 Open Journal of Metal, 2013, 3, 51-61 http://dx.doi.org/10.4236/ojmetal.2013.32A1007 Published Online July 2013 (http://www.scirp.org/journal/ojmetal) Apatite and Chitin Amendments Promote Microbial Activity and Augment Metal Removal in Marine Sediments Jinjun Kan1*, Anna Obraztsova2, Yanbing Wang3, Jim Leather2, Kirk G. Scheckel4, Kenneth H. Nealson3, Y. Meriah Arias-Thode2* 1Stroud Water Research Center, Avondale, USA 2SPAWAR Systems Center-Pacific, San Diego, USA 3Department of Earth Sciences, University of Southern California, Los Angeles, USA 4National Risk Management Research Laboratory, Environmental Protection Agency, Cincinnati, USA Email: *jkan@stroudcenter.org, meriah.ariasthode@navy.mil Received May 4, 2013; revised June 12, 2013; accepted June 19, 2013 Copyright © 2013 Jinjun Kan et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. ABSTRACT In situ amendments are a promising approach to enhance removal of metal contaminants from diverse environments including soil, groundwater and sediments. Apatite and chitin were selected and tested for copper, chromium, and zinc metal removal in marine sediment samples. Microbiological, molecular biological and chemical analyses were applied to investigate the role of these amendments in metal immobilization processes. Both apatite and chitin promoted micro- bial growth. These amendments induced corresponding bacterial groups including sulfide producers, iron reducers, and phosphate solubilizers; all that facilitated heavy metal immobilization and removal from marine sediments. Molecular biological approaches showed chitin greatly induced microbial population shifts in sediments and overlying water: chi- tin only, or chitin with apatite induced growth of bacterial groups such as Acidobacteria, Betaproteobacteria, Epsilon- proteobacteria, Firmicutes, Planctomycetes, Rhodospirillaceae, Spirochaetes, and Verrucomicrobia; whereas these bacteria were not present in the control. Community structures were also altered under treatments with increase of rela- tive abundance of Deltaproteobacteria and decrease of Actinobacteria, Alphaproteobacteria, and Nitrospirae. Many of these groups of bacteria have been shown to be involved in metal reduction and immobilization. Chemical analysis of pore- and overlying water also demonstrated metal immobilization primarily under chitin treatments. X-Ray absorp- tion spectroscopy (XAS) spectra showed more sorbed Zn occurred over time in both apatite and chitin treatments (from 9% - 27%). The amendments improved zinc immobilization in marine sediments that led to significant changes in the mineralogy: easily mobile Zn hydroxide phase was converted to an immobile Zn phosphate (hopeite). In-situ amendment of apatite and chitin offers a great bioremediation potential for marine sediments contaminated with heavy metals. Keywords: Apatite; Chitin; Amendments; Marine Sediment; DGGE; Microbial Community; Copper; Zinc; Chromium 1. Introduction In situ amendments have been shown as an effective ap- proach to enhance removal of metal or organic contami- nants under a variety of environments [1-6]. Currently, both inorganic and organic materials are widely used as promising sediment amendments; including geotextile mats, apatite, organoclay, and chitin. Inorganic amend- ments help to immobilize several toxic metals such as lead, zinc, and cadmium by forming stable compounds that render the metals no longer bioavailable [2,3,7]. Or- ganic amendments, in contrast, may induce indigenous microbes capable of bio-reduction and/or of direct or indirect immobilization of toxic metals [1,4,8,9]. Apatite, an inorganic phosphate mineral, has been ap- plied to facilitate the immobilization of several toxic metals including Cu, Zn and Pb [10-12]. For example, apatite amendments have a long remedial history at met- als contaminated soils sites [10,13,14]. Apatite’s use in sediment remediation is more recent [15]. The DoD (Navy) is currently evaluating apatite amendments at a *Corresponding author. C opyright © 2013 SciRes. OJMetal  J. KAN ET AL. 52 metal recycling facility to remediate high Zn levels in nearby sediments [16]. Phosphates released from apatite will sequester metals to form more stable metalphosphate complexes [3]. This process may be linked to activities of phosphate-solubilizing bacteria. Although marine en- vironments are not phosphate-limited, recently Uzair and Ahmed [17] have successfully isolated both attached and free-living marine bacteria that were able to solubilize phos- phate compounds when given suitable carbon sources. Meanwhile chitin, a homopolymer of N-acetylgluco- samine (NAG), is an abundant structural polysaccharide produced by many marine organisms. Several experi- ments have demonstrated that bacterial communities shift in response to amended chitin [18] and indigenous bacte- ria were capable of cleaving this polymer via the synthe- sis of specific enzymes that were induced by chitin [19]. Further, breakdown products (e.g. acetate and fructose) from chitin degradation may serve as ideal carbon sources for anaerobic and facultative microorganisms capable of metal reduction [20]. It is thus reasonable to expect that we hypothesize that amendment of apatite and/or chitin will stimulate bacterial growth, and subsequently en- hance metal transition, immobilization and sequestration in marine environments. Metabolically diverse indigenous microorganisms in- teract with metals in a variety of ways that lead to de- creased metal solubility and mobility [21-22]. Among these microbes, iron-reducing and sulfide-producing bac- teria (FeRB and SPB, respectively) are two biogeo- chemically important groups that have been shown to contain suitable physiology for metal precipitation and immobilization due to their metabolic end-products such as Fe(II) and HS− [23-29]. In addition to the effectiveness of bioremediation, one other critical perspective of inorganic or organic materi- als used as in situ amendment remediation alternatives, is to