated to the specific count of entero- cocci (this was checked on the eighteen strains, although the results are not reproduced here). 2.3.3. Genotypic Identification The isolates presumed to belong to the Lactococcus ge- nus—catalase negative, producing L-lactate, unable to grow on BEA—were cultured at 30˚C for 24 h in 5 mL of M17 broth. The total DNA was extracted using the Nucleospinb tissue kit (Machery-Nagel). Pure DNA was stored at −20˚C until use. A multiplex PCR assay was performed as described by Pu et al. [19], using the fol- lowing primers: 1RL, LacreR, LgR and PilpraR. PCR products were electrophoresed through 1% (w/v) agarose gel (Sigma-Aldrich) in TBE buffer (Tris—Borate— EDTA pH 8, Sigma-Aldrich) at 100V for 3 h. The DNA fragments were stained with ethidium bromide (Sigma- Aldrich), viewed under UV light (302 nm, Biorad) and photographed on a digital camera (Camedia C-5060). The band patterns were normalized and processed using the GelCompar 3.1 software (Applied Maths). The size of PCR products was determined with a DNA size marker (Sigma-Aldrich). Some isolates were subjected to gene sequencing, hav- ing been chosen as representative strains of each cluster obtained from the multiplex PCR dendrogram. The par- tial 16 S rRNA gene sequence analysis was performed by the company BACT UP (France) and the complete 16 S rRNA gene sequence analysis was performed by Idymik Company (France). 2.4. Statistical Analysis A repeatability test was performed with three Lactococ- cus strains. They were incubated at 30˚C in M17 broth. During their exponential growth phase, each of them was enumerated on ten M17 agar dishes. Standard errors were calculated with the Student number for a 5% threshold value for a (n − 1) degree of freedom. The standard-error was evaluated to 0.10 log(cfu)/mL for populations be- tween 7 and 9 log(cfu)/mL. Thus, two results were con- sidered as significantly different if the variation was su- perior to 0.10 log(cfu)/mL. Statistics—variance analysis, means, etc.—were performed by means of the XLSTAT software (2011, version 5.01). 3. Results 3.1. Growth Test of Reference Strains 3.1.1. Mediu m Specificity towards Each Strain None of the four Lc plantarum strains grew on modified Elliker (Table 3). Lactococcus colonies were difficult to count on FSDA, irrespective of the species. The enu- meration of lactococci on PCA + milk, Elliker, M17 Nal (+ nalidixic acid), Turner, KCA, and Chalmers did not produce significantly different results from those ob- tained on the control medium (M17 agar), differences Copyright © 2013 SciRes. FNS  Selective Lactococcus Enumeration in Raw Milk 53 Table 3. Results of growth tests of eighteen strains—Lc lactis, Lc cremoris, Lc hordniae, Lc plantarum, Lc garvieae, Lc diacety- lactis, Ln mesenteroides, Ln dextranicum, Ln cremoris, Ec durans, Lb plantarum, Lb paracasei, St thermophilus, E. coli, Ps fluorescens, S. aureus—on eight media—FSDA, modified Elliker, M17 Nal, PCA + milk, Elliker, modified Chalmers, modi- fied KCA, Turner—and on the control medium—M17. Lc lactis Lc cremoris Lc hordniae Lc plantarum Lc garvieae Lc diacetylactis Ln mesenteroides Ln dextranicum Ln cremoris Ec faecalis Ec faecium Ec durans Lb plantarum Lb paracasei St thermophilus E. coli Ps fluorescens S aureus FSDA A A A A A A Modified elliker A A A A A M17 M17 Nal A A PCA + milk Elliker Modified chalmers A A A A Modified KCA A A A A A A A Turner A A A A A A A A: absence of growth. Gray-coloured: growth. being less than or equal to 0.10 log(cfu)/mL (p < 0.05). Therefore, modified