Paper Menu >>
Journal Menu >>
 Vol.1, No.3, 95-101 (2010) Agricultural Sciences doi:10.4236/as.2010.13012 Copyright © 2010 SciRes. Openly accessible at http://www.scirp.org/journal/AS/ Molecular characterization of Cuban endemism Carica cubensis Solms using random amplified polymorphic DNA (RAPD) markers Jesús Rodríguez1, Pedro Rodríguez1, María E. González1, Pedro Martínez-Gómez2* 1Departamento de Fisología y Bioquímica Vegetal, Departamento de Genética, Instituto Nacional de Ciencias Agrícolas INCA, San José de las Lajas, Cuba 2Departamento de Mejora Vegetal, CEBAS-CSIC, Espinardo, Spain; *Corresponding Author: pmartinez@cebas.csic.es Received 28 July 2010; revised 30 August 2010; accepted 5 September 2010. ABSTRACT The objective of this work is to present an ap- propriate set of RAPD (random amplified poly- morphic DNA) markers using single and multi- plex PCR analysis suitable for the characteriza- tion of the endemic Cuban species Carica cu- bensis and the establishment of genetic rela- tionships with the cultivated species Carica pa- paya. RAPD markers presented a high level of polymorphism. In addition, the incorporation of more than one RAPD primer in the PCR analysis increased the number of obtained bands and the polymorphism of these bands. A total of 73 RAPD bands were detected (45 of them poly- morphic) with the nine RAPD markers assayed using single and multiplex PCR analysis. Re- sults demonstrated a reduced genetic variability within the tested Carica cubensis accessions. The observed clustering in this species could be better explained according to geographic proximity and can indicate the similar prece- dence of the isolated studied populations. C. cubensis seem to be subspecies of C. papaya adapted to the environmental conditions of the mountains of Cuba or a endemic species close to C. papaya. The implications of these results in the creation of effective germplasm core col- lection in Carica species have been also dis- cussed. Keywords: Carica Species; Germplasm; Molecular Markers; RAPDs; Breeding 1. INTRODUCTION Carica is a genus of family Caricaceae originating from Central and South America including more than forty different species being Carica papaya L. the most important species from the economically and agronomy point of view and the most cultivated species in these areas of Central and South America [1,2]. Cuba can be considered an area of putative endemism [3,4] of these species according to the origin and dispersion of the dif- ferent accessions analyzed. In this sense, one of these examples of related species is Carica cubensis Solms syn. Carica prosoposa L., an endemic fruit tree species from Cuba considered a papaya endemism in prelimi- nary studies [5]. This specie was described at the first time by Solms and Grafen in 1889 [6] indicating the presence of the specie in forested areas of Cuba since Baracoa to Portero of St. Andre. Carica cubensis is a shrub from 2 to 4 meters with a thick trunk and branches and leaves spongy no alternated and terminals. As well as Carica papaya, this species is at the same time dioecious with female and male plants, monocious with female and male flowers in the same plant, and polygamous with hermaphrodite flowers [7,8]. Flowers are white to yellow-green and can have three sex forms male (in the case of male plants) and female and hermaphrodite (in the case of female plants) Figure 1. Only the two last types give fruits. The fruit with a size around 80-100 grams is smaller than the cultivated papaya and contains an enzyme (papaine) with intensive digestive action that can be used for medicinal purpose against dyspepsia disease and with antitumoral proper- ties [5,9]. Carica cubensis is a wild related species of the cultivated papaya species (Carica papaya) with a lot of potential from the agronomic and breeding point of view. Some authors, however, included this Carica cubensis species inside the Carica papaya species. This species can be particularly useful in marginal lands, where they have been selected to withstand stress conditions and where they contribute to sustainable production with few inputs. 