limit/avoid the impacts on living organisms. Therefore, during our primary study, we tested how the inorganic and organic amendments impacted microbial communi- ties [30] and impacted the macro-invertebrate communi- ties [31]. Dose-response experiments of candidate mate- rials suggested that appropriate concentrations of apatite and chitin (5% and 0.5%, respectively) amendments had little or no negative impacts on water quality and ambient living organisms [31]. In this study, apatite and chitin were applied to micro- cosms containing pristine, coarse, sandy sediments from Yaquina Bay (YB), Oregon, and contaminated, fine- grained sediments from Mare Island (MI) Naval Ship- yard, California, with and without spiked heavy metals. Responses of the microbial communities (biomass and population structures) were monitored in the treated sedi- ments. Chemical analysis, including ICP-MS and X-ray absorption spectrometry was used to determine the ef- fects of the amendments on the sediment composition and zinc speciation. 2. Experimental 2.1. Sediment Description Pristine and uncontaminated, sandy, coarse sediments from Yaquina Bay (YB), OR, and historically heavy metal contaminated, fine-grained sediments from Mare Island (MI) Naval Shipyard, CA, were evaluated [29]. Major properties of both sediments including percentage of silt/clay, TOC (total organic carbon), and metals were listed in a previous study [31]. Because the MI sediments were aged, additional spikes of Cu, Zn, and Cr were in- corporated with the goal of metal concentrations in the overlying water of Cu = 0.25 ppm, and Zn and Cr = 1.0 ppm (Table 1). These concentrations were used as a sur- rogate for current metal contaminated sites. The metal additions were added with the assumption of no binding as TOC of coarse sand is less than 0.1%. The sediment types and treatments applied in this study are listed in Table 1. 2.2. Amendment Experiments with Apatite and Chitin Mesocosm preparation and experimental setups followed previously described methods [30-31]. Briefly, for each treatment, 150 g of sediment was added to each of 5 rep- licate beakers. A sixth replicate was used for monitoring daily water. Approximately 750 mL of uncontaminated and filtered (0.45 μm Millipor filter) natural seawater (salinity at 30 psu) collected from near the mouth of San Diego Bay, was added to each jar. A 3-day equilibration period preceded the experiments. All beakers were gently and continuously aerated. Based on the previous toxicological dose-response experiments and microbial response studies [30-31], apa- tite (5%) and chitin (0.5%) were selected as candidate amendment treatments. Concentrations of amendments and treatment setups are described in Table 1. Negative controls contained unspiked and unamended YB and MI sediments (Table 1; CUU and FUU, respectively). 2.3. Microbiological Sample Collection and Analyses Water and sediment core samples were collected for all analyses at certain time points as required. Because many metal-microbe interactions occur in the overlying water and surface sediments, overlying water and pore-waters were analyzed for bacterial populations. Detailed de- scriptions of sample collections and treat ments are pre- viously described [30]. Samples for molecular and mi- crobiology analyses were collected at Day 0 and Day 28. Copyright © 2013 SciRes. OJMetal 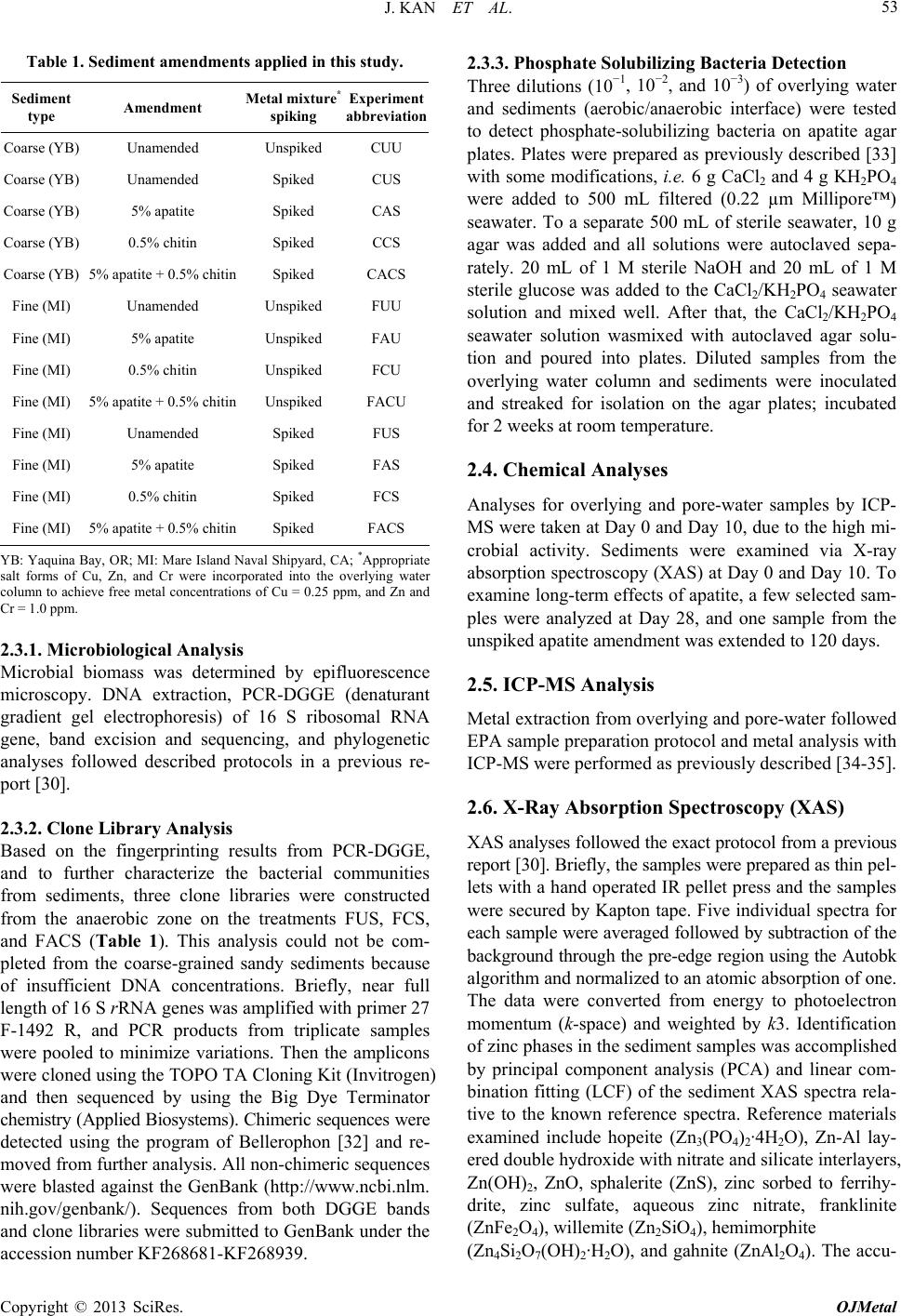 J. KAN ET AL. 53 Table 1. Sediment amendments applied in this study. Sediment type Amendment Metal mixture* spiking Experiment abbreviation Coarse (YB) Unamended Unspiked CUU Coarse (YB) Unamended Spiked CUS Coarse (YB) 5% apatite Spiked CAS Coarse (YB) 0.5% chitin Spiked CCS Coarse (YB) 5% apatite + 0.5% chitinSpiked CACS Fine (MI) Unamended Unspiked FUU Fine (MI) 5% apatite Unspiked FAU Fine (MI) 0.5% chitin Unspiked FCU Fine (MI) 5% apatite + 0.5% chitinUnspiked FACU Fine (MI) Unamended Spiked FUS Fine (MI) 5% apatite Spiked FAS Fine (MI) 0.5% chitin Spiked FCS Fine (MI) 5% apatite + 0.5% chitinSpiked FACS YB: Yaquina Bay, OR; MI: Mare Island Naval Shipyard, CA; *Appropriate salt forms of Cu, Zn, and Cr were incorporated into the overlying water column to achieve free metal concentrations of Cu = 0.25 ppm, and Zn and Cr = 1.0 ppm. 2.3.1. Microbiological Analysis Microbial biomass was determined by epifluorescence microscopy. DNA extraction, PCR-DGGE (denaturant gradient gel electrophoresis) of 16 S ribosomal RNA gene, band excision and sequencing, and phylogenetic analyses followed described protocols in a previous re- port [30]. 2.3.2. Clone Library Analysis Based on the fingerprinting results from PCR-DGGE, and to further characterize the bacterial communities from sediments, three clone libraries were constructed from the anaerobic zone on the treatments FUS, FCS, and FACS (Table 1). This analysis could not be com- pleted from the coarse-grained sandy sediments because of insufficient DNA concentrations. Briefly, near full length of 16 S rRNA genes was amplified with primer 27 F-1492 R, and PCR products from triplicate samples were pooled to minimize variations. Then the amplicons were cloned using the TOPO TA Cloning Kit (Invitrogen) and then sequenced by using the Big Dye Terminator chemistry (Applied Biosystems). Chimeric sequences were detected using the program of Bellerophon [32] and re- moved from further analysis. All non-chimeric sequences were blasted against the GenBank (http://www.ncbi.nlm. nih.gov/genbank/). Sequences from both DGGE bands and clone libraries were submitted to GenBank under the accession number KF268681-KF268939. 2.3.3. Phosphate Solubilizing Bacteria Detection Three dilutions (10−1, 10−2, and 10−3) of overlying water and sediments (aerobic/anaerobic interface) were tested to detect phosphate-solubilizing bacteria on apatite agar plates. Plates were prepared as previously described [33] with some modifications, i.e. 6 g CaCl2 and 4 g KH2PO4 were added to 500 mL filtered (0.22 µm Millipore™) seawater. To a separate 500 mL of sterile seawater, 10 g agar was added and all solutions were autoclaved sepa- rately. 20 mL of 1 M sterile NaOH and 20 mL of 1 M sterile glucose was added to the CaCl2/KH2PO4 seawater solution and mixed well. After that, the CaCl2/KH2PO4 seawater solution wasmixed with autoclaved agar solu- tion and poured into plates. Diluted samples from the overlying water column and sediments were inoculated and streaked for isolation on the agar plates; incubated for 2 weeks at room temperature. 2.4. Chemical Analyses Analyses for overlying and pore-water samples by ICP- MS were taken at Day 0 and Day 10, due to the high mi- crobial activity. Sediments were examined via X-ray absorption spectroscopy (XAS) at Day 0 and Day 10. To examine long-term effects of apatite, a few selected sam- ples were analyzed at Day 28, and one sample from the unspiked apatite amendment was extended to 120 days. 2.5. ICP-MS Analysis Metal extraction from overlying and pore-water followed EPA sample preparation protocol and metal analysis with ICP-MS were performed as previously described [34-35]. 2.6. X-Ray Absorption Spectroscopy (XAS) XAS analyses followed the exact protocol from a previous report [30]. Briefly, the samples were prepared as thin pel- lets with a hand operated IR pellet press and the samples were secured by Kapton tape. Five individual spectra for each sample were averaged followed by subtraction of the background through the pre-edge region using the Autobk algorithm and normalized to an atomic absorption of one. The data were converted from energy to photoelectron momentum (k-space) and weighted by k3. Identification of zinc phases in the sediment samples was accomplished by principal component analysis (PCA) and linear com- bination fitting (LCF) of the sediment XAS spectra rela- tive to the known reference spectra. Reference materials examined include hopeite (Zn3(PO4)2·4H2O), Zn-Al lay- ered double hydroxide with nitrate and silicate interlayers, Zn(OH)2, ZnO, sphalerite (ZnS), zinc sorbed to ferrihy- drite, zinc sulfate, aqueous zinc nitrate, franklinite (ZnFe2O4), willemite (Zn2SiO4), hemimorphite (Zn4Si2O7(OH)2·H2O), and gahnite (ZnAl2O4). The accu- Copyright © 2013 SciRes. OJMetal  J. KAN ET AL. 54 racy of LCF results was estimated to be 10% - 15% [36]. 3. Results and Discussion 3.1. Microbial Population Analysis Total chitin production by crustaceans in marine envi- ronments are estimated to reach up to 109 t per year [37], and most of the chitin (90%) is digested within 150 h in water column [38]. In marine sediments, chitin degrada- tion is slower and the breakdown process is primarily accomplished by indigenous bacteria with capability of digesting chitin extracellularly [39]. Therefore, it is ex- pected that high concentrations of chitin will favor growth and enrichment of certain bacterial groups. In this study, substantial increases of microbial cell numbers as- sociated with chitin amendments were observed after a 28-day incubation (Figure 1). Epifluorescence microsco- pic counts showed an increase in cell numbers in overly- ing waters from all treatments containing chitin: CCS, CACS, FCU and FCS. The numbers were from 0.38 × 106 cells/mL (control) to 3.27 × 106, 1.85 × 106, 1.08 × 106, 2.42 × 106 cells/mL, respectively. However, cell den- sity in the fine-grained sediments in the apatite plus chi- tin amendments, FACU and FACS, did not demonstrate statistically significant growth compared to the control. Chitin has been widely used as a bio-control material in soils in order to reduce parasitic diseases. Previous reports showed that chitin amendment was associated with an increase of bacterial populations with chitinolytic activities such as actinomycetes [40-41]. Major break- down products from chitin are acetate and fructose [20], both of which may serve as carbon sources for anaerobic and facultative microorganisms. Recently, Kanzog and colleagues [18] reported that both microbial numbers and chitinolytic activity significantly increased in the sedi- ment samples enriched with chitin in an experiment con- ducted in deep-sea sediment, indicating that chitin en- hanced microbial growth, enzymatic activity, and popu- lation structures, as well. Therefore, chitin may play an important role by providing a slow-release carbon source to maintain microbial activity. 3.2. Microbial Community Structure Several studies have already shown that microbial com- munity structures respond to variations of organic amend- ments such as chitin [18,40,41]. PCR-DGGE in combi- nation with DNA sequencing aided in identifying the predominant bacterial populations and to provide a com- prehensive picture of microbial community structure over time and under different amendment and metal con- ditions. Our study provides a “snapshot” for shift of bac- terial population structures under chitin amendments. Analysis of DGGE band profiles has indicated that Figure 1. Microscopic cell counts in overlying waters after 28 days incubation. X axis showed the treatments as listed in Table 1. *Significant difference compared to the control (unpaired t-test, p < 0.05). Data from treatments CAS, FAU, and FAS were not available. chitin amendments shifted the bacterial population struc- tures significantly in overlying waters (Figure 2). Phaeo- bacter (band 1) and Roseobacter (band 2) were the pre- dominant groups in the controls (CUU and FUU), while more distinct bacterial groups were present after chitin and/or apatite treatments in both sediment types (CCS, CACS, FCU, FACU, FCS and FACS, Figures 2 and 3). Most of these enriched bands (No. 5, 6, 8, 9, 10, 12, 14, 15, etc.) belonged to subgroups of Alphaproteobacteria (Figures 2 and 3). However, Betaproteobacteium (Band No. 3 Figure 3), was identified in most samples (CUU, CUS, CAS, FAU, FUS, and FCS). In fact, DGGE profiles from apatite and/or spiked metals were remarkably simi- lar to those from control sediments. Therefore, compared to chitin, addition of apatite or metals had less impact on the bacterial population structures (Figures 2 and 3). For sediments, more distinct and diverse bacterial groups were enriched in both aerobic/anaerobic interface and anaerobic zone sediments (Figure S1). As an exam- ple, in coarse-grained (YB) surface sediments, Bacter- oidetes (bands 19, 20), Spirochaetes (band 21), and Ro- seobacter from Alphaproteobacteria (band 22) were pre- sent in both chitin, and chitin + apatite treatments (CCS and CACS, Figures S1(a) and S2). In contrast, Del- taproteobacteria (bands 27, 28) and Bacteroidetes (bands 25, 26) dominated in anaerobic zone sediments from the same treatments (CCS and CACS, Figures S1(b) & S2). Due to the detection limit and sensitivity of DGGE ap- proach, treatments from fine sediments (MI) didn’t gen- erate distinguishable banding patterns and therefore, we further characterized bacterial community structures by clone library. As we expected, remarkable population shifts were observed from clone library analyses (Table 2 and Fig- ure 4). Actinobacteria, Alphaproteobacteria, Gam- maproteobacteria, Deltaproteobacteria, and Bacteroide- tes were predominant groups in unamended fine-grained sediments with spiked metals (FUS). After 28 days in- Copyright © 2013 SciRes. OJMetal  J. KAN ET AL. 55 Table 2. Bacterial population structures characterized by clone library. Relative abundance was based on the occur- rence of representative sequences. - = not detected. Relative abundance (%) Phylum FUS FCS FACS Impact Acidobacteria - - 1.9 Induced Actinobacteria 20.8 8.6 3.8 Decreased Alphaproteobacteria 22.9 14.3 13.2 Decreased Betaproteobacteria - 2.9 1.9 Induced Gammaproteobacteria 22.9 10 20.8 Decreased Amphritea 2.1 - 1.9 Marinobacterium 2.1 - - Oceanospirillum 14.6 1.4 - Paramoritella - - 3.8 Pseudomonas 2.1 - - Shewanella - - 1.9 Uncultured 2.1 8.6 13.2 Deltaproteobacteria 14.6 30 28.3 Decreased Desulfotalea - 1.4 3.8 Desulfovibrio - 5.7 3.8 Desulfuromonas - 5.7 - Desulfobacula - 1.4 1.9 Geobacter - 1.4 - Pelobacter 2.1 - - Uncultured 12.5 14.3 18.9 Epsilonproteobacteria - 4.3 9.4 Induced Bacteroidetes 10.4 10 11.3 Chloroflexi 2.1 1.4 1.9 Firmicutes - 2.9 5.7 Induced Nitrospirae 6.3 4.3 - Decreased Planctomycete - 5.7 - Induced Rhodospirillaceae - - 1.9 Induced Spirochaeta - 4.3 - Induced Verrucomicrobia - 1.4 - Induced cubation with chitin and chitin + apatite, distinct bacterial groups were induced, including Acidobacteria, Betapro- teobacteria, Epsilonproteobacteria, Firmicutes, Planc- tomycetes, Rhodospirillaceae, Spirochaetes, and Verru- comicrobia, which were not detected in the unamended treatment FUS (Table 2). To date, it is not easy to link the functionality of these bacterial groups with their ge- netic identification. However, high occurrence of these groups of microorganisms under heavy metal concentra- tions, e.g. Firmicutes indicated the potential direct or C U U C U S C C S C A C S F U U F C U F A C U F U S F C S F A C S C A S F A S F A U Figure 2. DGGE fingerprints of bacterial community from overlying waters (W). Labels referred to the treatments. Sample FAS failed to obtain PCR products. Bands 1-15 were selected and excised for sequencing. anoa haeum equitans A 318041 DGGE band 3 Uncultured betaproteobacterium GQ34109 DGGE band 8 Uncultured bacterium FN421572 Uncultured bacterium FJ202706 Uncultured bacterium EU183981 DGGE band 7 Uncultured Terasakiella sp. FJ753149 DGGE band 6 Donghicola eburneus DQ667965 Uncultured bacterium EF574092 Donghicola sp. EF587950 DGGE band 12 Uncultured marine bacterium AF177550 DGGE band 1 Phaeobacter sp. AB498882 DGGE band 13 Uncultured bacterium GU118445 Uncultured bacterium GU119459 DGGE band 15 Maritimibacter sp. EU052764 Uncultured alphaproteobacterium AY145603 Uncultured bacterium AJ319859 Psdeudoruegeria sp. FJ374173 Uncultured alphaproteobacterium GQ204861 DGGE band 9 DGGE band 14 Uncultured bacterium GU066436 DGGE band 10 Rhodobacteraceae bacterium AM990789 Rhodobacteraceae bacterium FJ460070 DGGE band 5 Uncultured alphaproteobacterium AM850968 DGGE band 2 DGGE band 4 DGGE band 11 Marine bacterium AF359535 Roseobacter sp. AY576690 0.05 100 100 90 71 99 79 93 70 99 99 79 90 89 Beta Alpha Figure 3. Phylogenetic analysis (DNA distance-NJ) of bac- terial DGGE band sequences obtained from overlying wa- ters. Bootstrap values were based on analyses of 1000 re- sampling of dataset. Nanoarchaeumequitans was used as an outgroup. Scale bar represent 0.05 substitutions per site. indirect involvement with metal immobilization or bio- remediation [42-43]. Further, over time Deltaproteobac- teria population increased in abundance, while that of Actinobacteria, Alphaproteobacteria, Nitrospirae de- creased (Table 2 and Figure 4). In fact, Deltaproteobac- teria became the most dominant groups of bacteria after Copyright © 2013 SciRes. OJMetal  J. KAN ET AL. 56 Figure 4. Clone library analyses of bacterial population structures from treatments FUS, FCS and FACS. 28 days incubation in both FCS (30%) and FAC (28.3%) (Table 2). Predominant sulfate-reducing bacteria (i.e. Desulfovibrio, Desulfobacula, Desulfotalea) and sulfur- reducing bacteria (i.e. Desulfuromonas) sequences were detected in Deltaproteobacteria class. These results agreed with previous observations on increasing sulfate-reduc- ing bacterial communities and activities in response to carbon source amendments [44]. Primarily due to their metabolic end-products such as sulfide, sulfate/sulfur re- ducing bacteria were able to facilitate metal sequestration and immobilization [24,45]. In addition, these bacteria have been noted for their capability of utilizing metals as electron acceptors via dissimilatory metal reduction [28- 29]. A good example is Geobacter, which was also re- trieved from our clone library analyses from FCS (Table 2). In line with Geobacter, Shewanella of the Gam- maproteobacteria class, another recognized metal re- ducer [46], was also present in the treatment with chitin + apatite (FACS). Both Geobacter and Shewanella have been proven to play critical roles in metal reduction, pre- cipitation and immobilization [1,23,24,27,45]. Thus, sul- fate/sulfur-reducing bacteria and metal-reducing bacteria responded to chitin and apatite amendments, and were likely responsible for the metal immobilization and se- questration. Although no significant bacterial cell counts and popu- lation shifts were observed in the treatment with apatite alone [30 and this study], apatite held great promise of inducing phosphate-solubilizing bacteria (PSB), which might facilitate the formation of phosphorites, natural apatite mineral deposits in environments [47-49]. Transi- tion metals in natural environments were expected to be sequestered with phosphates and form more stable metal- phosphate complexes and therefore decrease the bioavail ability of these metals [3]. In this study, we tested PSB on agar plates, and positive PSB colonies were present in both overlying water and sediments (Figure S3). The clearing zones indicated solubilization of calcium phos- phate from the media (Figure S3). To date, a variety of bacterial groups (e.g. Bacillus, Rhizobium, Pseudomonas, Serratia, Shewanella, Escherichia, Vibrio, and Proteus etc.) have been proven capable of solubilizing phosphate compounds [17,50-51]. Because phosphate is not a lim- iting factor in open oceans, only few studies have inves- tigated PSB from marine environments [33,51]. However, the presence of common groups of bacteria including Pseudomonas, Shewanella, and Vibrio from our clone libraries suggested that they might respond to apatite amendment and solubilize phosphate in marine sedi- ments. 3.3. ICP-MS Analysis of Spiked Metals In the presence of apatite only, less than 50% of Cu and Cr; and 0% of Zn were removed from the overlying wa- ter in the coarse-grained sediments (CAS; Table 3). A similar effect was observed in the fine-grained sediment as Cu and Cr were not removed at all; and in the Zn sample, ~25% of Zn was removed (FAS; Table 3). In the presence of chitin, between 60% - 100% of metal re- moval occurred in the overlying and pore-water (CCS and FCS; Table 3). The combined effect of apatite and chitin in metal removal was similar enough to chitin to demonstrate the important role of chitin in these proc- esses. These data are in agreement with the microbi- ological data where chitin enhanced microbiological ac- tivity versus apatite amendments. The chitin amendments likely stimulated sulfer-transforming bacteria [30,52] and FeS was observed in the mesocosms. In addition, there may have been metal sorption to the chitin particles. However, because chitin is a food source for bacteria, there is the potential when chitin is digested for metal re- solubilization. Therefore long-term chitin metal removal should be evaluated. Nevertheless, the role of apatite cannot be excluded as phosphate-solubilizing bacteria were detected. In addition, XAS data also showed the positive effects of apatite in sediments. 3.4. Effects of Apatite and Chitin Amendment on Zinc Mobilization Figure 5 showed an example X-ray absorption near-edge spectroscopy (XAS) spectra on FAU sediments over time post amendment addition and reference spectra of pri- mary components for linear combination fitting (LCF). Copyright © 2013 SciRes. OJMetal 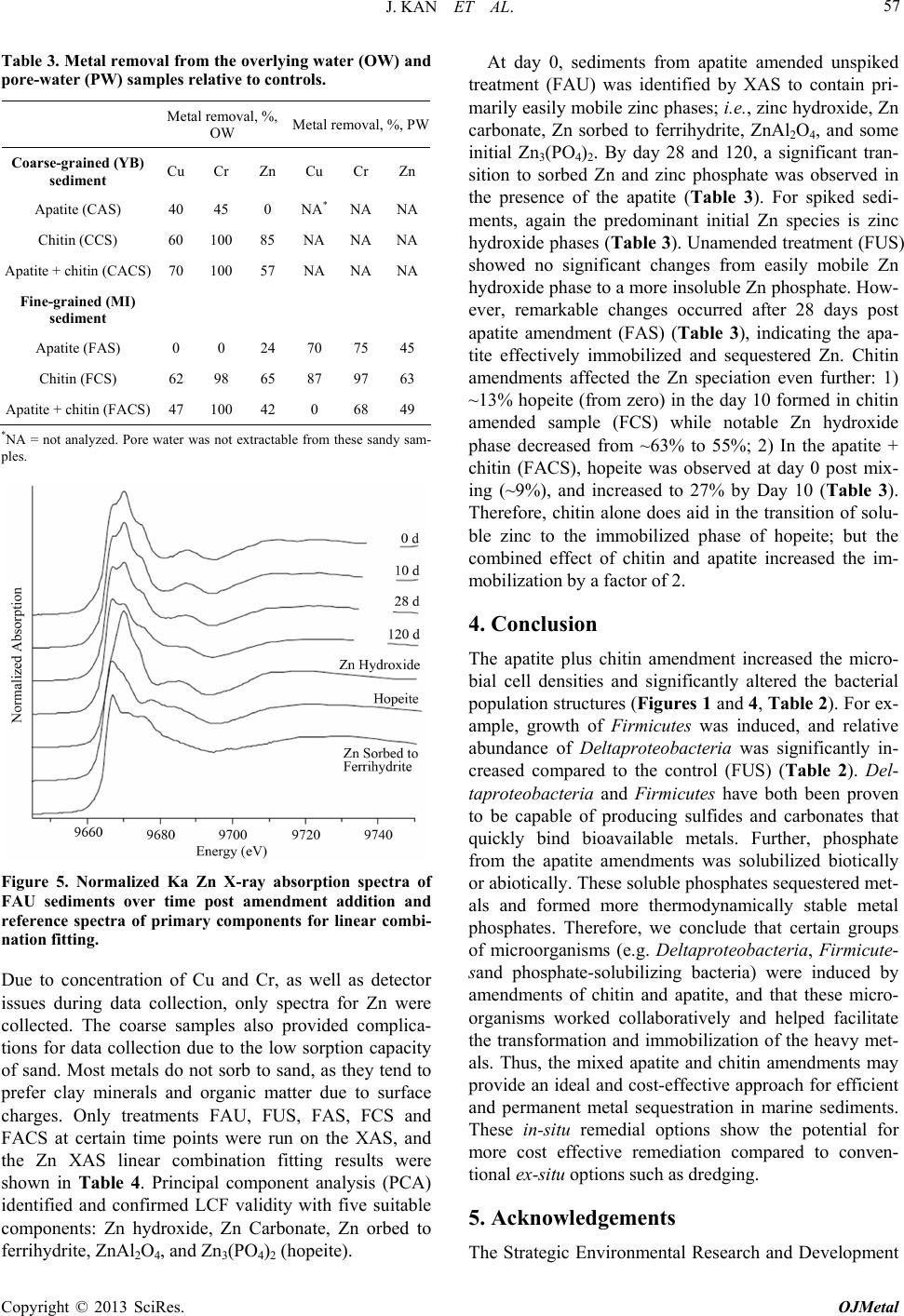 J. KAN ET AL. 57 Table 3. Metal removal from the overlying water (OW) and pore-water (PW) samples relative to controls. Metal removal, %, OW Metal removal, %, PW Coarse-grained (YB) sediment Cu Cr Zn Cu Cr Zn Apatite (CAS) 40 45 0 NA* NANA Chitin (CCS) 60 10085 NA NANA Apatite + chitin (CACS) 70 10057 NA NANA Fine-grained (MI) sediment Apatite (FAS) 0 0 24 70 75 45 Chitin (FCS) 62 98 65 87 97 63 Apatite + chitin (FACS) 47 10042 0 68 49 *NA = not analyzed. Pore water was not extractable from these sandy sam- ples. Figure 5. Normalized Ka Zn X-ray absorption spectra of FAU sediments over time post amendment addition and reference spectra of primary components for linear combi- nation fitting. Due to concentration of Cu and Cr, as well as detector issues during data collection, only spectra for Zn were collected. The coarse samples also provided complica- tions for data collection due to the low sorption capacity of sand. Most metals do not sorb to sand, as they tend to prefer clay minerals and organic matter due to surface charges. Only treatments FAU, FUS, FAS, FCS and FACS at certain time points were run on the XAS, and the Zn XAS linear combination fitting results were shown in Table 4. Principal component analysis (PCA) identified and confirmed LCF validity with five suitable components: Zn hydroxide, Zn Carbonate, Zn orbed to ferrihydrite, ZnAl2O4, and Zn3(PO4)2 (hopeite). At day 0, sediments from apatite amended unspiked treatment (FAU) was identified by XAS to contain pri- marily easily mobile zinc phases; i.e., zinc hydroxide, Zn carbonate, Zn sorbed to ferrihydrite, ZnAl2O4, and some initial Zn3(PO4)2. By day 28 and 120, a significant tran- sition to sorbed Zn and zinc phosphate was observed in the presence of the apatite (Table 3). For spiked sedi- ments, again the predominant initial Zn species is zinc hydroxide phases (Table 3). Unamended treatment (FUS) showed no significant changes from easily mobile Zn hydroxide phase to a more insoluble Zn phosphate. How- ever, remarkable changes occurred after 28 days post apatite amendment (FAS) (Table 3), indicating the apa- tite effectively immobilized and sequestered Zn. Chitin amendments affected the Zn speciation even further: 1) ~13% hopeite (from zero) in the day 10 formed in chitin amended sample (FCS) while notable Zn hydroxide phase decreased from ~63% to 55%; 2) In the apatite + chitin (FACS), hopeite was observed at day 0 post mix- ing (~9%), and increased to 27% by Day 10 (Table 3). Therefore, chitin alone does aid in the transition of solu- ble zinc to the immobilized phase of hopeite; but the combined effect of chitin and apatite increased the im- mobilization by a factor of 2. 4. Conclusion The apatite plus chitin amendment increased the micro- bial cell densities and significantly altered the bacterial population structures (Figures 1 and 4, Table 2). For ex- ample, growth of Firmicutes was induced, and relative abundance of Deltaproteobacteria was significantly in- creased compared to the control (FUS) (Table 2). Del- taproteobacteria and Firmicutes have both been proven to be capable of producing sulfides and carbonates that quickly bind bioavailable metals. Further, phosphate from the apatite amendments was solubilized biotically or abiotically. These soluble phosphates sequestered met- als and formed more thermodynamically stable metal phosphates. Therefore, we conclude that certain groups of microorganisms (e.g. Deltaproteobacteria, Firmicute- sand phosphate-solubilizing bacteria) were induced by amendments of chitin and apatite, and that these micro- organisms worked collaboratively and helped facilitate the transformation and immobilization of the heavy met- als. Thus, the mixed apatite and chitin amendments may provide an ideal and cost-effective approach for efficient and permanent metal sequestration in marine sediments. These in-situ remedial options show the potential for more cost effective remediation compared to conven- tional ex-situ options such as dredging. 5. Acknowledgements The Strategic Environmental Research and Development Copyright © 2013 SciRes. OJMetal  J. KAN ET AL. Copyright © 2013 SciRes. OJMetal 58 Table 4. Linear combination fitting of fine-grained sediments (Mare Island Naval Shipyard). Treatments FAU, FUS, FAS, FCS, and FACS were shown in Table 1. - = not detected. The R factor is the error (%) associated with the fits. Linear contribution fitting distribution (%) Treatments Amendment Metal mixture Reaction time Zn hydroxide1Zn carbonateSorbed Zn2 ZnAl2O4 Hopeite3 R factor (%) FAU 5% apatite Unspiked 0 days 59.1 7.4 9.5 18.8 5.4 3.00 10 days 59.1 7.9 8.5 18.5 6.2 2.95 28 days 43.4 8.4 18.2 12.9 17.2 1.50 120 days 25.3 4.4 29.9 7.2 33.3 0.35 FUS None Spiked 0 days 71.0 10.0 19.0 - - 1.61 10 days 70.0 10.0 18.0 - - 1.94 28 days 70.0 - 30.0 - - FAS 5% apatite Spiked 0 days 45.0 16.0 - 14.0 25.0 10 days 43.0 14.0 - 15.0 28.0 28 days 31.0 9.0 26.0 - 34.0 FCS 0.5% chitin Spiked 0 days 63.0 25.0 - 13.0 - 1.12 10 days 55.0 17.0 5.0 11.0 13.0 1.07 FACS 5% apatie + 0.5% chitinSpiked 0 days 68.0 13.0 - 10.0 9.0 1.97 10 days 52.0 15.0 - 8.0 27.0 0.97 1Includes Zn(OH)2 and related layered double hydroxides; 2Zn sorbed to ferrihydrite; 3Zn3(PO4)2. Program (SERDP) funded this research under project #ER-1551. The authors thank Gunther Rosen, Ryan Ha- lonen, Brandon Swope, and Ignacio Rivera for sampling and technical support. Although EPA contributed to this article, the research presented was not performed by or funded by EPA and was not subject to EPA’s quality system requirements. Consequently, the views, interpre- tations, and conclusions expressed in this article are solely those of the authors and do not necessarily reflect or represent EPA’s views or policies. REFERENCES [1] R. T. Anderson, H. A. Vrionis, I. Ortiz-Bernad, C. T. Resch, P. E. Long, R. Dayvault, K. Karp, S. Marutzky, D. R. Metzler, R. Peacock, D. C. White, M. Lowe and D. R. Lovley, “Stimulating the in Situ Activity of Geobacter Species to Remove Uranium from the Groundwater of a Uranium-Contaminated Aquifer,” Applied and Environ- mental Microbiology, Vol. 69, No. 10, 2003, pp. 5884- 5891. doi:10.1128/AEM.69.10.5884-5891.2003 [2] B. Sharma, K. H. Gardner, J. Melton, A. Hawkins and G. Tracey, “Evaluation of Activated Carbon as a Reactive Cap Sorbent for Sequestration of Polychlorinated Bi- phenyls in the Presence of Humic Acid,” Environmental Engineering Science, Vol. 26, No. 9, 2009, pp. 1371- 1379. doi:10.1089/ees.2008.0231 [3] S. Brown, R. Chaney, J. Hallfrisch, J. A. Ryan and W. R. Berti, “In Situ Soil Treatments to Reduce the Phyto and Bioavailability of Lead, Zinc, and Cadmium,” Journal of Environmental Quality, Vol. 33, No. 2, 2004, pp. 522-531. doi:10.2134/jeq2004.0522 [4] J. D. Istok, J. M. Senko, L. R. Krumholtz, D. Watson, M. A. Bogle, A. Peacock, Y. J. Chang and D. C. White, “In Situ Bioreduction of Technetium and Uranium in a Ni- trate Contaminated Aquifer,” Environmental Science & Technology, Vol. 38, No. 2, 2004, pp. 468-475. doi:10.1021/es034639p [5] T. P. Seager, and K. H. Gardner, “Barriers to Adoption of Novel Environmental Technologies: Contaminated Sedi- ments,” In: E. Levner, I. Linkov and J. M. Proth, Eds., Strategic Management of Marine Ecosystems, Springer, 2005, pp. 298-312. doi:10.1007/1-4020-3198-X_16 [6] C. J. Werth, R. A. Sanford, R. St John and G. C. Bar- nuevo, “Long-Term Management of Chlorinated Solvent Plumes Using a Slow-Release in Situ Electron Donor Source,” Abstract poster G-4. In: SERDP/ESTCP Meet- ing, Washington DC, 2005. [7] H. Paller and A.S. Knox, “Amendments for the in Situ Remediation of Contaminated Sediments: Evaluation of Potential Environmental Impacts,” Science of the Total Environment, Vol. 408, No. 20, 2010, pp. 4894-4900. doi:10.1016/j.scitotenv.2010.06.055 [8] Y. J. Chang, P. E. Long, R. Geyer, A. D. Peacock, C. T. Resch, K. Sublette, S. Pfiffner, A. Smithgall, R. T. An- derson, H. A. Vrionis, J. R. Stephen, R. Dayvault, I. Ortiz- Bernad, D. R. Lovley and D. C. White, “Microbial In- corporation of 13Clabeled Acetate at the Field Scale: De- tection of Microbes Responsible for Reduction of U(VI),” Environmental Science & Technology, Vol. 39, No. 23, 2005, pp. 9039-9048. doi:10.1021/es051218u [9] S. Lukas and J. T. Hollibaugh, “Response of Sediment Bacterial Assemblages to Selenate and Acetate Amend- ments,” Environmental Science & Technology, Vol. 35, No. 3, 2001, pp. 528-534. doi:10.1021/es001492i  J. KAN ET AL. 59 [10] Q. Y. Ma, T. J. Logan and S. J. Traina, “Lead Immobili- zation from Aqueous Solutions and Contaminated Soils Using Phosphate Rocks,” Environmental Science & Tech- nology, Vol. 29, No. 4, 1995, pp. 1118-1126. doi:10.1021/es00004a034 [11] S. Knox, D. I. Kaplan and M. H. Paller, “Phosphate Sources and Their Suitability for Remediation of Con- taminated Soils,” Science of the Total Environment, Vol. 357, No. 1-3, 2006, pp. 271-279. doi:10.1016/j.scitotenv.2005.07.014 [12] X. D. Cao, L. Q. Ma and A. Wahbi, “Immobilization of Cu, Zn, and Pb in Contaminated Soils Using Phosphate Rock and Phosphoric Acid,” Journal of Hazardous Mate- rials, Vol. 164, No. 2-3, 2009, pp. 555-564. doi:10.1016/j.jhazmat.2008.08.034 [13] Q. Y. Ma, S. Traina, T. Logan and J. Ryan, “In Situ Lead Immobilization by Apatite,” Environmental Science & Technology, Vol. 27, No. 9, 1993, pp. 1803-1810. doi:10.1021/es00046a007 [14] J. Wright, K. R. Rice, B. Murphy and J. Conca, “PIMS Using Apatite IITM: How It Works To Remediate Soil and Water,” In: R. E. Hinchee and B. Alleman, Eds., Sus- tainable Range Management, Battelle Press, Columbus, 2004. [15] U. Ghosh, R. G. Luthy, G. Cornelissen, D. Werner and C. A. Menzie, “In-Situ Sorbent Amendments: A New Direc- tion in Contaminated Sediment Management,” Environ- mental Science & Technology, Vol. 45, No. 4, 2011, pp. 1163-1168. doi:10.1021/es102694h [16] G. B. Williams, K. G. Scheckel, G. McDermott, D. Grat- son, D. Neptune and J. A. Ryan, “Speciation and Bio- availability of Zinc in Amended Sediments,” Chemical Speciation and Bioavailability, Vol. 23, No. 3, 2011, pp. 143-154. doi:10.3184/095422911X13103699236851 [17] B. Uzair and N. Ahmed, “Solubilization of Insoluble In- organic Phosphate Compounds by Attached and Free- Living Marine Bacteria,” Journal of Basic & Applied Sci- ences, Vol. 3, No. 2, 2007, pp. 59-63. [18] C. Kanzog, A. Ramette, N. V. Queric and M. Klages, “Response of Benthic Microbial Communities to Chitin Enrichment: An in Situ Study in the Deep Arctic Ocean,” Polar Biology, Vol. 32, No. 1, 2008, pp. 105-112. doi:10.1007/s00300-008-0510-4 [19] A. Boetius and K. Lochte, “Effect of Organic Enrich- ments on Hydrolytic Potentials and Growth of Bacteria in Deep-Sea Sediments,” Marine Ecology Progress Series, Vol. 140, 1996, pp. 239-250. doi:10.3354/meps140239 [20] B. L. Bassler, C. Yu, Y. C. Lee and S. Roseman, “Chitin Utilization by Marine Bacteria: Degradation and Catabo- lism of Chitin Oligosaccharides by Vibrio furnissii,” Jour- nal of Biological Chemistry, Vol. 266, No. 36, 1991, pp. 24276-24286. [21] L. Brierley, “Metal Immobilization Using Bacteria,” In: H. L. Ehrlich and C. L. Brierley, Eds., Microbial Mineral Recovery, McGraw-Hill, 1990, pp. 303-323. [22] B. M. Tebo, “Metal Precipitation by Marine Bacteria: Potential for Biotechnological Applications,” In: J. K. Setlow, Ed., Genetic Engineering, Plenum Press, 1995, pp. 231-261. [23] R. Lovley, “Dissimilatory Metal Reduction,” Annual Re- view of Microbiology, Vol. 47, 1993, pp. 263-290. doi:10.1146/annurev.mi.47.100193.001403 [24] L. J. Barnes, P. J. M. Scheeren and C. J. N. Buisman, “Microbial Removal of Heavy Metal and Sulfate from Contaminated Groundwaters,” In: J. L. Means and R. E. Hinchee, Eds., Emerging Technology for Bioremediation of Metals, CRC Press, 1994, pp. 38-49. [25] L. L. Barton and F. A. Tomei, “Characteristics and Ac- tivities of Sulfate-Reducing Bacteria,” In: L. L. Barton, Ed., Sulfate-Reducing Bacteria, Plenum Press, 1995, pp. 1-32. doi:10.1007/978-1-4899-1582-5_1 [26] R. T. Anderson and D. R. Lovley, “Ecology and Biogeo- chemistry of in Situ Groundwater Bioremediation,” In: J. G. Jones, Ed., Advances in Microbial Ecology, Plenum Press, 1997, pp. 289-350. doi:10.1007/978-1-4757-9074-0_7 [27] K. H. Nealson, “Sediment Bacteria: Who’s There, What Are They Doing, and What’s New?” Annual Review of Earth and Planetary Sciences, Vol. 25, 1997, pp. 403-434. doi:10.1146/annurev.earth.25.1.403 [28] B. M. Tebo and A. Y. Obraztsova, “Chromium(VI), Man- ganese(IV), Uranium(VI), and Iron(III): Electron Accep- tors for Growth for a Novel Spore Forming Sulfate Re- ducing Bacterium,” FEMS Microbiology Letters, Vol. 162, No. 1, 1998, pp. 193-198. doi:10.1111/j.1574-6968.1998.tb12998.x [29] Y. M. Arias and B. M. Tebo, “Comparative Studies of Cr(VI) Reduction by Sulfidogenic and Non-Sulfidogenic Microbial Communities,” Applied and Environmental Mi- crobiology, Vol. 69, No. 3, 2003, pp. 1847-1853. doi:10.1128/AEM.69.3.1847-1853.2003 [30] J. Kan, Y. Wang, A. Obraztsova, G. Rosen, J. Leather, K. G. Scheckel, K. H. Nealson and Y. M. Arias-Thode, “Ma- rine Microbial Community Response to Inorganic and Or- ganic Sediment Amendments in Laboratory Mesocosms,” Ecotoxicology and Environmental Safety, Vol. 74, No. 7, 2011, pp. 1931-1941. doi:10.1016/j.ecoenv.2011.06.011 [31] G. Rosen, J. Leather, J. Kan and Y. M. Arias-Thode, “Ecotoxicological Response of Marine Organisms to In- organic and Organic Sediment Amendments in Labora- tory Exposures,” Ecotoxicology and Environmental Safety, Vol. 74, No. 7, 2011, pp. 1921-1930. doi:10.1016/j.ecoenv.2011.06.023 [32] T. Huber, G. Faulkner and P. Hugenholtz, “Bellerophon; A Program to Detect Chimeric Sequences in Multiple Sequence Alignments,” Bioinformatics, Vol. 20, No. 14, 2004, pp. 2317-2319. doi:10.1093/bioinformatics/bth226 [33] K. Ayyakkannu and D. Chandramohan, “Occurrence and Distribution of Phosphate Solubilizing Bacteria and Phos- phatase in Marine Sediments at Porto Novo,” Marine Bi- ology, Vol. 11, No. 3, 1971, pp. 201-205. doi:10.1007/BF00401268 [34] J. Talbot and A. Weiss, “Laboratory Methods for ICP-MS Analysis of Trace Metals in Precipitation,” 1994. http://nsdi.epa.gov/glnpo/lmmb/methods/icmsmeth.pdf [35] S. E. Bufflap and H. E. Allen, “Comparison of Pore Wa- ter Sampling Techniques for Trace Metals,” Water Re- search, Vol. 29, No. 9, 1995, pp. 2051-2054. Copyright © 2013 SciRes. OJMetal  J. KAN ET AL. Copyright © 2013 SciRes. OJMetal 60 doi:10.1016/0043-1354(95)00032-G [36] G. Scheckel, J. A. Ryan, D. Allen and N. V. Lescano, “Determining Speciation of Pb in Phosphate-Amended Soils: Method Limitations,” Science of the Total Envi- ronment, Vol. 350, No. 1-3, 2005, pp. 261-272. doi:10.1016/j.scitotenv.2005.01.020 [37] H. M. Cauchie, “Chitin Production by Arthropods in the Hydrosphere,” Hydrobiology, Vol. 470, No. 1-3, 2002, pp. 63-96. doi:10.1023/A:1015615819301 [38] M. Poulicek and C. Jeuniaux, “Chitin Biodegradation in Marine Environments: An Experimental Approach,” Bio- chemical Systematics and Ecology, Vol. 19, No. 5, 1991, pp. 385-394. doi:10.1016/0305-1978(91)90055-5 [39] J. W. Deming and J. A. Baross, “The Early Diagenesis of Organic Matter: Bacterial Activity,” In: M. H. Engel and S. A. Macko, Eds., Organic Geochemistry: Principles and Application, Plenum Press, New York, 1993, pp. 119-144. doi:10.1007/978-1-4615-2890-6_5 [40] A. Bell, J. C. Hubbard, L. Liu, R. M. Davis and K. V. Subbarao, “Effects of Chitin and Chitosan on the Inci- dence and Severity of Fusarium Yellows in Celery,” Plant Disease, Vol. 82, No. 3, 1998, pp. 322-328. doi:10.1094/PDIS.1998.82.3.322 [41] J. Hallmann, R. Rodriguez-Kabana and J. W. Kloepper, “Chitin-Mediated Changes in Bacterial Communities of the Soil, Rhizosphere and within Roots of Cotton in Rela- tion to Nematode Control,” Soil Biology and Biochemis- try, Vol. 31, No. 4, 1999, pp. 551-560. doi:10.1016/S0038-0717(98)00146-1 [42] D. M. Akob, H. J. Mills and J. E. Kostka, “Metabolically Active Microbial Communities in Uranium-Contaminated Subsurface Sediment,” FEMS Microbiology Ecology, Vol. 59, No. 1, 2006, pp. 95-107. doi:10.1111/j.1574-6941.2006.00203.x [43] G. Garau, P. Castaldi, L. Santona, P. Deiana and P. Melis, “Influence of Red Mud, Zeolite and Lime on Heavy Metal Immobilization, Culturable Heterotrophic Micro- bial Populations and Enzyme Activities in a Contami- nated Soil,” Geoderma, Vol. 142, No. 1-2, 2007, pp. 47- 57. doi:10.1016/j.geoderma.2007.07.011 [44] J. Kleikemper, O. Pelz, M. H. Schroth and J. Zeyer, “Sul- fate-Reducing Bacterial Community Response to Carbon Source Amendments in Contaminated Aqufer Micro- cosms,” FEMS Microbiology Ecology, Vol. 42, No. 1, 2002, pp. 109-118. doi:10.1111/j.1574-6941.2002.tb01000.x [45] L. L. Barton and G. D. Fauque, “Biochemistry, Physiol- ogy and Biotechnology of Sulfate-Reducing Bacteria,” Advances in Applied Microbiology, Vol. 68, 2009, pp. 41-98. doi:10.1016/S0065-2164(09)01202-7 [46] C. Myers and K. H. Nealson, “Bacterial Manganese Re- duction and Growth with Manganese Oxide as the Sole Electron Acceptor,” Science, Vol. 240, No. 4857, 1988, pp. 1319-1321. doi:10.1126/science.240.4857.1319 [47] G. N. Baturin, “Some Unique Sedimentological and Geo- chemical Features of Deposits in Coastal Upwelling Re- gions,” In: J. Thiede and E. Suess, Eds., Coastal Upwell- ing, Plenum Press, 1983, pp. 11-27. doi:10.1007/978-1-4613-3709-6_2 [48] C. E. Reimers, M. Kastner and R. E. Garrison, “The Role of Bacterial Mats in Phosphate Mineralization with Par- ticular Reference to the Monterey Formation,” In: W. C. Burnett and S. R. Riggs, Eds., Phosphate Deposits of the World, Cambridge University Press, 1990, pp. 300-311. [49] N. Schulz and H. D. Schulz, “Large Sulfur Bacteria and the Formation of Phosphorite,” Science, Vol. 307, 2005, pp. 416-418. doi:10.1126/science.1103096 [50] H. Rodriguez and R. Fraga, “Review: Phosphate Solubi- lizing Bacteria and Their Role in Plant Growth Promo- tion,” Biotechnology Advances, Vol. 17, No. 4-5, 1999, pp. 319-339. doi:10.1016/S0734-9750(99)00014-2 [51] D. De Souza, S. Nair and D. Chandramohan, “Phosphate Solubilizing Bacteria around Indian Peninsula,” Indian Journal of Marine Sciences, Vol. 29, 2000, pp. 48-51. [52] A. Robinson-Lora and R. A. Brennan, “Efficient Metal Removal and Neutralization of Acid Mine Drainage by Crab-Shell Chitin under Batch and Continuous Flow Con- ditions,” Bioresource Technology, Vol. 100, No. 21, 2009, pp. 5063-5071. doi:10.1016/j.biortech.2008.11.063 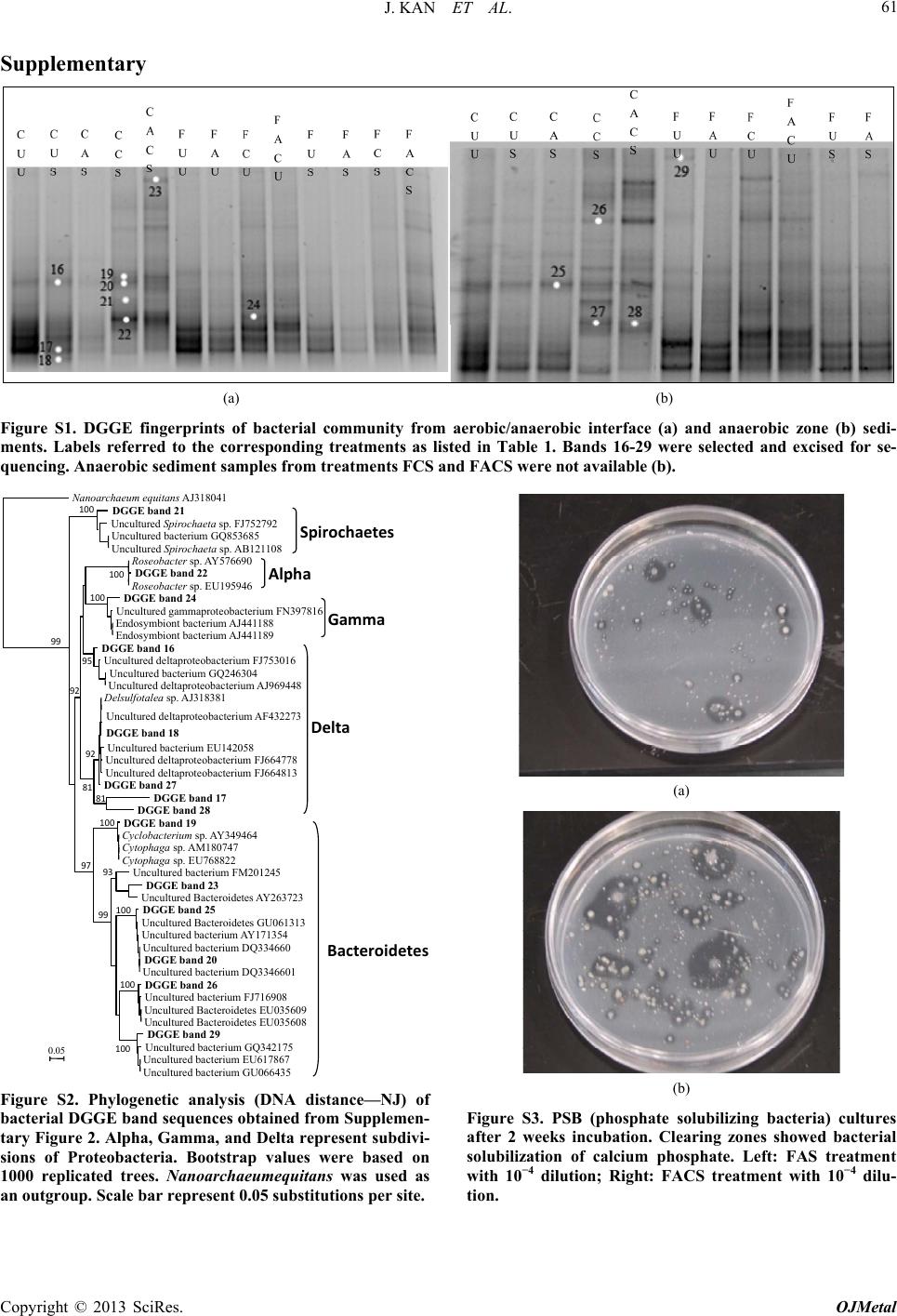 J. KAN ET AL. 61 Supplementary (a) (b) Figure S1. DGGE fingerprints of bacterial community from aerobic/anaerobic interface (a) and anaerobic zone (b) sedi- ments. Labels referred to the corresponding treatments as listed in Table 1. Bands 16-29 were selected and excised for se- quencing. Anaerobic sediment samples from treatments FCS and FACS were not available (b). Nanoa haeum equitans AJ318041 DGGE band 21 Uncultured Spirochaeta sp. FJ752792 Uncultured bacterium GQ853685 Uncultured Spirochaeta sp. AB121108 Roseobacter sp. AY576690 DGGE band 22 Roseobacter sp. EU195946 DGGE band 24 Uncultured gammaproteobacterium FN397816 Endosymbiont bacterium AJ441188 Endosymbiont bacterium AJ441189 DGGE band 16 Uncultured deltaproteobacterium FJ753016 Uncultured bacterium GQ246304 Uncultured deltaproteobacterium AJ969448 Delsulfotalea sp. AJ318381 Uncultured deltaproteobacterium AF432273 DGGE band 18 Uncultured bacterium EU142058 Uncultured deltaproteobacterium FJ664778 Uncultured deltaproteobacterium FJ664813 DGGE band 27 DGGE band 17 DGGE band 28 DGGE band 19 Cyclobacterium sp. AY349464 Cytophaga sp. AM180747 Cytophaga sp. EU768822 Uncultured bacterium FM201245 DGGE band 23 Uncultured Bacteroidetes AY263723 DGGE band 25 Uncultured Bacteroidetes GU061313 Uncultured bacterium AY171354 Uncultured bacterium DQ334660 DGGE band 20 Uncultured bacterium DQ3346601 DGGE band 26 Uncultured bacterium FJ716908 Uncultured Bacteroidetes EU035609 Uncultured Bacteroidetes EU035608 DGGE band 29 Uncultured bacterium GQ342175 Uncultured bacterium EU617867 Uncultured bacterium GU066435 0.05 99 100 95 100 100 92 92 81 81 97 100 99 93 100 100 100 Spirochaetes Alpha Gamma Delta Bacteroidete Figure S2. Phylogenetic analysis (DNA distance—NJ) of bacterial DGGE band sequences obtained from Supplemen- tary Figure 2. Alpha, Gamma, and Delta represent subdivi- sions of Proteobacteria. Bootstrap values were based on 1000 replicated trees. Nanoarchaeumequitans was used as an outgroup. Scale bar represent 0.05 substitutions per site. (a) (b) Figure S3. PSB (phosphate solubilizing bacteria) cultures after 2 weeks incubation. Clearing zones showed bacterial solubilization of calcium phosphate. Left: FAS treatment with 10−4 dilution; Right: FACS treatment with 10−4 dilu- tion. Copyright © 2013 SciRes. OJMetal
|