Elliker and FSDA media were set aside at this step of the study. The eighteen strains tested were all able to develop on M17, PCA + milk and Elliker media. E. coli and Ps fluo- rescens were inhibited on M17 Nal. On these four media, the colony’s morphotype was identical—small, circular, smooth, shiny and creamy, whatever the genus or the species. Therefore, these media did not appear suitable for the specific enumeration of lactococci. On modified Chalmers, if we consider the “non Lactococcus” strains, eight strains out of twelve were able to form colonies, their morphotype being identical to the one of lactococci. Consequently, this media was also set aside. On Turner and KCA, only four strains out of the twelve “non Lac- tococcus” strains were able to form pink or red Lacto- coccus type colonies: Ec faecium, Ec faecalis, E. coli and Ps fluorescens. Ec durans was only observed on Turner and S aureus on KCA. To sum up, growth on KCA and Turner gave the most convincing results for the selective enumeration of pre- sumed Lactococcus and Enterococcus genera. These two media were kept for further analysis whereas the others were set aside. Since enterococci were able to develop on these two media, we proposed to specifically count en- terococci on BEA medium and to subtract the entero- coccal count from the Turner or KCA count. 3.1.2. Species Recovery from RMM A sample of RMM was plated on BEA agar, Turner and KCA. Catalase negative colonies with a pink or a red colour were enumerated specifically on these last two media. The result obtained corresponded to the level of presumed lactococci + enterococci. The count result ob- tained on BEA was then subtracted from the KCA and the Turner results. This gave an estimation of the pre- sumed lactococcal microflora. The microbial results gathered from three RMM are displayed on Tabl e 4. The “KCA protocol” led to a fairly accurate estimation of the Lactococcus population, the discrepancy between objective and estimated data being inferior to 0.21 0.09 log(cfu)/mL. A variance analysis was performed on these data. No statistical difference was observed whatever the milk sample or strain consid- ered. On the contrary, the Turner medium systematically gave unusable results, enterococci and lactococci both being underestimated. Consequently this medium was not retained for the following analyses. 3.2. Protocol Design for Estimating the Lactococcus Population The microbial results obtained on six raw cow milks— Cf1, Cf2, Cf3, Gf1, Ct1, Ct2—are shown on Table 5. Between 50% and 100% of the catalase negative isolates were picked up from KCA Petri dishes. The phenotypic and genotypic characterisation of these isolates allowed presumed Lactococcus colonies to be separated from other contaminants—namely enterococci. All the Lacto- coccus isolates were Gram + cocci producing L-lactate and were inhibited by bile salts. On the basis of these Copyright © 2013 SciRes. FNS 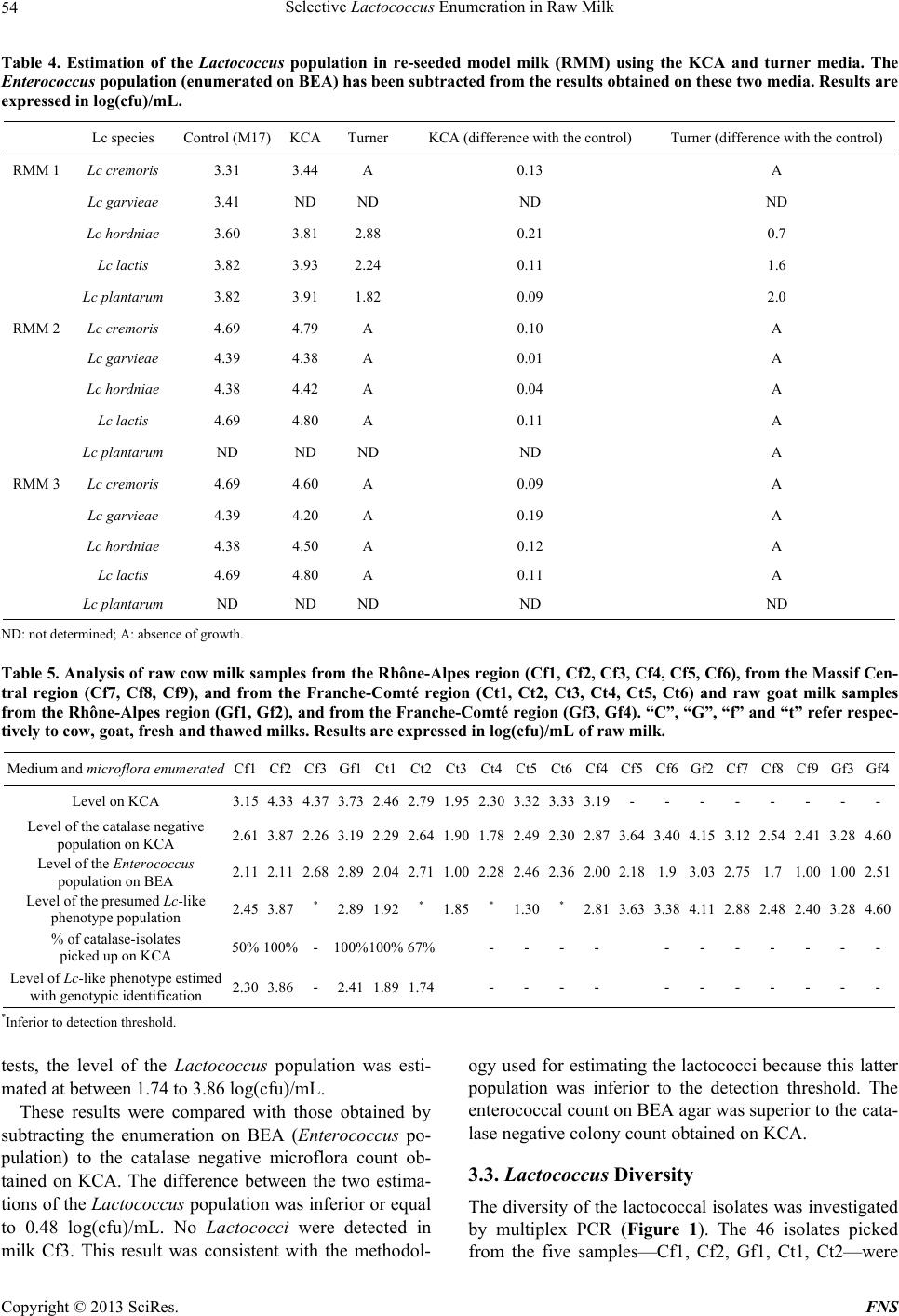 Selective Lactococcus Enumeration in Raw Milk 54 Table 4. Estimation of the Lactococcus population in re-seeded model milk (RMM) using the KCA and turner media. The Enterococcus population (enumerated on BEA) has been subtracted from the results obtained on these two media. Results are expressed in log(cfu)/mL. Lc species Control (M17) KCA TurnerKCA (difference with the control) Turner (difference with the control) RMM 1 Lc cremoris 3.31 3.44 A 0.13 A Lc garvieae 3.41 ND ND ND ND Lc hordniae 3.60 3.81 2.88 0.21 0.7 Lc lactis 3.82 3.93 2.24 0.11 1.6 Lc