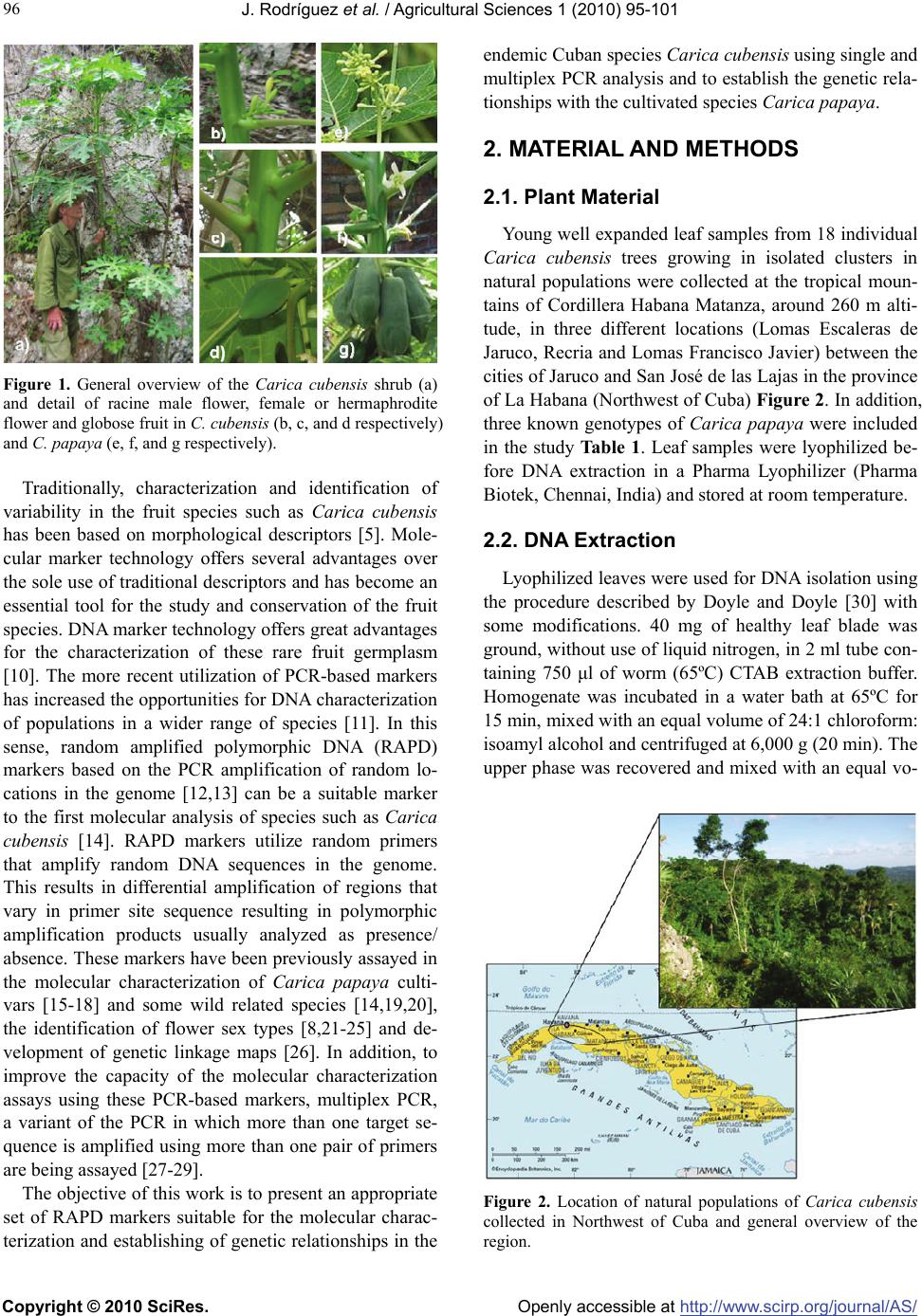 J. Rodríguez et al. / Agricultural Sciences 1 (2010) 95-101 Copyright © 2010 SciRes. Openly accessible at http://www.scirp.org/journal/AS/ 96 Figure 1. General overview of the Carica cubensis shrub (a) and detail of racine male flower, female or hermaphrodite flower and globose fruit in C. cubensis (b, c, and d respectively) and C. papaya (e, f, and g respectively). Traditionally, characterization and identification of variability in the fruit species such as Carica cubensis has been based on morphological descriptors [5]. Mole- cular marker technology offers several advantages over the sole use of traditional descriptors and has become an essential tool for the study and conservation of the fruit species. DNA marker technology offers great advantages for the characterization of these rare fruit germplasm [10]. The more recent utilization of PCR-based markers has increased the opportunities for DNA characterization of populations in a wider range of species [11]. In this sense, random amplified polymorphic DNA (RAPD) markers based on the PCR amplification of random lo- cations in the genome [12,13] can be a suitable marker to the first molecular analysis of species such as Carica cubensis [14]. RAPD markers utilize random primers that amplify random DNA sequences in the genome. This results in differential amplification of regions that vary in primer site sequence resulting in polymorphic amplification products usually analyzed as presence/ absence. These markers have been previously assayed in the molecular characterization of Carica papaya culti- vars [15-18] and some wild related species [14,19,20], the identification of flower sex types [8,21-25] and de- velopment of genetic linkage maps [26]. In addition, to improve the capacity of the molecular characterization assays using these PCR-based markers, multiplex PCR, a variant of the PCR in which more than one target se- quence is amplified using more than one pair of primers are being assayed [27-29]. The objective of this work is to present an appropriate set of RAPD markers suitable for the molecular charac- terization and establishing of genetic relationships in the endemic Cuban species Carica cubensis using single and multiplex PCR analysis and to establish the genetic rela- tionships with the cultivated species Carica papaya. 2. MATERIAL AND METHODS 2.1. Plant Material Young well expanded leaf samples from 18 individual Carica cubensis trees growing in isolated clusters in natural populations were collected at the tropical moun- tains of Cordillera Habana Matanza, around 260 m alti- tude, in three different locations (Lomas Escaleras de Jaruco, Recria and Lomas Francisco Javier) between the cities of Jaruco and San José de las Lajas in the province of La Habana (Northwest of Cuba) Figure 2. In addition, three known genotypes of Carica papaya were included in the study Table 1. Leaf samples were lyophilized be- fore DNA extraction in a Pharma Lyophilizer (Pharma Biotek, Chennai, India) and stored at room temperature. 2.2. DNA Extraction Lyophilized leaves were used for DNA isolation using the procedure described by Doyle and Doyle [30] with some modifications. 40 mg of healthy leaf blade was ground, without use of liquid nitrogen, in 2 ml tube con- taining 750 μl of worm (65ºC) CTAB extraction buffer. Homogenate was incubated in a water bath at 65ºC for 15 min, mixed with an equal volume of 24:1 chloroform: isoamyl alcohol and centrifuged at 6,000 g (20 min). The upper phase was recovered and mixed with an equal vo- Figure 2. Location of natural populations of Carica cubensis collected in Northwest of Cuba and general overview of the region.  J. Rodríguez et al. / Agricultural Sciences 1 (2010) 95-101 Copyright © 2010 SciRes. Openly accessible at http://www.scirp.org/journal/AS/ 97 97 Table 1. Carica cubensis and Carica papaya accessions assayed. Accession Species Origin Sex of the plant “Lomas Escaleras-1” Carica cubensis Seedling collected in Lomas Escaleras Plant without flower “Lomas Escaleras-2” Carica cubensis Seedling collected in Lomas Escaleras Plant without flower “Lomas Escaleras-3” Carica cubensis Seedling collected in Lomas Escaleras Female “Lomas Escaleras-4” Carica cubensis Seedling collected in Lomas Escaleras Plant without flower “Lomas Escaleras-5” Carica cubensis Seedling collected in Lomas Escaleras Plant without flower “Lomas Escaleras-6” Carica cubensis Seedling collected in Lomas Escaleras Plant without flower “Recria-1” Carica cubensis Seedling collected in Recria area Plant without flower “Recria-2” Carica cubensis Seedling collected in Recria area Plant without flower “Recria-3” Carica cubensis Seedling collected in Recria area Plant without flower “Recria-4” Carica cubensis Seedling collected in Recria area Plant without flower “Recria-5” Carica cubensis Seedling collected in Recria area Plant without flower “Recria-6” Carica cubensis Seedling collected in Recria area Plant without flower “Lomas Francisco-1” Carica cubensis Seedling collected in Lomas Fco Javier Male “Lomas Francisco-2” Carica cubensis Seedling collected in Lomas Fco Javier Male “Lomas Francisco-3” Carica cubensis Seedling collected in Lomas Fco Javier Female “Lomas Francisco-4” Carica cubensis Seedling collected in Lomas Fco Javier Plant without flower “Lomas Francisco-5” Carica cubensis Seedling collected in Lomas Fco Javier Female “Lomas Francisco-6” Carica cubensis Seedling collected in Lomas Fco Javier Male “Papaya” Carica papaya Open pollination of cultivar “Papaya” Female “Maradol Roja” Carica papaya Cuban commercial cultivar Plant without flower “1500” Carica papaya Cuban commercial cultivar Plant without flower lume of isopropanol at –20°C. The nucleic acid pellet was washed in 400 μl of 10 mM NH4Ac in 76% ethanol, dried, resuspended in 50 μl of TE, incubated with 0.5 μg of RNase-A at 37ºC for 30 min to digest RNA, and quantified using a Biophotometer (Eppendorf, Barcelona, Spain). 2.3. Random Amplified Polymorphic DNA (RAPD) Marker Application Nine RAPD universal primers (OPA-07, OPB-07, OPN-14, OPR-15, OPR-16; OPW-12, OPW-13, OPY-13 and OPZ-17) purchased from Operon Biotechnologies (Huntsville, USA) were assayed performing single (one primers) and multiplex (combination of two or three primers) PCR analysis Table 1. Amplifications were carried out in 20 µL total volume containing 1 × Buffer, 1 mM MgCl2, 0.16 mM of dNTP, 0.4 µmol of each primer, 1.0 unit of Taq DNA polymerase (New England Biolabs, Ipswich, USA), and 4 ng templates DNA. The amplification program consisted of a step of DNA melt- ing of 4 min at 94°C, followed by 35 cycles of 94°C for 1 min, 35°C for 1 min, and 72°C for 1 min, and a final elongation step of 72°C for 10 min. Amplified products were resolved in 2% agarose gels stained using Gel Red Nucleic Acid Gel Sating® (Biotium, Hatwad, CA, USA) and visualized with UV transmitted light. A 1 Kb DNA Ladder (Invitrogen Life Technologies, Barcelona, Spain) was used as molecular size standard. RAPD amplifica- tions were repeated at least twice in order to check the reproducibility of bands. 2.4. Data Analysis Polymorphic alleles were scored as present or absent (1/0). DNA band scoring was analyzed using GeneTools gel analysis software of SYNGENE (Beacon House, Nuffield Road, Cambridge, UK). The average polymor- phic information content (PIC) was calculated for RAPD markers across assay units by applying the formula given by Powell et al. (1996). Mean character difference distances were calculated for all pairwise comparisons with the MEGA4 test (http://www.megasoftware.net/) [31], which was used to construct UPGMA dendograms [32] depicting the phenetic relationship among the diff- erent accessions. Relative support for the branches in  J. Rodríguez et al. / Agricultural Sciences 1 (2010) 95-101 Copyright © 2010 SciRes. Openly accessible at http://www.scirp.org/journal/AS/ 98 each dendrogram was assessed with 2000 replicates of UPGMA bootstrap. 3. RESULTS AND DISCUSSION The nine RAPD primers (OPA-07, OPB-07, OPN-14, OPR-15, OPR-16; OPW-12, OPW-13, OPY-13 and OPZ-17) assayed generated polymorphic and reproduci- ble patterns in the Carica papaya and C. cubensis as- sayed genotypes. In addition, to improve the capacity and polymorphism of the molecular characterization assays using RAPD markers different multiplex PCR using more two or three RAPD primers have been as- sayed. Results showed that the incorporation of more than one RAPD primer in the PCR analysis increased the number of obtained bands and the polymorphism of these bands. The number of RAPD bands detected by each primer depended on species, primer and the single or multiple PCR analysis performed. A total of 73 RAPD bands were detected (45 of them polymorphic) with the nine RAPD markers assayed in C. cubensis using single and multiplex PCR analysis with a size range between 105 and 3600 bp. The total number of bands varied from 3 (for OPN-14) to 7 (for OPW-12 and OPZ-13/OPW-12/OPN-14 primer combination), with an average of 4.8 bands per analysis. In addition, the mean PIC score over all loci was 0.70, ranging from 0.33 in OPN-14 to 0.85 in OPZ-13/OPW- 12/OPN-14 primer combination Table 2. Even using lyophilized leaf samples results showed a high yield and good quality DNA with good results after the PCR analysis Figure 3. This fact is very important taking in account that the most crucial factor for the application of RAPD technology is the DNA quality and concentration [33,34]. In the case of C. papaya polymorphism was lower due to the reduced number of accessions assayed (only three) although proportionally higher than the results obtained in C. cubensis in which we assayed 18 accessions. A total of 64 RAPD bands were detected (28 of them po- lymorphic) with the nine RAPD markers assayed in C. cubensis using single and multiplex PCR analysis with a size range between 105 and 3600 bp. The total number of bands varied from 0 (for OPN-14, OPW-13, OPY-13) to 3 (for OPB-07, OPY-13/OPB-07/OPA-07 and OPW- 13/OPR-16/OPR-15 primer combination, with an aver- age of 4.2 bands per analysis. In addition, the mean PIC score over all loci was 0.33, ranging from 0.00 in OPN- 14, OPW-13 and OPY-13 to 0.63 in OPW-13/OPR-16/ OPR-15 primer combination Table 2. RAPD analysis showed higher number of bands (a- bundance) and higher polymorphism in comparison with other markers as has been described in papaya by Jo- bin-Décor [19] and other tropical species such as cacao [35] or coffee [36]. The low heterozygosities values showed in this study can be a consequence of the low number of Carica cubensis accessions assayed which can also be genetically close according with the geo- graphic proximity of the samples collected. For all the loci studied the expected heterozygosity was greater than the observed heterozygosity, implying the presence of null alleles or a deficit of heterozygotes due to non- random mating. The presence of common RAPD alleles indicated the closer genetic distance between the culti- vated papaya and C. cubensis in comparison with other wild papaya species before studied including C. cauli- flora, C. pubescens, C. parviflora or C. quercifolia [14, 19,20]. C. cubensis seem to be subspecies of C. papaya adapted to the environmental conditions of the moun- tains of Cuba or a Cuban endemic species close to C. papaya as was indicated previously by Leon and Alain [5]. Phenetic (taxonomy) relationships among Carica cubensis accessions were analyzed with several UPGMA dendograms Figure 4. Relationships moderately sup 1 2 3 4 5 6 7 8 9 10 11 12 13 14 M M 1 2 11 12 M 1 2 11 12 M 1 2 11 12 M 1 2 11 12 M 1 2 11 12 M 1 2 11 12 M 1 2 11 12 M 1 2 11 12 M 1 2 11 12 M 1 2 11 12 M 1 2 11 12 M 1 2 11 12 M λ1 2 3 4 5 6 7 8 9 10 11 12 13 14 DNA extraction RAPD screening RAPD analysis by single PCR OPA-07 OPX-03 OPA-10 OPG-13 OPZ-17 OPA-08 OPR-15 OPB-11 OPW-12 OPB-07 OPN-15 OPA-11 1 2 3 4 5 6 M M OPY-13 RAPD analysis by multiplex PCR 1 2 3 4 5 6 M OPY-13 + OPB-7 OPY-13 + OPB-7 + OPA-7 a) b) c) d) 1 2 3 4 5 6 7 8 9 10 11 12 13 14 M M 1 2 11 12 M 1 2 11 12 M 1 2 11 12 M 1 2 11 12 M 1 2 11 12 M 1 2 11 12 M 1 2 11 12 M 1 2 11 12 M 1 2 11 12 M 1 2 11 12 M 1 2 11 12 M 1 2 11 12 M 1 2 11 12 M 1 2 11 12 M 1 2 11 12 M 1 2 11 12 M 1 2 11 12 M 1 2 11 12 M λ1 2 3 4 5 6 7 8 9 10 11 12 13 14 DNA extraction RAPD screening RAPD analysis by single PCR OPA-07 OPX-03 OPA-10 OPG-13 OPZ-17 OPA-08 OPR-15 OPB-11 OPW-12 OPB-07 OPN-15 OPA-11 1 2 3 4 5 6 M M OPY-13 RAPD analysis by multiplex PCR 1 2 3 4 5 6 M OPY-13 + OPB-7 OPY-13 + OPB-7 + OPA-7 a) b) c) d) Figure 3. Agarose gel showing the DNA extraction of Carica samples (a); the first screening of RAPD markers in some genotypes (b); and the application of RAPD markers in all the genotypes using simple (c) and multi- plex (d) PCR analysis. Λ DNA quantification marker HindII from Invitrogen. M DNA ladder 1 Kb plus from Invitrogen.  J. Rodríguez et al. / Agricultural Sciences 1 (2010) 95-101 Copyright © 2010 SciRes. Openly accessible at http://www.scirp.org/journal/AS/ 99 99 Table 2. Universal primers (decamers) purchased from Operon Biotechnologies used as RAPD markers molecular characterization of the Carica cubensis accessions assayed. Carica cubensis Marker Number of bandsPolymorphic bandsSize (bp) PIC OPA-07 4 2 500-800 0.58 OPB-07 5 3 490-5490 0.83 OPN-14 3 1 970-2170 0.33 OPR-15 4 3 560-1110 0.83 OPR-16 5 3 750-3200 0.71 OPW-12 7 4 700-3250 0.75 OPW-13 4 2 500-1400 0.55 OPY-13 3 1 376-755 0.31 OPZ-17 6 4 400-915 0.76 OPY-13/OPB-07 4 3 95-755 0.79 OPY-13/OPB-07/OPA-07 5 3 95-1046 0.82 OPZ-13/OPW-12 6 4 500-2500 0.83 OPZ-13/OPW-12/OPN-14 7 5 300-3600 0.85 OPW-13/OPR-16 5 4 750-3500 0.80 OPW-13/OPR-16/OPR-15 5 3 750-3500 0.80 Figure 4. Dendograms obtained by UPGMA cluster analysis based on mean character differences among the Carica cubensis and Carica papaya accessions assayed using RAPD markers by single and multiplex PCR anlaysis. Numbers in the branches represent bootstrap values. ported established two groups in relation to the two dif- ferent studied species. In addition, bootstrap values of UPGMA dendogram obtained with the utilization of RAPDs were slightly higher. In the case of C. cubensis in the most of cases accessions studied from a location are closer than accessions from other locations. This clustering is explained by geographic proximity and can indicate the similar precedence of the isolated studied populations. However, no clustering and RAPD markers association was observed in our samples according to the sex characterization as has been before observed in C. papaya species [8,21-23]. These results also can support both establish hypothesis C. cubensis as a subspecies of C. papaya or a Cuban endemic species close to C. pa- paya. One of the goals of conservation programs in these rare fruit species such as Carica cubensis is to charac- terize and maintain existing level of variation and ge- netic resources [10]. Genetic resources not only provide the required raw material for suitable genetic crop im- provement, but offer a unique gene combination to en- sure adaptability and productivity [37,38]. Designing of core collection using suitable DNA markers involves an appropriate use of diversity, offering breeders an oppor- tunity to work with a manageable number of accessions. C. cubensis is a better adapted species to the Cuban con- ditions and could also be a source of new genes for low temperature resistance or disease resistance including the Papaya Ring Spot Virus (PRSV) [39,40]. Other impor- tant characteristic of this species is the smaller fruit (around 100 grams) which will be a new good commer- cial trait.  J. Rodríguez et al. / Agricultural Sciences 1 (2010) 95-101 Copyright © 2010 SciRes. Openly accessible at http://www.scirp.org/journal/AS/ 100 Designing of core collection involves an appropriate use of diversity, offering breeders an opportunity to work with a manageable number of accessions. Therefore, the availability of an optimized protocol for DNA charac- terization to estimate the genetic diversity and to ensure genetically representative non-redundant samples is of great interest [41]. In this sense, universal RAPD mark- ers can be a most suitable in the molecular characteriza- tion of C. cubensis accessions and to construct efficient core collections as has been described by Bortolini [42] in white clover complementary to the application of more recent developed markers in C. papaya of domi- nant SPAR (Single primer amplification reaction) [17] and SCAR (sequence characterized amplified region) markers [25], and codominant SSR (Single sequence repeat) markers [43,44]. Other additional advantages of RAPD markers are that do not require labour intensive and expensive, no genomic/cDNA libraries are need for development of probes and very small quantity of DNA is need in the PCR reactions even using lyophilized leaf samples. 4. ACKNOWLEDGEMENTS This work is part of an International Collaboration between the Instituto Nacional de Ciencias Agrícolas (INCA) of La Habana (Cuba) and the CEBAS-CSIC of Murcia (Spain). Authors want to thanks the help of the Herbario Nacional de Cuba in the identification of the Carica cubensis accessions assayed. REFERENCES [1] Badillo, V. (1971) Monographia de la familia Caricaceae. Maracay, Venezuela. [2] Manshardt, R.M. (1992) Papaya. In: Biotechnology of Perennial Fruit Crops, Hammerschlag, F.A. and Litz, R.E., Eds., Cambridge University Press, Oxford, 489-511. [3] Heinrichs, J., Lindner, M., Groth, H., Hentschel, J., Feldberg, K., Renker, C., Engel, J.J., von Konrat, M., Long, D.G. and Schneider, H. (2006) Goodbye or welcome Gondwana? Insights into the phylogenetic biogeography of the leafy liverwort Plagiochila with a description of Proskauera, gen. nov. (Plagiochilaceae, Jungermanniales). Plant Systematic and Evolution, 258, 227-250. [4] Feldberg, K., Hentschel, J. and Wilson, R. (2007) Phy- logenetic biogeography of the leafy liverwort Herbertus based on nuclear and chloroplast DNA sequence data: Correlation between genetic variation and geographical distribution. Journal of Biogeography, 34, 688-698. [5] Leon, H. and Alain, H. (1953) Familia caricaceae. In: Flora de Cuba, Ed., Museo de Historia Natural del Cole- gio de La sale, La Habana, 352-354. [6] Solms, L. and Grafen, H. (1889) Die heimath un der ursprung des cultivirten melonebaumes, Carica papaya L. Botanische Zeitung, 49, 791-798. [7] Sing, I. and Sirohi, S.C. (1977) Sex expression studies in papaya. Plant Journal Research, 2, 150-152. [8] Reddy, G.M. (2006) Seedling leaf morphology in identi- fication of sex types and confirmation through RAPD markers in Carica papaya L. Journal of Genetic and Breeding, 60, 1-10. [9] Oviedo, R. and Ventosa, I. (2006) Fichas del Herbario Nacional de Cuba. Academia Nacional de Ciencias de Cuba. [10] Martínez-Gómez, P., Majourhat, K., Zeinalabedini, M., Erogul, D., Khayam-Nekoui, M., Hafidi, A., Piqueras, A. and Gradziel, T.M. (2007) Use of biotechnology for pre- serving rare fruit germplasm. Bioremediation, Biodiver- sity and Bioavailability, 1, 31-40. [11] Powell, W., Morgante, M., Andre, C., Hanafey, M., Vogel, J., Tingey, S. and Rafalski, A. (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Molecular Breeding, 2, 225-238. [12] Welsh, J. and McClelland, M. (1990) Finger printing genomes using PCR with arbitraries primers. Nucleic Acid Research, 18, 7213-7218. [13] Messaoud, C., Afif, M., Boulila, A., Rejeb, M.N. and Boussaid, M. (2007) Genetic variation of Tunisian Myr- tus communis L. (Myrtaceae) populations assessed by isozymes and RAPDs. Annals of Forest Science, 63, 845-853. [14] Sharon, D., Hillel, J., Vainstein, A. and Lavi, U. (1992) Application of DNA fingerprintings for identification and genetic analysis of Carica papaya and other Carica spe- cies. Euphytica, 62, 119-126. [15] Stiles, J.I., Lemine, C., Sondur, S., Morshidi, M.B. and Manshardt, M. (1993) Randomly amplified polymorphic DNA for evaluating genetic relationships among papaya cultivars. Theoretical and Applied Genetics, 85, 697-701. [16] Vitoria, A.P., de Souza, G.A., Bressan-Smith, R.E., Pinto, F.D., Pascal, B., Guimaraes, P.S., Daben, R.F. and Gon- zaga, M. (2004) DNA fingerprint of Carica papaya L. genotypes by RAPD markers. Journal of New Seed, 6, 51-65. [17] Saxena, S., Chandra, R., Srivastava, A.P., Mishra, M. and Ranade, S.A. (2005) Analysis of genetic diversity among papaya cultivars using single primer amplification reac- tion (SPAR). Journal of Horticultural Science & Bio- technology, 80, 291-296. [18] da Silva, F.F., Pereira, G.M., Campos, W., Damasceo- Junior, P.C., Santana-Pereira, T.M., Cancela-Ramos, H.G., Pio-Viana, A. and Ferregeti, G.A. (2007a) Monitoring of the genetic variability in papaya parent “Formosa” of “UENF/CALIMAN 01” hybrid via RAPD. Crop Breed- ing and Applied Biotechnology, 7, 36-42. [19] Jobin-Décor, M.P., Graham, G.C., Henry, R.J. and Drew, R.A. (1997) RAPD and isozyme analysis of relationships between Carica papaya and wild species. Genetic Re- sources and Crop Evolution, 44, 471-477. [20] Magdalita, P.M., Drew, R.A., Adkins, S.W and Godwin, I.D. (1997) Morphological, molecular and cytological analysis of Carica papaya L. and C. cauliflora inter- specific hybrids. Theoretical and Applied Genetics, 95, 224-229. [21] Parasmis, A.S., Gupta, V.S., Tambankar, S.A. and Ran- jekar, P.K. (2000) A highly reliable sex diagnosis PCR assay for mass screaning of papaya seedlings. Molecular Breeding, 6, 337-344.  J. