 Open Journal of Statistics, 2013, 3, 26-40 http://dx.doi.org/10.4236/ojs.2013.34A004 Published Online August 2013 (http://www.scirp.org/journal/ojs) Copy Mean: A New Method to Impute Intermittent Missing Values in Longitudinal Studies Christophe Genolini1,2*, René Écochard3,4,5, Hélène Jacqmin-Gadda6 1UMR U1027, INSERM, Université Paul Sabatier, Toulouse, France 2CeRSM (EA 2931), UFR STAPS, Université de Paris Ouest Nanterre La Défense, Nanterre, France 3Hospices Civils de Lyon, Service de Biostatistique, Lyon, France 4Université Lyon 1, Villeurbanne, France 5CNRS, UMR5558, Laboratoire de Biométrie et Biologie Evolutive, Villeurbanne, France 6Université de Bordeaux, ISPED, Centre INSERM U897-Epidemiology-Biostatistique, Bordeaux, France Email: *christophe.genolini@u-paris10.fr Received April 23, 2013; revised May 23, 2013; accepted May 30, 2013 Copyright © 2013 Christophe Genolini et al. This is an open access article distributed under the Creative Commons Attribution Li- cense, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. ABSTRACT Longitudinal studies are those in which the same variable is repeatedly measured at different times. These studies are more likely than others to suffer from missing values. Since the presence of missing values may have an important im- pact on statistical analyses, it is important that they should be dealt with properly. In this paper, we present “Copy Mean”, a new method to impute intermittent missing values. We compared its efficiency in eleven imputation methods dedicated to the treatment of missing values in longitudinal data. All these methods were tested on three markedly dif- ferent real datasets (stationary, increasing, and sinusoidal pattern) with complete data. For each of them, we generated nine types of incomplete datasets that include 10%, 30%, or 50% of missing data using either a Missing Completely at Random, a Missing at Random, or a Missing Not at Random missingness mechanism. Our results show that Copy Mean has a great effectiveness, exceeding or equaling the performance of other methods in almost all configurations. The ef- fectiveness of linear interpolation is highly data-dependent. The Last Occurrence Carried Forward method is strongly discouraged. Keywords: Imputation; Longitudinal Data; Intermittent Missing Values 1. Introduction Longitudinal studies are those in which the same variable is repeatedly measured at different times. They are more likely than others to suffer from missing values [1-3]. Indeed, it is frequent that subjects miss a clinical visit or fill out incompletely a questionnaire. The missing data have been classified into three main categories [1]: Missing Completely at Random (MCAR) when the mis- singness probability is independent on the variables, Missing at Random (MAR) when the missingness prob- ability depends only on the observed variables, and Miss- ing Not at Random (MNAR) when the missingness probability may depend on unobserved variables. When the main analysis involves statistical modeling of the change over time of the longitudinal variable using, for instance, mixed models, the model parameters are generally estimated by the maximum likelihood and it is well-known that the maximum likelihood estimation is robust to MAR data [2,4,5]. However, selection models and pattern-mixture models have been proposed when the data are MNAR or when a sensitivity analysis to this assumption is performed [2,4-7]. This paper focuses on situations where the main anal- ysis does not involve modeling and on likelihood- based methods such as descriptive studies, exploratory analyses, non-parametric clustering, etc. These kinds of analyses are very sensitive to missing data, even when the miss- ingness mechanism is MAR; then imputation methods are very useful. Twisk [8] and Engels [3] compared several imputation methods for longitudinal studies. Twisk proposed a clas- sification of imputation methods into two categories: “Cross-sectional” methods that impute missing values at time t using information available at time t and “longitu- dinal” methods that impute the missing values of an in- dividual i using all the non-missing values of i. Engels *Corresponding author. C opyright © 2013 SciRes. OJS  C. GENOLINI ET AL. 27 suggested four categories: 1) “No personal data” methods do not use information available on individual subjects; 2) “baseline data” methods use the information present at baseline but no time-dependent information; 3) “before data only” methods consider all the information available before the occurrence of the missing value; and 4) “be- fore and after” methods impute the missing values using all available information. Regarding the evaluation of performance, Engels pro- posed different indices to compare the performance of imputation methods. These indices are mainly based on the difference between the imputed values and the actual values [3]. The present article aims at comparing different impu- tation methods for missing values in longitudinal studies. Section 2 provides the general framework and the meth- odology: a formal definition of the concept of missing- ness, a presentation of the imputation methods, and the criteria used to measure performance. This section re- views the classical methods and presents an original me- thod called Copy Mean. Section 3 presents the design of the simulation study and Section 4 presents the results. A discussion is provided in Section 5. 2. Methods 2.1. Notations Let us consider a set S of n subjects. For each subject, an outcome variable Y is measured at t different times. The value of Y for subject i at a specific time l is noted il . For subject i, the sequence y it .12 ,,, iii yy y is called a trajectory. For a specific time l, vector .12 ,,, llln l yy y y is called a cross-sectional meas- urement. When il is missing, the value obtained by using a given imputation method IM is noted M il . 2.2. Classification of Missingness In their founding documents, Rubin and Little distin- guished three kinds of missingness [9,10]. They consid- ered trajectories without missingness TRUE (unavailable data) and trajectories with missing values OBS Y (avail- able measured longitudinal data). Then R denotes the Boolean matrix of the location of a missing value and Y ISS Y the missing part of TRUE Y. Thus, TRUE OBS YY ISS . The classification of Little and Rubin is then based on a potential link between R and TRUE YS Y , OB Y, and ISS Y: MCAR: A value is Missing Comp letely at Random if the probability that il y be missing il Py is inde- pendent of Y: . TRUE il MAR: A value is Missing at Random if the probabil- ity that il y be missing is independent of Constap ntPy ISS Y, but may depend on the observed values OBS Y. For exam- ple, if patients who performed badly at time 1l decide to miss time l, the missing data will be MAR: il OBS Py FY. MNAR: A value is Missing Not at Random if the probability that il y be missing depends on ISS Y. Typically, the probability for an observation il y to be missing at time l depends on the current value of Y at time l. For example, if patients who suppose they would perform badly at time l refuse to be tested at time l, the data will be MNAR: il MISS The impact of the mechanism of missingness on the imputation of the missing values was examined by Mo- lenberghs [11]. In the particular case of longitudinal data, the missingness mechanisms were classified according to the position of the missing values within the trajectory: Py FY. Intermittent missing data are missing within a trajec- tory. Formally, il y is an intermittent missing value if there exists a and b, alb , such that ia y and y are not missing. ib Monotone missing data are missing either at the be- ginning or at the end of a trajectory. This includes the case of left-or right-censored follow-ups. If a value is missing, then all the following (respectively, preced- ing) values are also missing. Formally, il y is a (right) monotone missing value if, for all dl, id y is missing. Some imputation techniques, such as the Linear Inter- polation or the Copy Mean (see Sections 2.3.3 and 2.3.4), are not compatible with these two missingness mecha- nisms. In this article, we will focus on intermittent miss- ing data, either MCAR, MAR, or MNAR. 2.3. Imputation Methods Herein, 12 imputation methods are compared. They were grouped according to the information necessary for their implementation and are summarized in Table 1. 2.3.1. No Information Only the complete-case method does not require infor- mation. 1) Complete case method: This method removes any trajectory with one or several missing values [10]. Par- ticularly radical, it is the easiest way to implement. Nev- ertheless, it has serious drawbacks [12] including major loss of information and biases as soon as data are not MCAR. 2.3.2. Cross-Sectional Imputation These methods use only data collected at a given time (time at which the value is missing). The imputation of a missing value at time l is made according to the values from the other individuals observed at time l, i.e. the cross-sectional measurement .12 ,,, lllnl yy y y. 2) The Cross Mean method replaces by the mean il Table 1. Imputation methods and their characteristics. Copyright © 2013 SciRes. OJS  C. GENOLINI ET AL. 28 Imputation method Cross-sectional Longitudinal External information 1) Complete case 2) Cross Mean √ 3) Cross Median √ 4) Cross Hot Deck √ 5) Traj Mean √ 6) Traj Median √ 7) Traj Hot Deck √ 8) LOCF √ 9) Linear Interpolation √ 10) Spline Interpolation √ 11) Copy Mean √ √ 12) Linear Regression, Internal √ √ 13) Linear Regression, External √ √ √ of the values observed at time l. 3) The Cross Median method replaces by the median of the values observed at time l. il y 4) The Cross Hot Deck method replaces il by a value randomly chosen among all values observed at time l. y 2.3.3. Longitudinal Imputation These methods use only the non-missing data of the same subject. The imputation is made independently of the data from other individuals, only the trajectory .12 ,,, iii it yy y is used. 5) The Traj Mean replaces by the average of the values of trajectory . il y .i 6) The Traj Median replaces by the median of the values of trajectory . y il y y .i 7) The Traj Hot Deck replaces il by a value chosen randomly among the values of trajectory . y .i 8) The Last Occurrence Carried Forward (LOCF) replaces by the previous non-missing value. y il 9) The Linear Interpolation replaces il by drawing a line between the two non-missing values that immedi- ately precede and follow the missing one. Let ia and be the closest preceding and following non-missing yy y ib y values of ; then il y LI ib ia il iayy yyla ba . 10) The Spline Interpolation replaces il by draw- ing a cubic spline between the two non-missing values that immediately precede and follow the missing one. For mathematical details, see Fritsch and Carlson [13]. y 2.3.4. Cross-Sectional and Longitudinal Imputation (Cross & Long) These methods use both longitudinal information and cross-sectional information . .i y l 11) Copy Mean is an original method. It is included in the R package kml [14-16]. Howerver, its efficiency has not been compared to other method until today. It com- bines linear interpolation and imputation using the popu- lation’s mean trajectory. Formally, let il be the miss- ing value and and be the closest preceding and y. y ia yib y following non-missing values1. Let .1 . ,, t yy y de- note the mean trajectory of a population S. I il is the value obtained by imputing using linear interpola- il y tion. Let . I l y be the value obtained by applying a linear interpolation between a and b on the mean trajectory: . LI ib ia lia y yyla ba . Then the average variation l V at time l is the difference between .l and . I l y, i.e. .. I lll Vyy . From there, the Copy Mean imputes il by adding the average variation y l V LI to the result of the linear interpolation: il il l CM yAV. Figure 1 shows an ex- ample of a trajectory imputed using the Copy Mean. 12) Linear Regression, Internal: the principle is, for each l, to construct a model that predicts the values of .l using the other variables il with yy l l . Since variables .l y may also contain missing values, the process is iterative by gradual approximation: Initially, all the missing values are imputed (by one of the methods described above). A model regressing .1 y as a function of .2.3. ,,, t is built. Missing values in .1 y are replaced by the values predicted by the model. yy y A model regressing .2 y as a function of .1.3. ,,, t is built. Missing values in .2 y are replaced by the values predicted by the model. yy y In the same way, all the .l y are imputed using a pre- dictive model. Then the process is iterated: a new model is con- structed for .1 whose values are again calculated, then for .2 and so on. Each iteration allows a little more precision in estimating the missing values. y y After a predetermined number of iterations, the proc- ess stops. In this article, the initialization process was done using Cross Mean and the process was iterated 10 times. 1All these notations are illustrated Figure 1. Copyright © 2013 SciRes. OJS  C. GENOLINI ET AL. Copyright © 2013 SciRes. OJS 29 located in Aix-en-Provence, Dijon, and Lyon (France), Milano and Verona (Italy), DÃ1 4sseldorf (Germany), Liège (Belgium) and Madrid (Spain). Urine preg- nanediol-3a-glucuronide was measured before ovulation. This variable is a continuous in the range [0.05; 26.6] mg/L (Overall mean: 11.5 mg/L; overall standard devia- tion: 18.3). The trajectories of this variable have the characteristic of being non-stationary and increasing. Of the 102 trajectories, two (1.96% of total) had missing values. These trajectories were removed from the present study. Because some imputation methods require the use of covariates, we chose five covariates more or less cor- related with the longitudinal variable under study: weight, size, age at menarche, number of children, and current age. Figure 1. Copy Mean imputation. The individual trajectory l is in black, the mean trajectory y is in red. The dot- ted lines are the values imputed by linear interpolation. The dashed lines are values imputed by Copy Mean. 2.4. Cross-Sectional and Longitudinal Imputation using Covariables (External) Fish: The second dataset (Figure 2(b)) comes from a study on an automatic pattern recognition system applied to the monitoring of fish migration [18]. It included 350 individuals. The main variable is continuous in the range [−1.83; 1.95] (overall mean: 0.16; overall standard devia- tion: 0.89). The trajectories present some large variations and are close to sinusoidal functions. The dataset has no missing values but the covariates were not accessible; thus, methods that use covariates were not tested on this dataset. Finally, it is possible to use all the information, including some covariates measured at baseline: 13) Linear Regression, External: the principle is the same as the internal linear regression (iterative process on all cross-sectional variables) but the predictive model for .l is a function of both other trajectories y.l y and some covariates. 3. Simulation Alcohol: The third dataset (Figure 2(c)) comes from the Quebec Longitudinal Study of Child Development led by the GRIP [19]. In this study, 1831 participants were interviewed retrospectively; thus, the data show a very low rate of missingness. The monthly alcohol con- sumption was rated on a four-point scale (0 to 4, overall mean: 1.18; overall standard deviation: 1.09). The main feature of this study is the stability of the values over time. Three trajectories had missing values (0.16% of total); they were removed from the study. The covariates selected were: sex, happiness scores, income, tobacco consumption, and expenditure on tobacco. 3.1. Data Generation The present simulation study was performed using three existing datasets with complete data. Several incomplete datasets were obtained by generating missing values ac- cording to different schemes. To be as general as possi- ble, we worked on three datasets with very different cha- racteristics. 3.1.1. The Three Datasets Pregnanediol: The first dataset (Figure 2(a)) comes from a study on human menstrual cycles [17]. The initial aim of the study was a search for biomarkers for accurate prediction of ovulation. One hundred and two women were recruited from eight natural family planning clinics 3.1.2. Generation of Missing Values Several methods may be used to generate missing values Figure 2. Graphical representations of the three dataset. Individual trajectories are in black. The overall mean trajectories re in red. (a) Pregnanediol; (b) Fish; (c) Alcohol. a  C. GENOLINI ET AL. 30 [20]. In the present article, for each of 3 complete data- sets, we generated 9 (3 × 3) types of incomplete datasets that included 10%, 30%, or 50% missing data using ei- ther a MCAR, a MAR, or a MNAR missingness mecha- nism. This process was repeated 500 times. Thus, 13,500 datasets (3 × 9 × 500) were simulated. The incomplete datasets on pregnanediol and alcohol were analyzed with the 12 imputation methods. The incomplete datasets on fish were analyzed with only the 11 methods that do not require external data. To generate intermittent missing values in a complete dataset, we defined a probability function 1 il PR that il be missing for l in y 2, 1t (the first and last values were always observed ones). In the MCAR case, this probability is independent of Y: 0 1 MCAR il logiR b y y t P. In the MAR case, the prob- ability depends on il where is the last observed value preceding il : il y 01 1 AR il P Ril logitbby . Finally, in the MNAR case, the probability depends on the current value : il y 01 1 NAR il P Ril bbylogit . 3.2. Imputation Quality Comparison Criteria To assess the quality of the different imputation methods, we considered the deviation which is the difference be- tween the true and the imputed value [3] The deviation then leads to three criteria: 1) the Bias is the mean of the deviation; 2) the Mean Absolute Deviation (MAD) is the average of the absolute deviations; and, 3) the Root Mean Square Deviation (RMSD) is the square root of the mean of the square of the deviation. When il is the real value that method IM imputed as y M il , the Bias is M il il y m , the MAD is M il il yy m and the RMSD is 2 IM il il yy m , m being the total number of miss- ing values. 3.3. Methods and Softwares All the analyses were performed with R software [21]. Classical and new imputation methods have been pro- grammed and published in package Longitudinal Data on CRAN [22]. The spline imputation method was pro- grammed using stats package [13,23]. Imputations need- ing linear regression used function mice (mice package) with method “predictive mean matching” [24]. 4. Results During data construction, three mechanisms of missing- ness (MCAR, MAR, and MNAR), three percentages of missing data (10%, 30%, and 50%) and three types of data (Pregnanediol, Fish, and Alcohol) were considered. The analysis of the results showed that the missingness mechanism and the type of dataset had impacts on the performance of the methods but not the percentage of missing data. Thus, for brevity, only the tables relative to 30% missing data will be presented in the main text. The full results are given in the Appendix. 4.1. Mean Absolute Deviation Results The Mean Absolute Deviation (MAD) is the average of the absolute deviations between the real values and the imputed values. Table 2 presents the mean result for each method according to the missingness mechanism and the type of dataset. For better readability, the results were standardized: in each case (each column) the per- formance of the best method (the lowest MAD) was set to 1 so that all other results are multiples of this reference value. In Table 2, the performances of the “good meth- ods” are highlighted in bold. The “good methods” are those whose values are between 1 and 1.2. The threshold of 1.2 was chosen arbitrarily. With Pregnanediol data, Copy Mean, Linear Interpola- tion, LOCF, Traj Median and Traj Mean, were the best. With Fish data, the most effective methods were Copy Mean, Linear Regression Internal, Cross Median, and Cross Mean. All methods that use only longitudinal in- formation performed poorly with this data set character- ized by a strong non-linear trend with low inter-subject variability (see Figure 2(b)). With Alcohol data, Linear Interpolation and Copy Mean gave the best results. There were no marked differences between MCAR, MAR, and MNAR. Only the Spline Interpolation method performed poorly with MAR on Alcohol dataset. This was probably due to the fact that, with MAR, long series of contiguous missing values are more likely; in such a case, the Spline Interpolation method imputes by poly- nomials with values far from the original curve. 4.2. Root Mean Square Deviation Results Table 3 presents the root mean square deviation results. Here too, the results were standardized. The performance of the best method (the lowests RMSD) was set to 1 so that all other results are multiples of this reference value. In Table 3, the hight performance values (1.4 or lower) are highlighted in bold. The threshold of 1.4 was chosen arbitrarily. The results with the Root Mean Square De- viation were close to those obtained with the MAD crite- rion. They are detailed in the Appendix. 4.3. Bias Results Table 4 presents the results for bias. The “good methods” (between −0.03 and 0.03) are highlighted in bold. The hresholds of −0.03 and +0.03 were arbitrarily chosen. t Copyright © 2013 SciRes. OJS 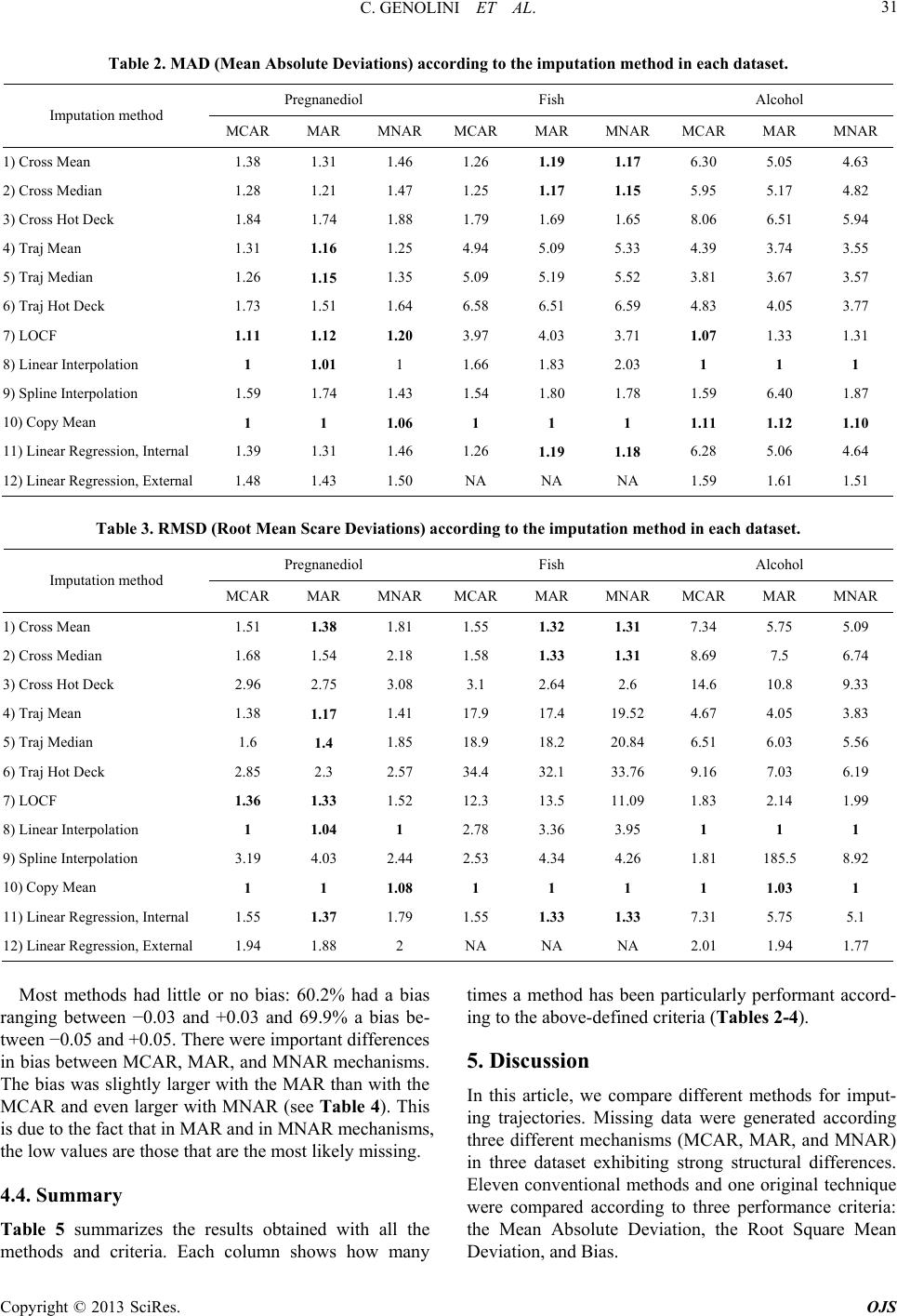 C. GENOLINI ET AL. 31 Table 2. MAD (Mean Absolute Deviations) according to the imputation method in each dataset. Pregnanediol Fish Alcohol Imputation method MCAR MAR MNAR MCAR MAR MNAR MCAR MAR MNAR 1) Cross Mean 1.38 1.31 1.46 1.26 1.19 1.17 6.30 5.05 4.63 2) Cross Median 1.28 1.21 1.47 1.25 1.17 1.15 5.95 5.17 4.82 3) Cross Hot Deck 1.84 1.74 1.88 1.79 1.69 1.65 8.06 6.51 5.94 4) Traj Mean 1.31 1.16 1.25 4.94 5.09 5.33 4.39 3.74 3.55 5) Traj Median 1.26 1.15 1.35 5.09 5.19 5.52 3.81 3.67 3.57 6) Traj Hot Deck 1.73 1.51 1.64 6.58 6.51 6.59 4.83 4.05 3.77 7) LOCF 1.11 1.12 1.20 3.97 4.03 3.71 1.07 1.33 1.31 8) Linear Interpolation 1 1.01 1 1.66 1.83 2.03 1 1 1 9) Spline Interpolation 1.59 1.74 1.43 1.54 1.80 1.78 1.59 6.40 1.87 10) Copy Mean 1 1 1.06 1 1 1 1.11 1.12 1.10 11) Linear Regression, Internal 1.39 1.31 1.46 1.26 1.19 1.18 6.28 5.06 4.64 12) Linear Regression, External 1.48 1.43 1.50 NA NA NA 1.59 1.61 1.51 Table 3. RMSD (Root Mean Scare Deviations) according to the imputation method in each dataset. Pregnanediol Fish Alcohol Imputation method MCAR MAR MNAR MCAR MAR MNAR MCAR MAR MNAR 1) Cross Mean 1.51 1.38 1.81 1.55 1.32 1.31 7.34 5.75 5.09 2) Cross Median 1.68 1.54 2.18 1.58 1.33 1.31 8.69 7.5 6.74 3) Cross Hot Deck 2.96 2.75 3.08 3.1 2.64 2.6 14.6 10.8 9.33 4) Traj Mean 1.38 1.17 1.41 17.9 17.4 19.52 4.67 4.05 3.83 5) Traj Median 1.6 1.4 1.85 18.9 18.2 20.84 6.51 6.03 5.56 6) Traj Hot Deck 2.85 2.3 2.57 34.4 32.1 33.76 9.16 7.03 6.19 7) LOCF 1.36 1.33 1.52 12.3 13.5 11.09 1.83 2.14 1.99 8) Linear Interpolation 1 1.04 1 2.78 3.36 3.95 1 1 1 9) Spline Interpolation 3.19 4.03 2.44 2.53 4.34 4.26 1.81 185.5 8.92 10) Copy Mean 1 1 1.08 1 1 1 1 1.03 1 11) Linear Regression, Internal 1.55 1.37 1.79 1.55 1.33 1.33 7.31 5.75 5.1 12) Linear Regression, External 1.94 1.88 2 NA NA NA 2.01 1.94 1.77 Most methods had little or no bias: 60.2% had a bias ranging between −0.03 and +0.03 and 69.9% a bias be- tween −0.05 and +0.05. There were important differences in bias between MCAR, MAR, and MNAR mechanisms. The bias was slightly larger with the MAR than with the MCAR and even larger with MNAR (see Table 4). This is due to the fact that in MAR and in MNAR mechanisms, the low values are those that are the most likely missing. 4.4. Summary Table 5 summarizes the results obtained with all the methods and criteria. Each column shows how many times a method has been particularly performant accord- ing to the above-defined criteria (Tables 2-4). 5. Discussion In this article, we compare different methods for imput- ing trajectories. Missing data were generated according three different mechanisms (MCAR, MAR, and MNAR) in three dataset exhibiting strong structural differences. Eleven conventional methods and one original technique were compared according to three performance criteria: the Mean Absolute Deviation, the Root Square Mean Deviation, and Bias. Copyright © 2013 SciRes. OJS 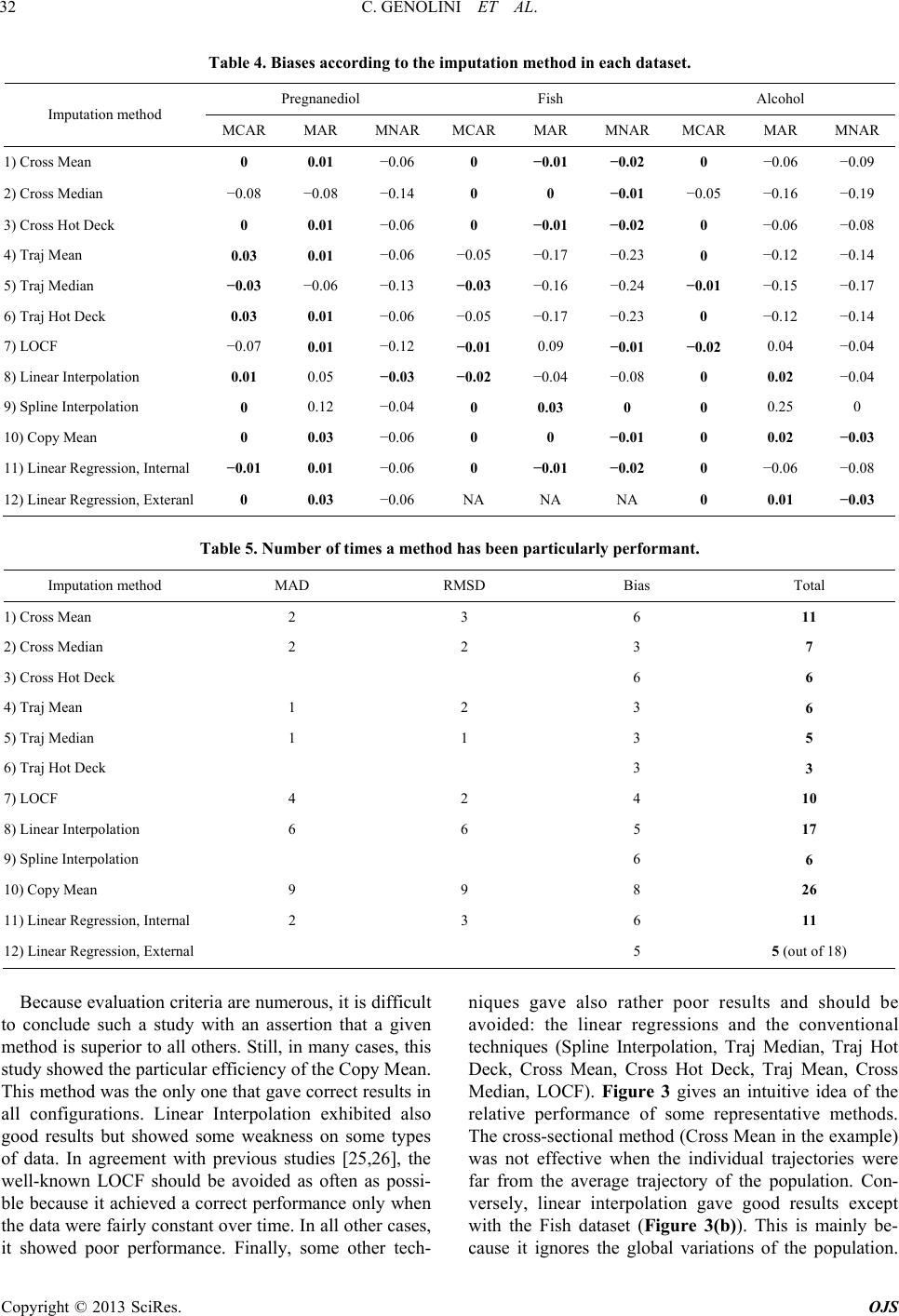 C. GENOLINI ET AL. 32 Table 4. Biases according to the imputation method in each dataset. Pregnanediol Fish Alcohol Imputation method MCAR MAR MNAR MCAR MAR MNAR MCAR MAR MNAR 1) Cross Mean 0 0.01 −0.06 0 −0.01 −0.02 0 −0.06 −0.09 2) Cross Median −0.08 −0.08 −0.14 0 0 −0.01 −0.05 −0.16 −0.19 3) Cross Hot Deck 0 0.01 −0.06 0 −0.01 −0.02 0 −0.06 −0.08 4) Traj Mean 0.03 0.01 −0.06 −0.05 −0.17 −0.23 0 −0.12 −0.14 5) Traj Median −0.03 −0.06 −0.13 −0.03 −0.16 −0.24 −0.01 −0.15 −0.17 6) Traj Hot Deck 0.03 0.01 −0.06 −0.05 −0.17 −0.23 0 −0.12 −0.14 7) LOCF −0.07 0.01 −0.12 −0.01 0.09 −0.01 −0.02 0.04 −0.04 8) Linear Interpolation 0.01 0.05 −0.03 −0.02 −0.04 −0.08 0 0.02 −0.04 9) Spline Interpolation 0 0.12 −0.04 0 0.03 0 0 0.25 0 10) Copy Mean 0 0.03 −0.06 0 0 −0.01 0 0.02 −0.03 11) Linear Regression, Internal −0.01 0.01 −0.06 0 −0.01 −0.02 0 −0.06 −0.08 12) Linear Regression, Exteranl 0 0.03 −0.06 NA NA NA 0 0.01 −0.03 Table 5. Number of times a method has been particularly performant. Imputation method MAD RMSD Bias Total 1) Cross Mean 2 3 6 11 2) Cross Median 2 2 3 7 3) Cross Hot Deck 6 6 4) Traj Mean 1 2 3 6 5) Traj Median 1 1 3 5 6) Traj Hot Deck 3 3 7) LOCF 4 2 4 10 8) Linear Interpolation 6 6 5 17 9) Spline Interpolation 6 6 10) Copy Mean 9 9 8 26 11) Linear Regression, Internal 2 3 6 11 12) Linear Regression, External 5 5 (out of 18) Because evaluation criteria are numerous, it is difficult to conclude such a study with an assertion that a given method is superior to all others. Still, in many cases, this study showed the particular efficiency of the Copy Mean. This method was the only one that gave correct results in all configurations. Linear Interpolation exhibited also good results but showed some weakness on some types of data. In agreement with previous studies [25,26], the well-known LOCF should be avoided as often as possi- ble because it achieved a correct performance only when the data were fairly constant over time. In all other cases, it showed poor performance. Finally, some other tech- niques gave also rather poor results and should be avoided: the linear regressions and the conventional techniques (Spline Interpolation, Traj Median, Traj Hot Deck, Cross Mean, Cross Hot Deck, Traj Mean, Cross Median, LOCF). Figure 3 gives an intuitive idea of the relative performance of some representative methods. The cross-sectional method (Cross Mean in the example) was not effective when the individual trajectories were far from the average trajectory of the population. Con- versely, linear interpolation gave good results except with the Fish dataset (Figure 3(b)). This is mainly be- cause it ignores the global variations of the population. Copyright © 2013 SciRes. OJS 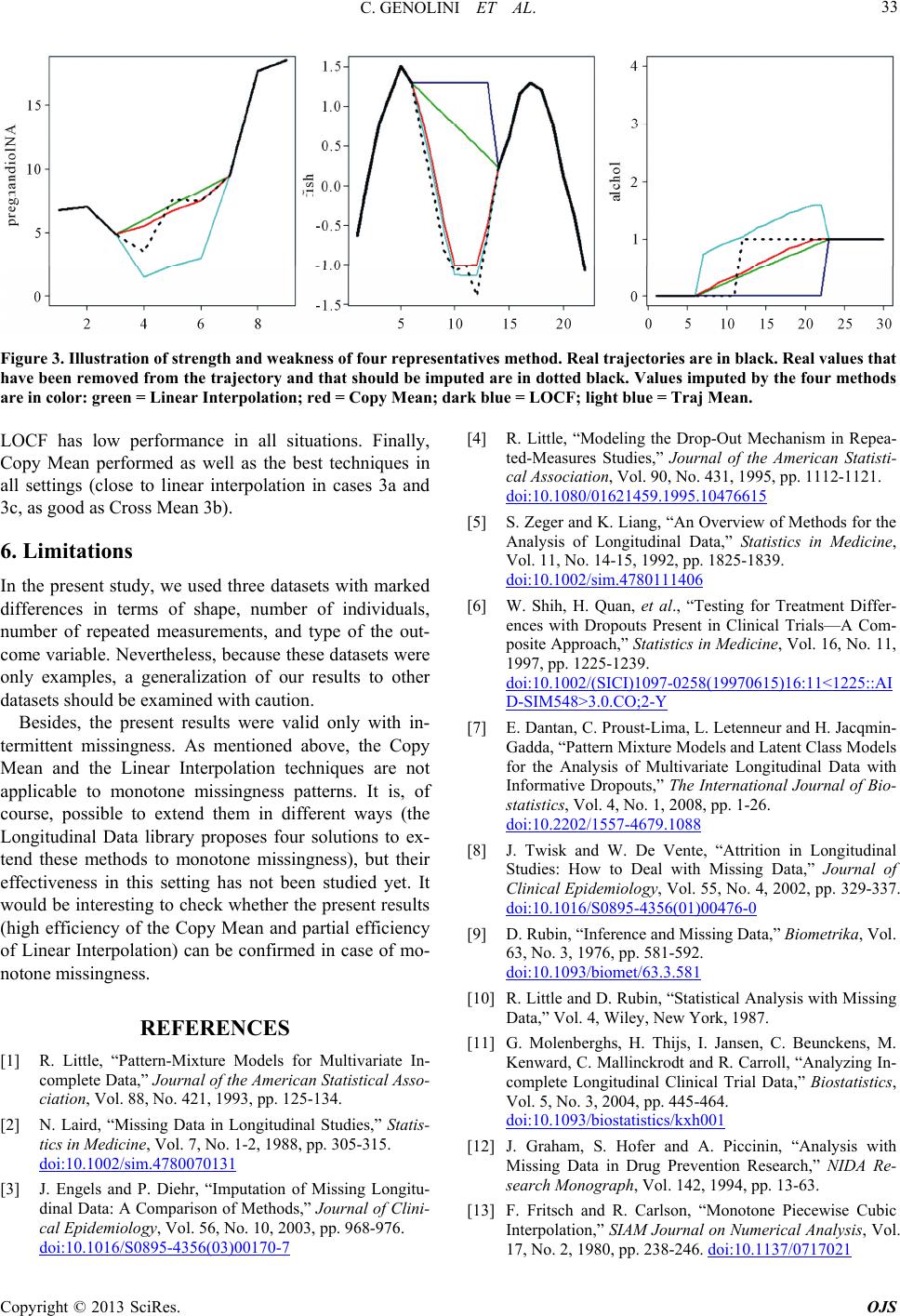 C. GENOLINI ET AL. 33 Figure 3. Illustration of strength and weakness of four representatives method. Real trajectories are in black. Real values that have been removed from the trajectory and that should be imputed are in dotted black. Values imputed by the four methods are in color: green = Linear Interpolation; red = Copy Mean; dark blue = LOCF; light blue = Traj Mean. LOCF has low performance in all situations. Finally, Copy Mean performed as well as the best techniques in all settings (close to linear interpolation in cases 3a and 3c, as good as Cross Mean 3b). 6. Limitations In the present study, we used three datasets with marked differences in terms of shape, number of individuals, number of repeated measurements, and type of the out- come variable. Nevertheless, because these datasets were only examples, a generalization of our results to other datasets should be examined with caution. Besides, the present results were valid only with in- termittent missingness. As mentioned above, the Copy Mean and the Linear Interpolation techniques are not applicable to monotone missingness patterns. It is, of course, possible to extend them in different ways (the Longitudinal Data library proposes four solutions to ex- tend these methods to monotone missingness), but their effectiveness in this setting has not been studied yet. It would be interesting to check whether the present results (high efficiency of the Copy Mean and partial efficiency of Linear Interpolation) can be confirmed in case of mo- notone missingness. REFERENCES [1] R. Little, “Pattern-Mixture Models for Multivariate In- complete Data,” Journal of the American Statistical Asso- ciation, Vol. 88, No. 421, 1993, pp. 125-134. [2] N. Laird, “Missing Data in Longitudinal Studies,” Statis- tics in Medicine, Vol. 7, No. 1-2, 1988, pp. 305-315. doi:10.1002/sim.4780070131 [3] J. Engels and P. Diehr, “Imputation of Missing Longitu- dinal Data: A Comparison of Methods,” Journal of Clini- cal Epidemiology, Vol. 56, No. 10, 2003, pp. 968-976. doi:10.1016/S0895-4356(03)00170-7 [4] R. Little, “Modeling the Drop-Out Mechanism in Repea- ted-Measures Studies,” Journal of the American Statisti- cal Association, Vol. 90, No. 431, 1995, pp. 1112-1121. doi:10.1080/01621459.1995.10476615 [5] S. Zeger and K. Liang, “An Overview of Methods for the Analysis of Longitudinal Data,” Statistics in Medicine, Vol. 11, No. 14-15, 1992, pp. 1825-1839. doi:10.1002/sim.4780111406 [6] W. Shih, H. Quan, et al., “Testing for Treatment Differ- ences with Dropouts Present in Clinical Trials—A Com- posite Approach,” Statistics in Medicine, Vol. 16, No. 11, 1997, pp. 1225-1239. doi:10.1002/(SICI)1097-0258(19970615)16:11<1225::AI D-SIM548>3.0.CO;2-Y [7] E. Dantan, C. Proust-Lima, L. Letenneur and H. Jacqmin- Gadda, “Pattern Mixture Models and Latent Class Models for the Analysis of Multivariate Longitudinal Data with Informative Dropouts,” The International Journal of Bio- statistics, Vol. 4, No. 1, 2008, pp. 1-26. doi:10.2202/1557-4679.1088 [8] J. Twisk and W. De Vente, “Attrition in Longitudinal Studies: How to Deal with Missing Data,” Journal of Clinical Epidemiology, Vol. 55, No. 4, 2002, pp. 329-337. doi:10.1016/S0895-4356(01)00476-0 [9] D. Rubin, “Inference and Missing Data,” Biometrika, Vol. 63, No. 3, 1976, pp. 581-592. doi:10.1093/biomet/63.3.581 [10] R. Little and D. Rubin, “Statistical Analysis with Missing Data,” Vol. 4, Wiley, New York, 1987. [11] G. Molenberghs, H. Thijs, I. Jansen, C. Beunckens, M. Kenward, C. Mallinckrodt and R. Carroll, “Analyzing In- complete Longitudinal Clinical Trial Data,” Biostatistics, Vol. 5, No. 3, 2004, pp. 445-464. doi:10.1093/biostatistics/kxh001 [12] J. Graham, S. Hofer and A. Piccinin, “Analysis with Missing Data in Drug Prevention Research,” NIDA Re- search Monograph, Vol. 142, 1994, pp. 13-63. [13] F. Fritsch and R. Carlson, “Monotone Piecewise Cubic Interpolation,” SIAM Journal on Numerical Analysis, Vol. 17, No. 2, 1980, pp. 238-246. doi:10.1137/0717021 Copyright © 2013 SciRes. OJS  C. GENOLINI ET AL. 34 [14] C. Genolini and B. Falissard, “Kml: k-Means for Longitu- dinal Data,” Computational Statistics, Vol. 25, No. 2, 2010, pp. 317-328. doi:10.1007/s00180-009-0178-4 [15] C. Genolini and B. Falissard, “Kml: A Package to Cluster Longitudinal Data,” Computer Methods and Programs in Biomedicine, Vol. 104, No. 3, 2011, pp. e112-e121. doi:10.1016/j.cmpb.2011.05.008 [16] C. Genolini, J. Pingault, T. Driss, S. Côté, R. Tremblay, F. Vitaro, C. Arnaud and B. Falissard, “KmL3D: A Non-Pa- rametric Algorithm for Clustering Joint Trajectories,” Computer Methods and Programs in Biomedicine, Vol. 109, No. 1, 2012, pp. 104-111. [17] R. Ecochard, H. Boehringer, M. Rabilloud and H. Marret, “Chronological Aspects of Ultrasonic, Hormonal, and Other Indirect Indices of Ovulation,” BJOG: An Interna- tional Journal of Obstetrics & Gynaecology, Vol. 108, No. 8, 2001, pp. 822-829. doi:10.1111/j.1471-0528.2001.00194.x [18] D. Lee, J. Archibald, R. Schoenberger, A. Dennis and D. Shiozawa, “Contour Matching for Fish Species Recogni- tion and Migration Monitoring,” Applications of Compu- tational Intelligence in Biology, Vol. 122, 2008, pp. 183- 207. [19] R. Tremblay, R. Pihl, F. Vitaro, and P. Dobkin, “Predict- ing Early Onset of Male Antisocial Behavior from Pre- school Behavior,” Archives of General Psychiatry, Vol. 51, No. 9, 1994, p. 732. doi:10.1001/archpsyc.1994.03950090064009 [20] O. François and P. Leray, “Generation of Incompliete Test-Data Usinng Bayesinan Networks,” International Joint Conference on Neural Networks, Orlando, 12-17 August 2007, pp. 2391-2396. [21] R Development Core Team, “A Language and Environ- ment for Statistical Computing,” R Foundation for Statis- tical Computing, Vienna, 2012. [22] C. Genolini, “Longitudinal Data,” R Package Version 2.3., 2012. [23] G. Forsythe, M. Malcolm and C. Moler, “Computer Me- thods for Mathematical Computations,” Prentice Hall Professional Technical Reference, 1977. [24] S. Buuren and K. Groothuis-Oudshoorn, “Mice: Multi- variate Imputation by Chained Equations in r,” Journal of Statistical Software, Vol. 45, No. 3, 2011. [25] G. Gadbury, C. Coffey and D. Allison, “Modern Statis- tical Methods for Handling Missing Repeated Measure- ments in Obesity Trial Data: Beyond LOCF,” Obesity Reviews, Vol. 4, No. 3, 2003, pp. 175-184. doi:10.1046/j.1467-789X.2003.00109.x [26] S. Fielding, G. Maclennan, J. Cook and C. Ramsay, “A Review of RCTS in Four Medical Journals to Assess the Use of Imputation to Overcome Missing Data in Quality of Life Outcomes,” Trials, Vol. 9, No. 1, 2008, p. 51. doi:10.1186/1745-6215-9-51 Copyright © 2013 SciRes. OJS 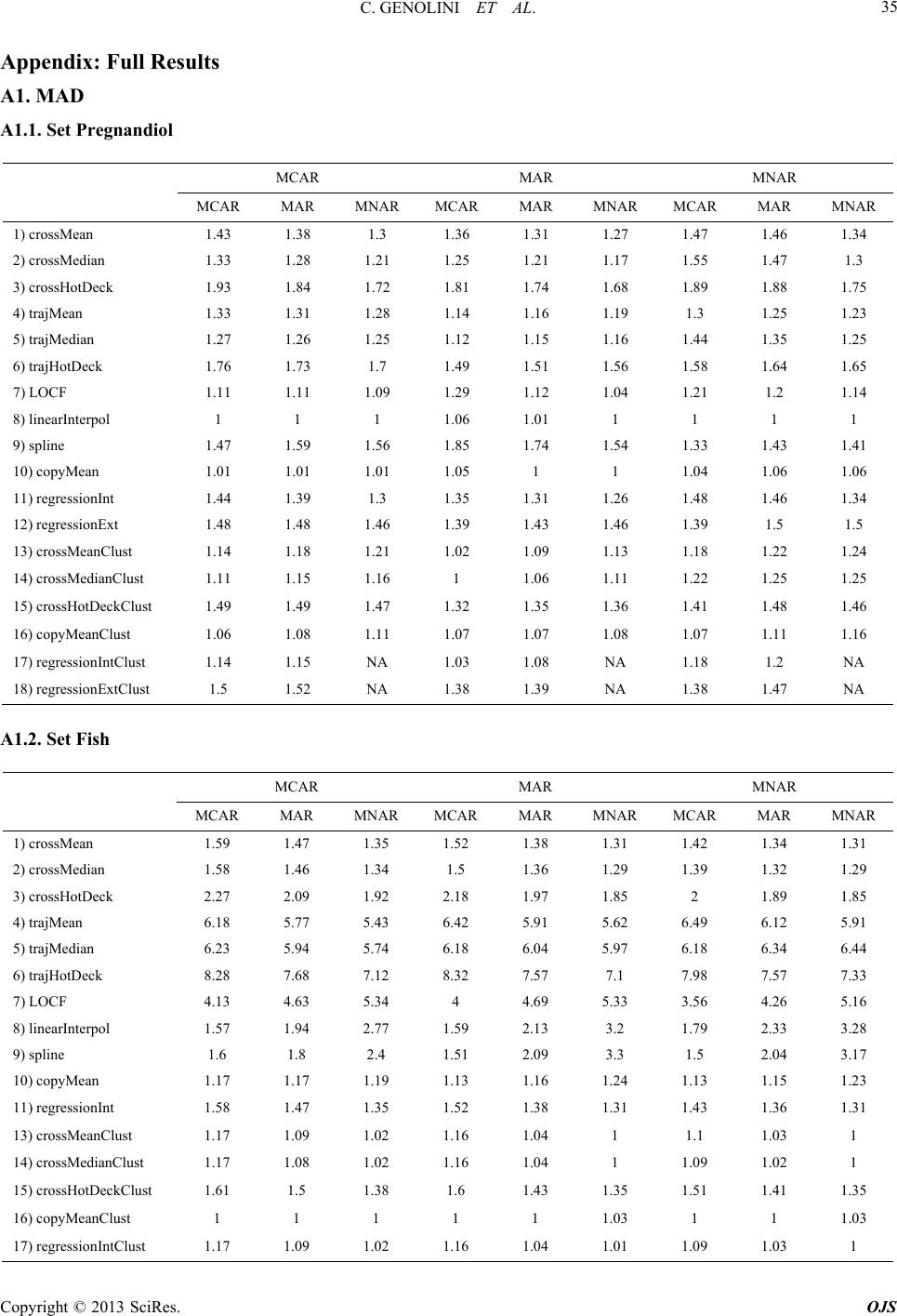 C. GENOLINI ET AL. 35 Appendix: Full Results A1. MAD A1.1. Set Pregnandiol MCAR MAR MNAR MCAR MAR MNAR MCAR MAR MNAR MCAR MAR MNAR 1) crossMean 1.43 1.38 1.3 1.36 1.31 1.27 1.47 1.46 1.34 2) crossMedian 1.33 1.28 1.21 1.25 1.21 1.17 1.55 1.47 1.3 3) crossHotDeck 1.93 1.84 1.72 1.81 1.74 1.68 1.89 1.88 1.75 4) trajMean 1.33 1.31 1.28 1.14 1.16 1.19 1.3 1.25 1.23 5) trajMedian 1.27 1.26 1.25 1.12 1.15 1.16 1.44 1.35 1.25 6) trajHotDeck 1.76 1.73 1.7 1.49 1.51 1.56 1.58 1.64 1.65 7) LOCF 1.11 1.11 1.09 1.29 1.12 1.04 1.21 1.2 1.14 8) linearInterpol 1 1 1 1.06 1.01 1 1 1 1 9) spline 1.47 1.59 1.56 1.85 1.74 1.54 1.33 1.43 1.41 10) copyMean 1.01 1.01 1.01 1.05 1 1 1.04 1.06 1.06 11) regressionInt 1.44 1.39 1.3 1.35 1.31 1.26 1.48 1.46 1.34 12) regressionExt 1.48 1.48 1.46 1.39 1.43 1.46 1.39 1.5 1.5 13) crossMeanClust 1.14 1.18 1.21 1.02 1.09 1.13 1.18 1.22 1.24 14) crossMedianClust 1.11 1.15 1.16 1 1.06 1.11 1.22 1.25 1.25 15) crossHotDeckClust 1.49 1.49 1.47 1.32 1.35 1.36 1.41 1.48 1.46 16) copyMeanClust 1.06 1.08 1.11 1.07 1.07 1.08 1.07 1.11 1.16 17) regressionIntClust 1.14 1.15 NA 1.03 1.08 NA 1.18 1.2 NA 18) regressionExtClust 1.5 1.52 NA 1.38 1.39 NA 1.38 1.47 NA A1.2. Set Fish MCAR MAR MNAR MCAR MAR MNAR MCAR MAR MNAR MCAR MAR MNAR 1) crossMean 1.59 1.47 1.35 1.52 1.38 1.31 1.42 1.34 1.31 2) crossMedian 1.58 1.46 1.34 1.5 1.36 1.29 1.39 1.32 1.29 3) crossHotDeck 2.27 2.09 1.92 2.18 1.97 1.85 2 1.89 1.85 4) trajMean 6.18 5.77 5.43 6.42 5.91 5.62 6.49 6.12 5.91 5) trajMedian 6.23 5.94 5.74 6.18 6.04 5.97 6.18 6.34 6.44 6) trajHotDeck 8.28 7.68 7.12 8.32 7.57 7.1 7.98 7.57 7.33 7) LOCF 4.13 4.63 5.34 4 4.69 5.33 3.56 4.26 5.16 8) linearInterpol 1.57 1.94 2.77 1.59 2.13 3.2 1.79 2.33 3.28 9) spline 1.6 1.8 2.4 1.51 2.09 3.3 1.5 2.04 3.17 10) copyMean 1.17 1.17 1.19 1.13 1.16 1.24 1.13 1.15 1.23 11) regressionInt 1.58 1.47 1.35 1.52 1.38 1.31 1.43 1.36 1.31 13) crossMeanClust 1.17 1.09 1.02 1.16 1.04 1 1.1 1.03 1 14) crossMedianClust 1.17 1.08 1.02 1.16 1.04 1 1.09 1.02 1 15) crossHotDeckClust 1.61 1.5 1.38 1.6 1.43 1.35 1.51 1.41 1.35 16) copyMeanClust 1 1 1 1 1 1.03 1 1 1.03 17) regressionIntClust 1.17 1.09 1.02 1.16 1.04 1.01 1.09 1.03 1 Copyright © 2013 SciRes. OJS 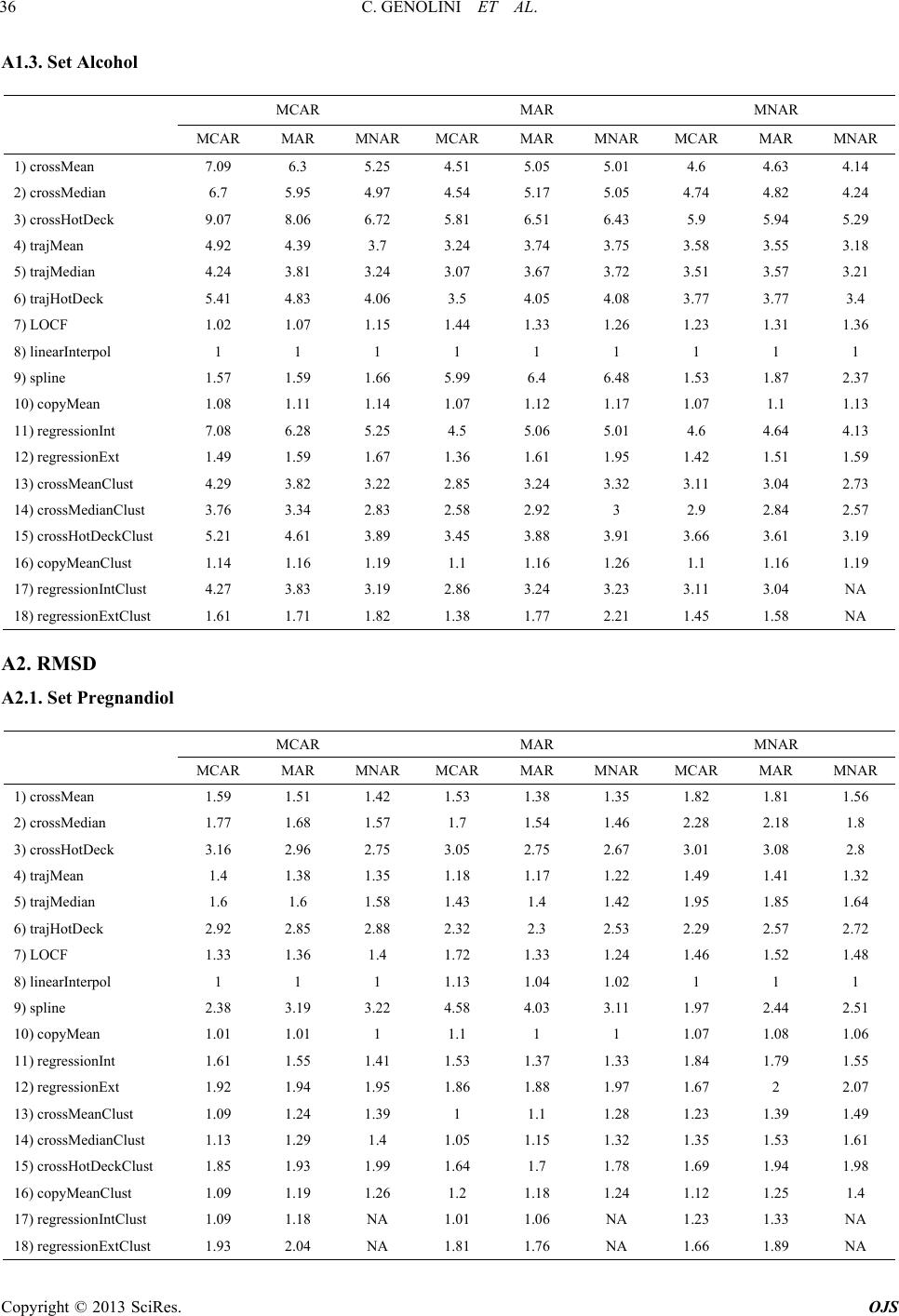 C. GENOLINI ET AL. 36 A1.3. Set Alcohol MCAR MAR MNAR MCAR MAR MNAR MCAR MAR MNAR MCAR MAR MNAR 1) crossMean 7.09 6.3 5.25 4.51 5.05 5.01 4.6 4.63 4.14 2) crossMedian 6.7 5.95 4.97 4.54 5.17 5.05 4.74 4.82 4.24 3) crossHotDeck 9.07 8.06 6.72 5.81 6.51 6.43 5.9 5.94 5.29 4) trajMean 4.92 4.39 3.7 3.24 3.74 3.75 3.58 3.55 3.18 5) trajMedian 4.24 3.81 3.24 3.07 3.67 3.72 3.51 3.57 3.21 6) trajHotDeck 5.41 4.83 4.06 3.5 4.05 4.08 3.77 3.77 3.4 7) LOCF 1.02 1.07 1.15 1.44 1.33 1.26 1.23 1.31 1.36 8) linearInterpol 1 1 1 1 1 1 1 1 1 9) spline 1.57 1.59 1.66 5.99 6.4 6.48 1.53 1.87 2.37 10) copyMean 1.08 1.11 1.14 1.07 1.12 1.17 1.07 1.1 1.13 11) regressionInt 7.08 6.28 5.25 4.5 5.06 5.01 4.6 4.64 4.13 12) regressionExt 1.49 1.59 1.67 1.36 1.61 1.95 1.42 1.51 1.59 13) crossMeanClust 4.29 3.82 3.22 2.85 3.24 3.32 3.11 3.04 2.73 14) crossMedianClust 3.76 3.34 2.83 2.58 2.92 3 2.9 2.84 2.57 15) crossHotDeckClust 5.21 4.61 3.89 3.45 3.88 3.91 3.66 3.61 3.19 16) copyMeanClust 1.14 1.16 1.19 1.1 1.16 1.26 1.1 1.16 1.19 17) regressionIntClust 4.27 3.83 3.19 2.86 3.24 3.23 3.11 3.04 NA 18) regressionExtClust 1.61 1.71 1.82 1.38 1.77 2.21 1.45 1.58 NA A2. RMSD A2.1. Set Pregnandiol MCAR MAR MNAR MCAR MAR MNAR MCAR MAR MNAR MCAR MAR MNAR 1) crossMean 1.59 1.51 1.42 1.53 1.38 1.35 1.82 1.81 1.56 2) crossMedian 1.77 1.68 1.57 1.7 1.54 1.46 2.28 2.18 1.8 3) crossHotDeck 3.16 2.96 2.75 3.05 2.75 2.67 3.01 3.08 2.8 4) trajMean 1.4 1.38 1.35 1.18 1.17 1.22 1.49 1.41 1.32 5) trajMedian 1.6 1.6 1.58 1.43 1.4 1.42 1.95 1.85 1.64 6) trajHotDeck 2.92 2.85 2.88 2.32 2.3 2.53 2.29 2.57 2.72 7) LOCF 1.33 1.36 1.4 1.72 1.33 1.24 1.46 1.52 1.48 8) linearInterpol 1 1 1 1.13 1.04 1.02 1 1 1 9) spline 2.38 3.19 3.22 4.58 4.03 3.11 1.97 2.44 2.51 10) copyMean 1.01 1.01 1 1.1 1 1 1.07 1.08 1.06 11) regressionInt 1.61 1.55 1.41 1.53 1.37 1.33 1.84 1.79 1.55 12) regressionExt 1.92 1.94 1.95 1.86 1.88 1.97 1.67 2 2.07 13) crossMeanClust 1.09 1.24 1.39 1 1.1 1.28 1.23 1.39 1.49 14) crossMedianClust 1.13 1.29 1.4 1.05 1.15 1.32 1.35 1.53 1.61 15) crossHotDeckClust 1.85 1.93 1.99 1.64 1.7 1.78 1.69 1.94 1.98 16) copyMeanClust 1.09 1.19 1.26 1.2 1.18 1.24 1.12 1.25 1.4 17) regressionIntClust 1.09 1.18 NA 1.01 1.06 NA 1.23 1.33 NA 18) regressionExtClust 1.93 2.04 NA 1.81 1.76 NA 1.66 1.89 NA Copyright © 2013 SciRes. OJS  C. GENOLINI ET AL. 37 A2.2. Set Fish MCAR MAR MNAR MCAR MAR MNAR MCAR MAR MNAR MCAR MAR MNAR 1) crossMean 2.43 2.07 1.89 2.16 1.89 1.81 1.96 1.85 1.82 2) crossMedian 2.47 2.1 1.92 2.17 1.9 1.83 1.96 1.85 1.82 3) crossHotDeck 4.83 4.12 3.74 4.34 3.77 3.58 3.83 3.66 3.6 4) trajMean 27.33 23.79 22.68 27.33 24.94 24.26 28.47 27.47 26.89 5) trajMedian 27.88 25.25 25.44 25.31 25.97 27.46 25.88 29.34 31.78 6) trajHotDeck 53.2 45.78 42.33 52.83 45.82 42.73 51.07 47.52 45.27 7) LOCF 12 16.46 24.22 11.88 19.28 26.09 9.36 15.61 23.6 8) linearInterpol 2.28 3.69 8.52 2.23 4.79 11.55 2.81 5.56 11.57 9) spline 2.36 3.37 9.55 2.25 6.19 22.35 2.29 6 20.04 10) copyMean 1.33 1.33 1.54 1.25 1.43 1.75 1.24 1.41 1.71 11) regressionInt 2.38 2.06 1.89 2.17 1.89 1.81 1.96 1.87 1.83 13) crossMeanClust 1.2 1.05 1 1.16 1 1 1.07 1 1.01 14) crossMedianClust 1.22 1.06 1.01 1.18 1.02 1.01 1.08 1.01 1.02 15) crossHotDeckClust 2.28 1.99 1.84 2.21 1.88 1.81 2.06 1.9 1.83 16) copyMeanClust 1 1 1.08 1 1.06 1.18 1 1.06 1.18 17) regressionIntClust 1.2 1.05 1 1.16 1 1.01 1.07 1 1 A2.3. Set Alcohol MCAR MAR MNAR MCAR MAR MNAR MCAR MAR MNAR MCAR MAR MNAR 1) crossMean 8.38 7.39 6.03 5.13 5.75 5.73 4.86 5.09 4.49 2) crossMedian 9.92 8.75 7.16 6.35 7.5 7.41 6.2 6.74 5.87 3) crossHotDeck 16.75 14.77 12.04 9.69 10.84 10.8 8.88 9.33 8.22 4) trajMean 5.28 4.7 3.91 3.43 4.05 4.17 3.73 3.83 3.44 5) trajMedian 7.35 6.55 5.41 4.98 6.03 6.19 5.2 5.56 5 6) trajHotDeck 10.39 9.22 7.59 6.09 7.03 7.16 5.94 6.19 5.53 7) LOCF 1.75 1.84 1.96 2.38 2.14 2.02 1.78 1.99 2.06 8) linearInterpol 1 1.01 1 1 1 1 1 1 1 9) spline 1.54 1.82 2.67 179.22 185.59 174.71 4.26 8.92 15.19 10) copyMean 1 1.01 1 1.03 1.03 1.04 1 1 1.01 11) regressionInt 8.37 7.36 6.02 5.12 5.75 5.73 4.86 5.1 4.48 12) regressionExt 1.95 2.03 2.08 1.73 1.94 2.28 1.63 1.77 1.84 13) crossMeanClust 3.85 3.41 2.86 2.52 2.86 2.89 2.75 2.73 2.4 14) crossMedianClust 4.59 4.07 3.39 2.96 3.37 3.49 3.2 3.24 2.94 15) crossHotDeckClust 7.43 6.53 5.38 4.46 5.03 5.07 4.39 4.48 3.93 16) copyMeanClust 1.01 1 1.01 1.04 1.04 1.08 1.01 1.02 1.04 17) regressionIntClust 3.84 3.41 2.85 2.52 2.87 2.88 2.74 2.74 NA 18) regressionExtClust 2.03 2.13 2.32 1.65 2.09 2.65 1.61 1.83 NA Copyright © 2013 SciRes. OJS 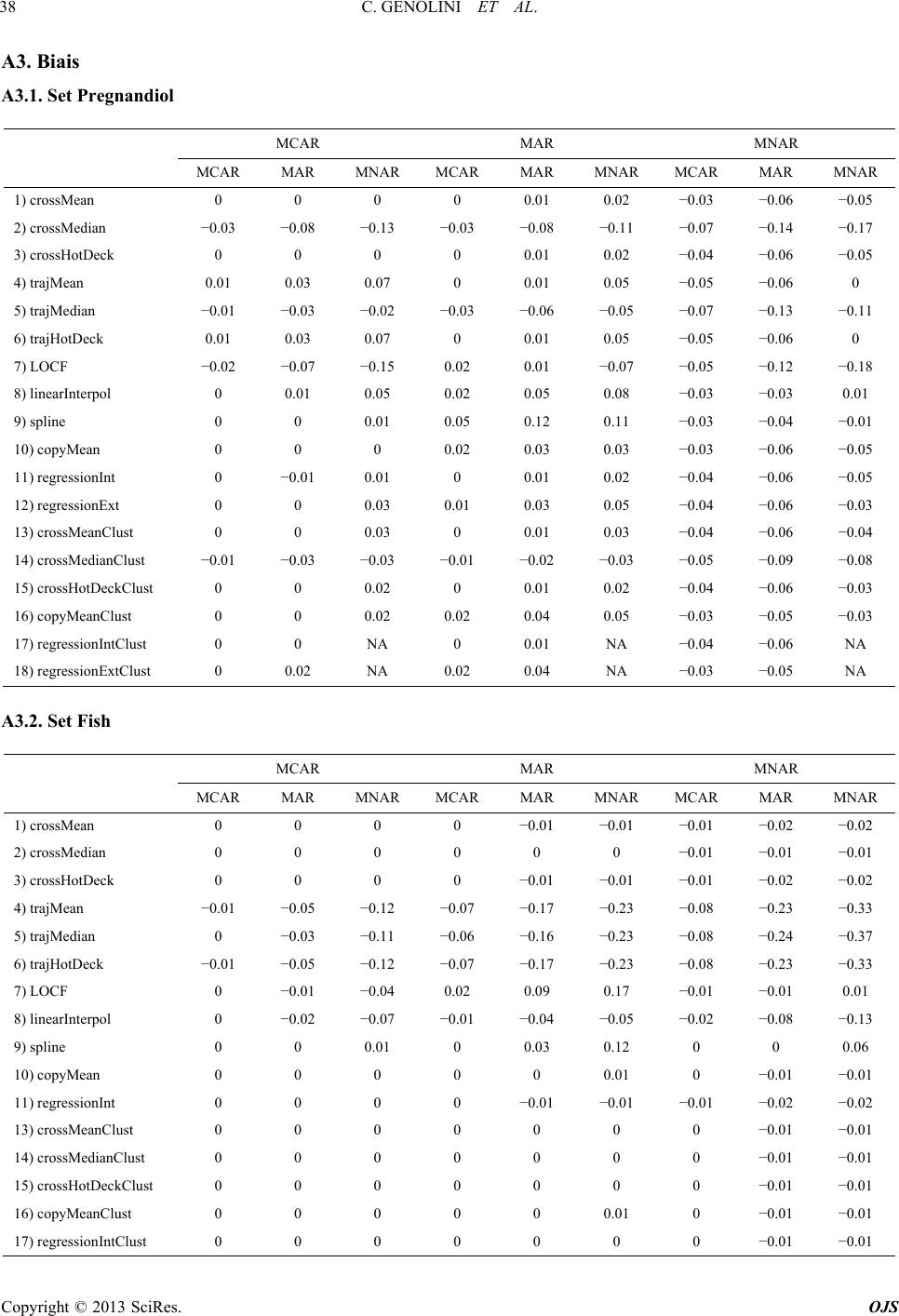 C. GENOLINI ET AL. 38 A3. Biais A3.1. Set Pregnandiol MCAR MAR MNAR MCAR MAR MNAR MCAR MAR MNAR MCAR MAR MNAR 1) crossMean 0 0 0 0 0.01 0.02 −0.03 −0.06 −0.05 2) crossMedian −0.03 −0.08 −0.13 −0.03 −0.08 −0.11 −0.07 −0.14 −0.17 3) crossHotDeck 0 0 0 0 0.01 0.02 −0.04 −0.06 −0.05 4) trajMean 0.01 0.03 0.07 0 0.01 0.05 −0.05 −0.06 0 5) trajMedian −0.01 −0.03 −0.02 −0.03 −0.06 −0.05 −0.07 −0.13 −0.11 6) trajHotDeck 0.01 0.03 0.07 0 0.01 0.05 −0.05 −0.06 0 7) LOCF −0.02 −0.07 −0.15 0.02 0.01 −0.07 −0.05 −0.12 −0.18 8) linearInterpol 0 0.01 0.05 0.02 0.05 0.08 −0.03 −0.03 0.01 9) spline 0 0 0.01 0.05 0.12 0.11 −0.03 −0.04 −0.01 10) copyMean 0 0 0 0.02 0.03 0.03 −0.03 −0.06 −0.05 11) regressionInt 0 −0.01 0.01 0 0.01 0.02 −0.04 −0.06 −0.05 12) regressionExt 0 0 0.03 0.01 0.03 0.05 −0.04 −0.06 −0.03 13) crossMeanClust 0 0 0.03 0 0.01 0.03 −0.04 −0.06 −0.04 14) crossMedianClust −0.01 −0.03 −0.03 −0.01 −0.02 −0.03 −0.05 −0.09 −0.08 15) crossHotDeckClust 0 0 0.02 0 0.01 0.02 −0.04 −0.06 −0.03 16) copyMeanClust 0 0 0.02 0.02 0.04 0.05 −0.03 −0.05 −0.03 17) regressionIntClust 0 0 NA 0 0.01 NA −0.04 −0.06 NA 18) regressionExtClust 0 0.02 NA 0.02 0.04 NA −0.03 −0.05 NA A3.2. Set Fish MCAR MAR MNAR MCAR MAR MNAR MCAR MAR MNAR MCAR MAR MNAR 1) crossMean 0 0 0 0 −0.01 −0.01 −0.01 −0.02 −0.02 2) crossMedian 0 0 0 0 0 0 −0.01 −0.01 −0.01 3) crossHotDeck 0 0 0 0 −0.01 −0.01 −0.01 −0.02 −0.02 4) trajMean −0.01 −0.05 −0.12 −0.07 −0.17 −0.23 −0.08 −0.23 −0.33 5) trajMedian 0 −0.03 −0.11 −0.06 −0.16 −0.23 −0.08 −0.24 −0.37 6) trajHotDeck −0.01 −0.05 −0.12 −0.07 −0.17 −0.23 −0.08 −0.23 −0.33 7) LOCF 0 −0.01 −0.04 0.02 0.09 0.17 −0.01 −0.01 0.01 8) linearInterpol 0 −0.02 −0.07 −0.01 −0.04 −0.05 −0.02 −0.08 −0.13 9) spline 0 0 0.01 0 0.03 0.12 0 0 0.06 10) copyMean 0 0 0 0 0 0.01 0 −0.01 −0.01 11) regressionInt 0 0 0 0 −0.01 −0.01 −0.01 −0.02 −0.02 13) crossMeanClust 0 0 0 0 0 0 0 −0.01 −0.01 14) crossMedianClust 0 0 0 0 0 0 0 −0.01 −0.01 15) crossHotDeckClust 0 0 0 0 0 0 0 −0.01 −0.01 16) copyMeanClust 0 0 0 0 0 0.01 0 −0.01 −0.01 17) regressionIntClust 0 0 0 0 0 0 0 −0.01 −0.01 Copyright © 2013 SciRes. OJS 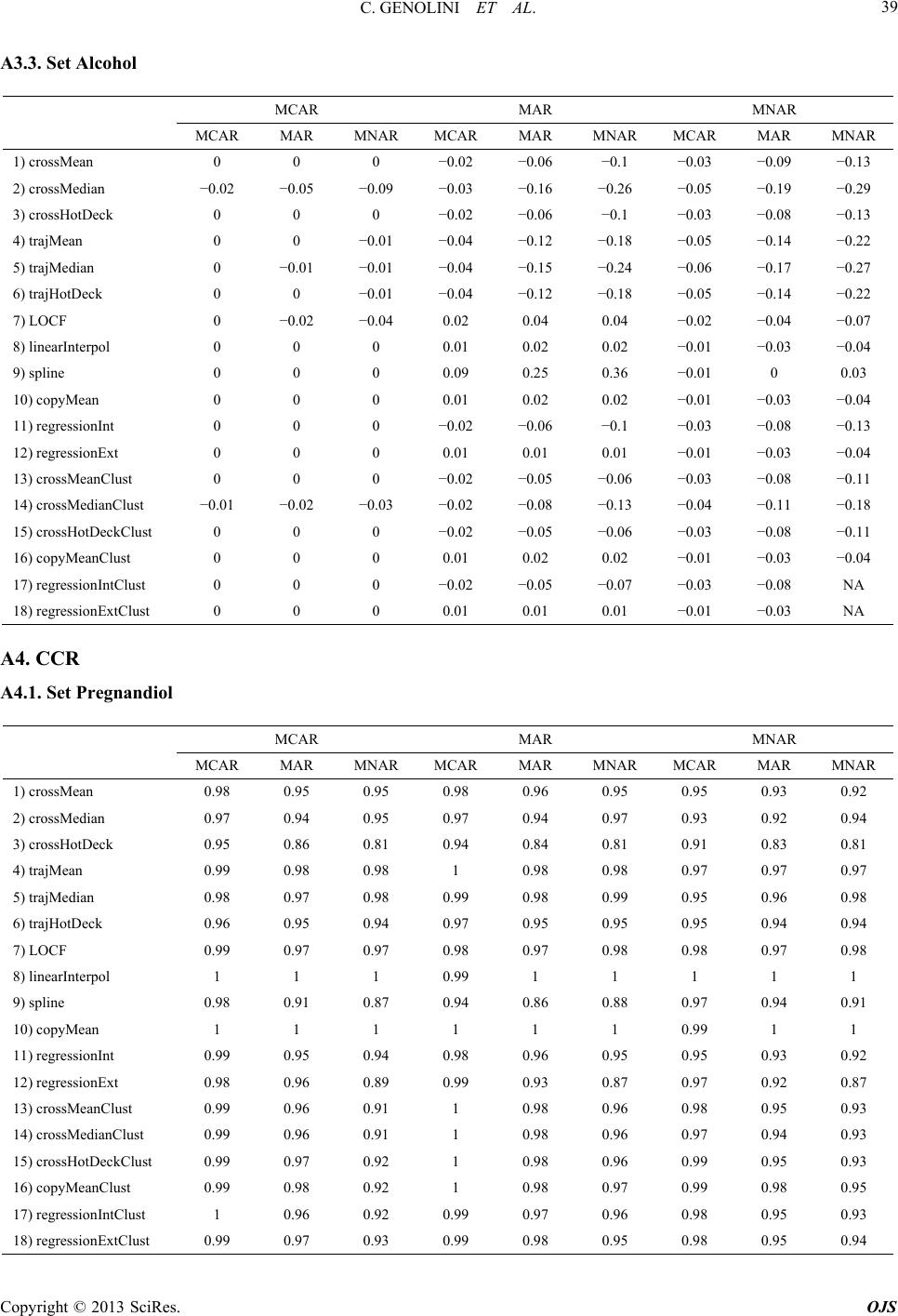 C. GENOLINI ET AL. 39 A3.3. Set Alcohol MCAR MAR MNAR MCAR MAR MNAR MCAR MAR MNAR MCAR MAR MNAR 1) crossMean 0 0 0 −0.02 −0.06 −0.1 −0.03 −0.09 −0.13 2) crossMedian −0.02 −0.05 −0.09 −0.03 −0.16 −0.26 −0.05 −0.19 −0.29 3) crossHotDeck 0 0 0 −0.02 −0.06 −0.1 −0.03 −0.08 −0.13 4) trajMean 0 0 −0.01 −0.04 −0.12 −0.18 −0.05 −0.14 −0.22 5) trajMedian 0 −0.01 −0.01 −0.04 −0.15 −0.24 −0.06 −0.17 −0.27 6) trajHotDeck 0 0 −0.01 −0.04 −0.12 −0.18 −0.05 −0.14 −0.22 7) LOCF 0 −0.02 −0.04 0.02 0.04 0.04 −0.02 −0.04 −0.07 8) linearInterpol 0 0 0 0.01 0.02 0.02 −0.01 −0.03 −0.04 9) spline 0 0 0 0.09 0.25 0.36 −0.01 0 0.03 10) copyMean 0 0 0 0.01 0.02 0.02 −0.01 −0.03 −0.04 11) regressionInt 0 0 0 −0.02 −0.06 −0.1 −0.03 −0.08 −0.13 12) regressionExt 0 0 0 0.01 0.01 0.01 −0.01 −0.03 −0.04 13) crossMeanClust 0 0 0 −0.02 −0.05 −0.06 −0.03 −0.08 −0.11 14) crossMedianClust −0.01 −0.02 −0.03 −0.02 −0.08 −0.13 −0.04 −0.11 −0.18 15) crossHotDeckClust 0 0 0 −0.02 −0.05 −0.06 −0.03 −0.08 −0.11 16) copyMeanClust 0 0 0 0.01 0.02 0.02 −0.01 −0.03 −0.04 17) regressionIntClust 0 0 0 −0.02 −0.05 −0.07 −0.03 −0.08 NA 18) regressionExtClust 0 0 0 0.01 0.01 0.01 −0.01 −0.03 NA A4. CCR A4.1. Set Pregnandiol MCAR MAR MNAR MCAR MAR MNAR MCAR MAR MNAR MCAR MAR MNAR 1) crossMean 0.98 0.95 0.95 0.98 0.96 0.95 0.95 0.93 0.92 2) crossMedian 0.97 0.94 0.95 0.97 0.94 0.97 0.93 0.92 0.94 3) crossHotDeck 0.95 0.86 0.81 0.94 0.84 0.81 0.91 0.83 0.81 4) trajMean 0.99 0.98 0.98 1 0.98 0.98 0.97 0.97 0.97 5) trajMedian 0.98 0.97 0.98 0.99 0.98 0.99 0.95 0.96 0.98 6) trajHotDeck 0.96 0.95 0.94 0.97 0.95 0.95 0.95 0.94 0.94 7) LOCF 0.99 0.97 0.97 0.98 0.97 0.98 0.98 0.97 0.98 8) linearInterpol 1 1 1 0.99 1 1 1 1 1 9) spline 0.98 0.91 0.87 0.94 0.86 0.88 0.97 0.94 0.91 10) copyMean 1 1 1 1 1 1 0.99 1 1 11) regressionInt 0.99 0.95 0.94 0.98 0.96 0.95 0.95 0.93 0.92 12) regressionExt 0.98 0.96 0.89 0.99 0.93 0.87 0.97 0.92 0.87 13) crossMeanClust 0.99 0.96 0.91 1 0.98 0.96 0.98 0.95 0.93 14) crossMedianClust 0.99 0.96 0.91 1 0.98 0.96 0.97 0.94 0.93 15) crossHotDeckClust 0.99 0.97 0.92 1 0.98 0.96 0.99 0.95 0.93 16) copyMeanClust 0.99 0.98 0.92 1 0.98 0.97 0.99 0.98 0.95 17) regressionIntClust 1 0.96 0.92 0.99 0.97 0.96 0.98 0.95 0.93 18) regressionExtClust 0.99 0.97 0.93 0.99 0.98 0.95 0.98 0.95 0.94 Copyright © 2013 SciRes. OJS 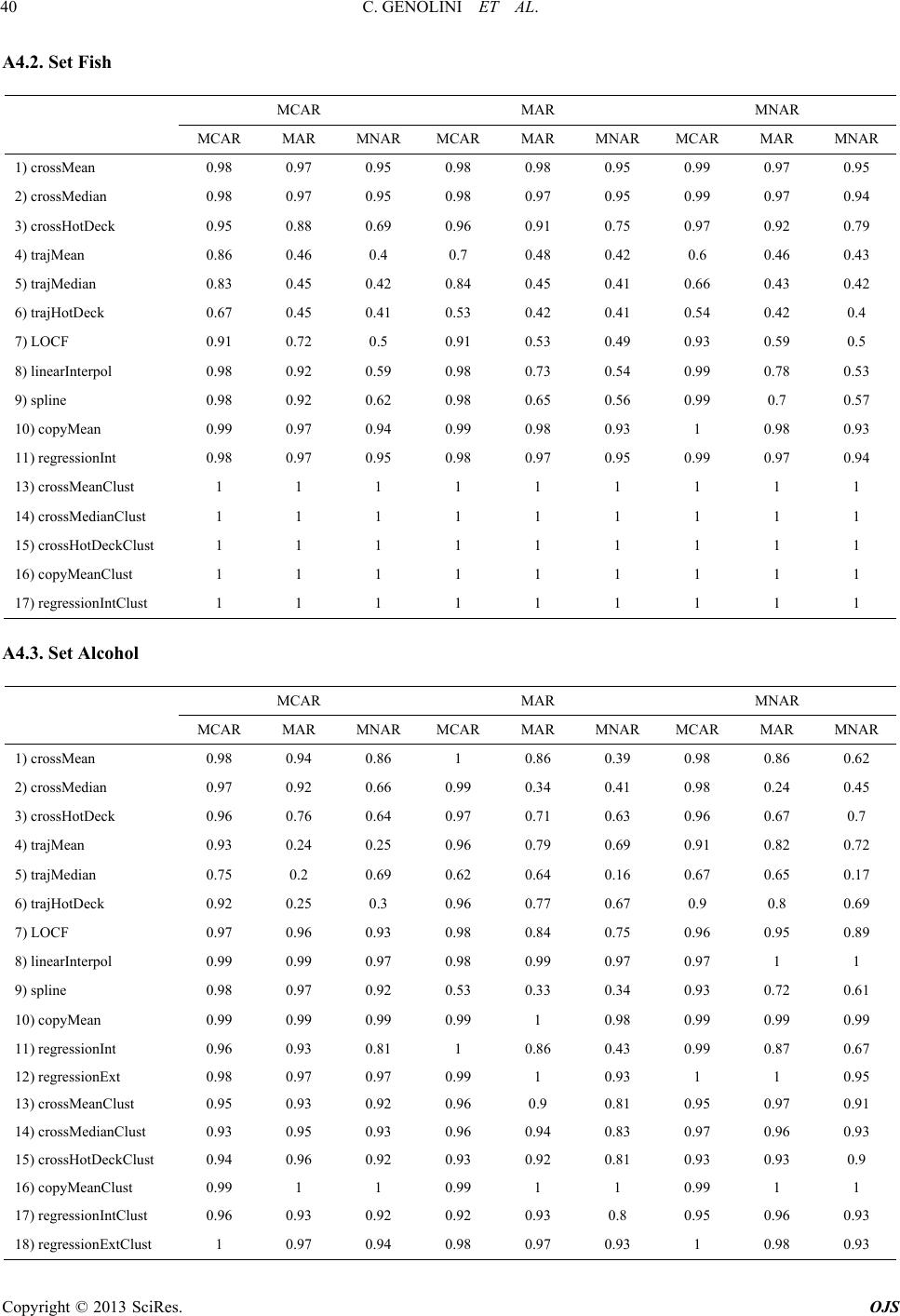 C. GENOLINI ET AL. Copyright © 2013 SciRes. OJS 40 A4.2. Set Fish MCAR MAR MNAR MCAR MAR MNAR MCAR MAR MNAR MCAR MAR MNAR 1) crossMean 0.98 0.97 0.95 0.98 0.98 0.95 0.99 0.97 0.95 2) crossMedian 0.98 0.97 0.95 0.98 0.97 0.95 0.99 0.97 0.94 3) crossHotDeck 0.95 0.88 0.69 0.96 0.91 0.75 0.97 0.92 0.79 4) trajMean 0.86 0.46 0.4 0.7 0.48 0.42 0.6 0.46 0.43 5) trajMedian 0.83 0.45 0.42 0.84 0.45 0.41 0.66 0.43 0.42 6) trajHotDeck 0.67 0.45 0.41 0.53 0.42 0.41 0.54 0.42 0.4 7) LOCF 0.91 0.72 0.5 0.91 0.53 0.49 0.93 0.59 0.5 8) linearInterpol 0.98 0.92 0.59 0.98 0.73 0.54 0.99 0.78 0.53 9) spline 0.98 0.92 0.62 0.98 0.65 0.56 0.99 0.7 0.57 10) copyMean 0.99 0.97 0.94 0.99 0.98 0.93 1 0.98 0.93 11) regressionInt 0.98 0.97 0.95 0.98 0.97 0.95 0.99 0.97 0.94 13) crossMeanClust 1 1 1 1 1 1 1 1 1 14) crossMedianClust 1 1 1 1 1 1 1 1 1 15) crossHotDeckClust 1 1 1 1 1 1 1 1 1 16) copyMeanClust 1 1 1 1 1 1 1 1 1 17) regressionIntClust 1 1 1 1 1 1 1 1 1 A4.3. Set Alcohol MCAR MAR MNAR MCAR MAR MNAR MCAR MAR MNAR MCAR MAR MNAR 1) crossMean 0.98 0.94 0.86 1 0.86 0.39 0.98 0.86 0.62 2) crossMedian 0.97 0.92 0.66 0.99 0.34 0.41 0.98 0.24 0.45 3) crossHotDeck 0.96 0.76 0.64 0.97 0.71 0.63 0.96 0.67 0.7 4) trajMean 0.93 0.24 0.25 0.96 0.79 0.69 0.91 0.82 0.72 5) trajMedian 0.75 0.2 0.69 0.62 0.64 0.16 0.67 0.65 0.17 6) trajHotDeck 0.92 0.25 0.3 0.96 0.77 0.67 0.9 0.8 0.69 7) LOCF 0.97 0.96 0.93 0.98 0.84 0.75 0.96 0.95 0.89 8) linearInterpol 0.99 0.99 0.97 0.98 0.99 0.97 0.97 1 1 9) spline 0.98 0.97 0.92 0.53 0.33 0.34 0.93 0.72 0.61 10) copyMean 0.99 0.99 0.99 0.99 1 0.98 0.99 0.99 0.99 11) regressionInt 0.96 0.93 0.81 1 0.86 0.43 0.99 0.87 0.67 12) regressionExt 0.98 0.97 0.97 0.99 1 0.93 1 1 0.95 13) crossMeanClust 0.95 0.93 0.92 0.96 0.9 0.81 0.95 0.97 0.91 14) crossMedianClust 0.93 0.95 0.93 0.96 0.94 0.83 0.97 0.96 0.93 15) crossHotDeckClust 0.94 0.96 0.92 0.93 0.92 0.81 0.93 0.93 0.9 16) copyMeanClust 0.99 1 1 0.99 1 1 0.99 1 1 17) regressionIntClust 0.96 0.93 0.92 0.92 0.93 0.8 0.95 0.96 0.93 18) regressionExtClust 1 0.97 0.94 0.98 0.97 0.93 1 0.98 0.93
|