plantarum 3.82 3.91 1.82 0.09 2.0 RMM 2 Lc cremoris 4.69 4.79 A 0.10 A Lc garvieae 4.39 4.38 A 0.01 A Lc hordniae 4.38 4.42 A 0.04 A Lc lactis 4.69 4.80 A 0.11 A Lc plantarum ND ND ND ND A RMM 3 Lc cremoris 4.69 4.60 A 0.09 A Lc garvieae 4.39 4.20 A 0.19 A Lc hordniae 4.38 4.50 A 0.12 A Lc lactis 4.69 4.80 A 0.11 A Lc plantarum ND ND ND ND ND ND: not determined; A: absence of growth. Table 5. Analysis of raw cow milk samples from the Rhône-Alpes region (Cf1, Cf2, Cf3, Cf4, Cf5, Cf6), from the Massif Cen- tral region (Cf7, Cf8, Cf9), and from the Franche-Comté region (Ct1, Ct2, Ct3, Ct4, Ct5, Ct6) and raw goat milk samples from the Rhône-Alpes region (Gf1, Gf2), and from the Franche-Comté region (Gf3, Gf4). “C”, “G”, “f” and “t” refer respec- tively to cow, goat, fresh and thawed milks. Results are expressed in log(cfu)/mL of raw milk. Medium and microflora enumeratedCf1 Cf2 Cf3 Gf1 Ct1 Ct2 Ct3 Ct4 Ct5 Ct6Cf4Cf5Cf6 Gf2 Cf7 Cf8 Cf9Gf3Gf4 Level on KCA 3.15 4.33 4.37 3.732.462.791.952.303.323.333.19- - - - - - - - Level of the catalase negative population on KCA 2.61 3.87 2.26 3.19 2.29 2.641.90 1.78 2.49 2.30 2.87 3.64 3.40 4.15 3.12 2.54 2.41 3.28 4.60 Level of the Enterococcus population on BEA 2.11 2.11 2.68 2.89 2.04 2.71 1.00 2.28 2.46 2.36 2.00 2.181.93.03 2.75 1.7 1.00 1.00 2.51 Level of the presumed Lc-like phenotype population 2.45 3.87 * 2.89 1.92* 1.85 * 1.30 * 2.81 3.63 3.38 4.11 2.88 2.48 2.40 3.28 4.60 % of catalase-isolates picked up on KCA 50% 100% - 100%100%67% - - - - - - - - - - - Level of Lc-like phenotype estimed with genotypic identification 2.30 3.86 - 2.411.891.74 - - - - - - - - - - - *Inferior to detection threshold. tests, the level of the Lactococcus population was esti- mated at between 1.74 to 3.86 log(cfu)/mL. These results were compared with those obtained by subtracting the enumeration on BEA (Enterococcus po- pulation) to the catalase negative microflora count ob- tained on KCA. The difference between the two estima- tions of the Lactococcus population was inferior or equal to 0.48 log(cfu)/mL. No Lactococci were detected in milk Cf3. This result was consistent with the methodol- ogy used for estimating the lactococci because this latter population was inferior to the detection threshold. The enterococcal count on BEA agar was superior to the cata- lase negative colony count obtained on KCA. 3.3. Lactococcus Diversity The diversity of the lactococcal isolates was investigated by multiplex PCR (Figure 1). The 46 isolates picked from the five samples—Cf1 Cf2, Gf1, Ct1, Ct2—were , Copyright © 2013 SciRes. FNS  Selective Lactococcus Enumeration in Raw Milk 55 100908070 200.00 400.00 600.00 800.00 1000 1200 1400 1600 1800 2000 4.00E+3 Cluster 1 - Lc Lactis [1] Cluster 2 - Lc Lactis [1] Cluster 3 - Streptococcus spp. [31] Custer 4 - unidentified [2] * Cluster 5 - Lc Lactis [9] Cluster 6 - Enterococcus spp. [1] Cluster 7 - Lc raffinolactis [1] * Figure 1. Dendrogram drawn by UPGMA of correlation value of normalized multiplex-PCR patterns from lactococci. Each pattern is identified by a cluster number and by a number between brackets referring to the number of strains which dis- played this profile. The seven clusters from 1 to 7 are defined at a coefficient of similarity of 80% materialized by a bold ver- tical line. Strains coming from goat’s raw milk are identified by a *. arranged in 7 clusters (80% similarity threshold). Only one profile is observed in each cluster. Cluster 6 is an example of an enterococcal profile. The isolate picked from a raw goat milk sample Cf2 was identified as En- terococcus spp by DNA sequencing. Nine isolates picked from samples Cf1, Ct1 and Ct2 form cluster 5. A 228 pb band is observed. This size is specific to the Lactococcus lactis species, a result confirmed by DNA partial se- quencing of the 16 S rRNA genes of one isolate coming from the Ct1 sample. The remaining 36 isolates could not be identified by multiplex PCR. One representative of each cluster was thus identified by sequencing. With the exception of cluster 4, formed by 2 isolates collected from raw goat milk, all these strains were able to be identified. Clusters 1 and 2 were associated with Lc lactis. These two isolates were taken from the Cf1 sample. Cluster 7 included one strain identified as Lc raffinolactis. This isolate came from raw goat milk (sample Gf1). It is noteworthy that these three isolates did not present the typical DNA bands observed for Lc lactis (238 pb) and Lc raffinolactis (860 pb), although sequencing identifica- tion was doubtless (>97%). Cluster 3 included 31 isolates coming from the milk Cf2. Among them, one isolate was assigned to Streptococcus ssp. by partial 16S rRNA gene sequencing. This result was partially confirmed by a se- quencing test made by another lab which established the genus assignment. This isolate was inhibited by bile salts, a characteristic of lactococci. The presence of strepto- cocci on the surface of KCA, although not surprising, led us to limit the protocol here developed to the evaluation of the Lactococcus and the Streptococcus population. Thereafter in this article, this microflora will be design- nated as “Lactococcus-like” microflora, implying that the distinction between lactococci and streptococci by the KCA culture-dependant methodology is not possible. 3.4. Lactococcus-Like Microflora Levels in Raw Milk The Lactococcus-like population ranged from 1.30 to 3.87 log(cfu)/mL for raw cow milk and from 2.89 to 4.60 log(cfu)/mL for raw goat milk. In four raw cow milk samples, this population was inferior to the detection threshold. In this case, the enterococcal microflora pro- bably overwhelmed lactococci. No differences were ob- served considering the geographical origin of the raw milk. 4. Discussion The bacteria from the Lactococcus genus are used as starters to ferment many different dairy products. They are also present at low levels in raw milk, coming from plants and biofilms present in the milking machine [17]. Lactococci have been studied extensively for many years and the yearly number of publications varies between less than 300 to more than 400 articles. Surprisingly, whereas the knowledge—genetic, metabolic, transcryp- tomic, etc.—of this microorganism has been greatly im- proved, no attempts have been made to develop media for its specific enumeration in a complex food matrix. Moreover, knowledge has specifically focused on Lc lactis, and little is known about the other species or sub- species. In this work, we tried to take stock of the level and the diversity of lactococci in raw milk. The prelimi- nary step in reaching this objective involved developing a methodology that would allow the Lactococcus popula- tion to be estimated in raw milk, whatever the species or the subspecies present. Many media have been proposed in the past for the enumeration of lactococci. In spite of substantial efforts, none of them proved to be selective. Among them, M17, PCA + milk and Elliker enable the growth of mesophilic aero tolerant LAB [10,20]. The addition of 0.04 g·L−1 nalidixic acid to these three media allows the Gram negative bacteria to be depressed. But other Gram posi- tive bacteria are still able to grow, an observation also made by Corroler et al. [21]. The use of thallium acetate in the modified Elliker medium [19], another Gram Copyright © 2013 SciRes. FNS  Selective Lactococcus Enumeration in Raw Milk 56 negative inhibitor, proved to hinder the growth of the four Lactococcus plantarum strains we tested. Even if few references exist on this species, Demarigny et al. [22] regularly found this bacterium in natural whey starters. The observations we made on FSDA were identical to those of Grattepanche et al. [23]. If the protease positive colony morphotype—round, regular, smooth—was easily observed, protease negative colonies were never detected. Modified Chalmers did not enable us to clearly distin- guish the different lactic acid bacteria genera on the basis of their morphotype. In fact, two media were found to be of interest, Turner agar and KCA. Among other compo- nents, these plate count agar include TTC, a redox poten- tial indicator. Acid fermentation by lactococci generally leads to the concomitant decrease in the redox potential. Colonies then appear red/pink. In pure culture, KCA and Turner agar gave similar enumerations to those obtained with the reference M17 medium, whatever the Lacto- coccus species or subspecies. But in mixed-culture, the Turner agar underestimated the Lactococcus population. Among several hypotheses, we can propose as a possible explanation that microorganisms competed for nutrients. Indeed, Lactococcus is a nutritionally demanding bacte- rium. For instance for Lc lactis, the amount of amino acid auxotrophies varies between 6 to 8 [24]. The Turner composition is less rich than KCA (less nitrogen and glucidic sources). The resulting nutritional stress that can occur on Turner may explain the discrepancy between control (M17) and test (Turner) data. KCA proved, then, to be the most suitable medium. On KCA, a catalase test must be added to distinguish catalase negative colonies supposed to be assigned to Lactococcus or Enterococ cus. At the same time, Enterococcus bacteria have to be counted on BEA. This result is then subtracted from the result obtain on KCA. This gives a rather good estima- tion of the Lactococcus population in pure or mixed cul- tures. Six raw cow milk samples were used to evaluate the diversity of the Lactococcus species. The isolates col- lected from these raw milk samples were subjected to multiplex PCR [19] and partial 16 S rRNA gene se- quence analysis. The multiplex PCR allowed 9 isolates out of 44 to be accurately identified. The relevance of this method when applied on wild strains appears here debatable. Indeed, the work of Pu et al. [19] only focused on selected starter strains. While the multiplex PCR gave pertinent results on these strains, our own observations would appear to cast doubt on the pertinence of this methodology when applied to wild bacteria. Although many reasons for this could be put forward, at this time, we have no satisfactory explanation to offer. The partial gene sequencing did, however, bring other information. Lc lactis was detected without any doubt in three raw cow milk samples and Lc raffinolactis in one raw goat milk sample. Two isolates picked from one raw goat milk could not be identified by DNA sequencing. More- over, sequencing allowed us to discover Streptococcus strains which had first been assigned to Lactococcus on the basis of their phenotype. Streptococcus has already been observed in raw cow milk. Thus, the presence of this genus is not surprising. Franciosi et al. [25] identified St. dysgalactiae, St. parau- beris, St. suis, St. macedonicus in raw cow milk. Desma- sures et Beuvier [26] also reported Streptococcus ssp. in raw cow and goat milk. Jans et al. [27] designated the Strep- tococcus bovis/Streptococcus equinus complex (SBSEC) “as the predominant LAB in spontaneously fermented African milk products, […] Mexican, Greek, and Italian cheese”. Some of the streptococcal strains show a lot of simili- tudes with lactococci, among them, the bile salt inhibi- tion. In our methodology, it would seem to be impossible to distinguish Lactococcus from Streptococcus strains. This could lead us to question the actual identification of the enumerated microflora already published in the lit- erature. Indeed, our results suggest that some Strepto- coccus strains may have been confused with the Lacto- coccus genus. The mean level of Lactococcus-like mi- croflora in eleven raw cow milk samples was estimated at 3.18 log(cfu)/mL with a minimum of 1.30 log(cfu)/mL and a maximum of 3.38 log(cfu)/mL. In four raw goat milk samples, the mean was superior: 4.14 log(cfu)/mL with a minimum of 2.89 log(cfu)/mL and a maximum of 4.60 log(cfu)/mL. These results are consistent with those published on raw cow and goat milk in the literature [26]. No differences were observed between raw milk coming from the three different regions. Published data taking into account the geographical origin of milk samples is still lacking. Moreover, the number of samples analysed in this study was probably not sufficient to assess the possible influence of this factor. The study of lactococcal diversity brings out, unsure- prisingly, the presence of Lc lactis in the majority of raw cow milk [25,26,28]. On the other hand, Lc raffinolactis, found in raw goat milk, is reported to a lesser extent in the literature on raw goat milk [22,29]. 5. Conclusion Our methodology is getting close to being able to enume- rate lactococci. The results obtained were congruent with those in the literature. However, it was not possible to separate streptococci, which are phenotypically similar to lactococci, unless the whole catalase negative isolates on KCA were to be gene sequenced. This is unsuitable in a dairy lab for routine analyses. Our results confirm the impossibility of developing a methodology dedicated to selectively enumerating the lactococcal microflora. How- ever, it would be interesting to extend this work to en- Copyright © 2013 SciRes. FNS  Selective Lactococcus Enumeration in Raw Milk 57 compass more raw milk samples and to examine in par- ticular the streptococcal microflora. 6. Acknowledgements The authors would like to thank Carl Holland for English corrections. This project was supported by the RMT (“Réseau Mixte Technologique”) “Fromages de Terroir” and the CASDAR project “FloracQ” (Ministère de l’Agriculture et de la Pêche, Chambre d’agriculture du cantal). REFERENCES [1] L. Quigley, O. O’Sullivan, T. P. Beresford, R. P. Ross, G. F. Fitzgerald and P. D. Cotter, “Molecular Approaches to Analysing the Microbial Composition of Raw Milk and Raw Milk Cheese,” International Journal of Food Mi- crobiology, Vol. 150, No. 2-3, 2011, pp. 81-94. doi:10.1016/j.ijfoodmicro.2011.08.001 [2] M. Vacheyrou, A.-C. Normand, P. Guyot, C. Cassagne, R. Piarroux and Y. Bouton, “Cultivable Microbial Commu- nities in Raw Cow Milk and Potential Transfers from Sta- bles of Sixteen French Farms,” International Journal of Food Microbiology, Vol. 146, No. 3, 2011, pp. 253-262. doi:10.1016/j.ijfoodmicro.2011.02.033 [3] S. Buchin, V. Delague, G. Duboz, J. L. Berdague, E. Beuvier, S. Pochet and R. Grappin, “Influence of Pas- teurization and Fat Composition of Milk on the Volatile Compounds and Flavor Characteristics of a Semi-Hard Cheese,” Journal of Dairy Science, Vol. 81, No. 12, 1997, pp. 3097-3108. doi:10.3168/jds.S0022-0302(98)75874-6 [4] E. Beuvier, K. Berthaud, S. Cegarra, A. Dasen, S. Pochet, S. Buchin and G. Duboza, “Ripening and Quality of Swiss-Type Cheese Made from Raw, Pasteurized or Mi- crofiltered Milk,” International Dairy Journal, Vol. 7, No. 5, 1997, pp. 311-323. doi:10.1016/S0958-6946(97)00015-0 [5] Y. Bouton, P. Guyot and R. Grappin, “Preliminary Char- acterization of Microflora of Comté Cheese,” Journal of Applied Microbiology, Vol. 85, No. 1, 1998, pp. 123-131. doi:10.1046/j.1365-2672.1998.00476.x [6] Y. Demarigny, E. Beuvier, S. Buchin, S. Pochet and R Grappin, “Influence of Raw Milk Microflora on the Char- acteristics of Swiss-Type Cheeses: II. Biochemical and Sensory Characteristics,” Le Lait, Vol. 77, No. 1, 1997, pp. 151-167. doi:10.1051/lait:1997110 [7] N. Desmasures, “Etude de laits de Haute Qualité: Carac- téristiques Microbiologiques et Aptitude à la Transforma- tion en Camembert au lait cru,” Ph.D. Dissertation, Uni- versity of Caen, Caen, 1995. [8] E. Beuvier and S. Buchin, “Raw Milk Cheeses,” In: P. F. Fox, P. L. H. McSweeney, T. M. Cogan and T. P. Guinee, Eds., Cheese: Chemistry, Physics and Microbiology, 3rd Edition, Tec & Doc Editions, Vol. 1, Paris, 2004, pp. 319- 345. doi:10.1016/S1874-558X(04)80072-1 [9] E. Casalta, R. Zennaro, M.-X. Maroselli and R. Legouar, “Effect of Specific Starters on Microbiological, Bio- chemical and Sensory Characteristics of Venaco, a Cor- sican Soft Cheese,” Sciences de Aliments, Vol. 17, No. 1, 1997, pp. 79-94. [10] B. E. Terzaghi and W. E. Sandine, “Improved Medium for Lactic Streptococci and Their bacteriophages,” Ap- plied and Environmental Microbiology, Vol. 29, No. 6, 1975, pp. 807-813. [11] N. Turner, W. E. Sandine, P. R. Elliker and E. A. Day, “Use of Tetrazolium Dyes in an Agar Medium for Dif- ferentiation of Streptococcus lactis and Streptococcus cremoris,” Journal of Dairy Science, Vol. 46, No. 5, 1963, pp. 380-385. doi:10.3168/jds.S0022-0302(63)89059-1 [12] G. Waes, “The Enumeration of Aromabacteria in BD Starters,” Netherlands Milk and Dairy Journal, Vol. 22, 1968, pp. 29-39. [13] A. M. Huggins and W. E. Sandine, “Differenciation of Fast and Slow Milk Coagulating Isolates in Strains of Streptococci,” Journal of Dairy Science, Vol. 67, No. 8, 1984, pp. 1674-1679. doi:10.3168/jds.S0022-0302(84)81491-5 [14] N. Desmasures and M. Gueguen, “Monitoring the Micro- biology of High Quality Milk by Monthly Sampling over 2 Years,” Journal of Dairy Research, Vol. 64, No. 2, 1997, pp. 271-280. doi:10.1017/S0022029996002130 [15] J. F. Chamba, G. Bonnaz and P. Bourg, “Comparaisons de Diverses Méthodes de Dénombrement de la Flore Acidifiante du lait cru,” Le Lait, Vol. 61, No. 609-610, 1981, pp. 555-567. doi:10.1051/lait:1981609-61035 [16] V. Vanos and L. Cox, “Rapid Routine Method for the Detection of Lactic Acid Bacteria among Competitive Flora,” Food Microbiology, Vol. 3, No. 3, 1986, pp. 223- 234. doi:10.1016/0740-0020(86)90003-1 [17] M. Dalmasso, C. Hennequin, D. Duc and Y. Demarigny, “Influence of Backslopping on the Acidifications Curves of ‘Tomme’ Type Cheeses Made during 10 Successive Days,” Journal of Food Engineering, Vol. 92, No. 1, 2009, pp. 50-55. doi:10.1016/j.jfoodeng.2008.10.019 [18] P. R. Elliker, A. W. Anderson and G. Hannesson, “An Agar Culture Medium for Lactic Acid Streptococci and Lactobacilli,” Journal of Dairy Science, Vol. 39, No. 11, 1956, pp. 1611-1612. doi:10.3168/jds.S0022-0302(56)94896-2 [19] Z. Y. Pu, M. Dobos, G. K. Y. Limsowtin and I. B. Powell, “Integrated Polymerase Chain Reaction-Based Procedures for the Detection and Identification of Species and Sub- species of the Gram-Positive Bacterial Genus Lactococ- cus,” Journal of Applied Microbiology, Vol. 93, No. 2, 2002, pp. 353-361. doi:10.1046/j.1365-2672.2002.01688.x [20] V. Michel, A. Hauwuy and J. F. Chamba, “La Flore Microbienne de laits Crus Vache: Diversité et Influence des Conditions de Production,” Le Lait, Vol. 81, No. 5, 2001, pp. 575-592. doi:10.1051/lait:2001151 [21] D. Corroler, I. Mangin, N. Desmasures and M. Gueguen, “An Ecological Study of Lactococci Isolated from Raw Milk in the Camembert Cheese Registered Designation of Origin Area,” Applied and Environmental Microbiology, Vol. 64, No. 2, 1998, pp. 4729-4735. Copyright © 2013 SciRes. FNS  Selective Lactococcus Enumeration in Raw Milk Copyright © 2013 SciRes. FNS 58 [22] Y. Demarigny, C. Sabatier, N. Laurent, S. Prestoz, V. Rigobello and M. J. Blachier, “Microbiological Diversity in Natural Whey Starters Used to Make Traditional Ro- camadour Goat Cheese and Possible Relationships with Its Bitterness,” Italian Journal of Food Science, Vol. 18. No. 3, 2006, pp. 251-266. [23] F. Grattepanche, P. Audet and C. Lacroix, “Milk Fer- mentation by Functional Mixed Culture Producing Nisin Z and Exopolysaccharides in a Fresh Cheese Model,” In- ternational Dairy Journal, Vol. 17, No. 2, 2007, pp. 123- 132. [24] V. Monnet, D. Atlan, C. Beal, M. C. Champomier-Verges, M. P. Chapot-Chartier, H. Chouayekh, M. Cogain-Bous- quet, M. Deghorain, P. Gaudu, C. Gilbert, E. Guedon, I. Guillouard, P. Goffin, J. Guzzu, P. Hols, V. Juillard, V. Ladero, N. Lindley, S. Lortal, P. Loubiere, E. Maguin, C. Monnet, F. Rul, R. Tourdot-Marechal and M. Yvon, “Mé- tabolisme et Ingénierie Métabolique,” In: G. Corrieu and F. M. Luquet, Eds., Bactéries Lactiques—De la Géné- tique aux Ferments, Tec & Doc Editions, Paris, 2008, pp. 271-509. [25] E. Franciosi, L. Settanni, A. Cavazza and E. Poznanski, “Biodiversity and Technological Potential of Wild Lactic Acid Bacteria from Raw Cows’ Milk,” International Dairy Journal, Vol. 19, No. 1, 2009, pp. 3-11. doi:10.1016/j.idairyj.2008.07.008 [26] N. Desmasures and E. Beuvier, “Ce qu’il Faut Savoir Avant d’Intervenir sur les Microflores des laits,” In: Microflore du lait cru, CNAOL and GIS Alpes Jura, 2011, pp. 13-74. [27] C. Jans, C. Lacroix and L. Meile, “A Novel Multiplex PCR/RFLP Assay for the Identification of Streptococcus bovis/Streptococcus Equinus Complex Members from Dairy Microbial Communities Based on the 16S rRNA Gene,” FEMS Microbiology Letters, Vol. 326, No. 2, 2012, pp. 144-150. doi:10.1111/j.1574-6968.2011.02443.x [28] A. Mallet, M. Guéguen, F. Kauffmann, C. Chesneau, A. Sesboué and N. Desmasures “Quantitative and Qualita- tive Microbial Analysis of Raw Milk Reveals Substantial Diversity Influenced by Herd Management Practices,” International Dairy Journal, Vol. 27, No. 1-2, 2012, pp. 13-21. doi:10.1016/j.idairyj.2012.07.009 [29] A. Badis, D. Guetarni, B. Moussa Boudjemac, D. E. Hen- nic and M. Kihalc, “Identification and Technological Pro- perties of Lactic Acid Bacteria Isolated from Raw Goat Milk of Four Algerian Races,” Food Microbiology, Vol. 21, No. 5, 2004, pp. 579-588. doi:10.1016/j.fm.2003.11.006
|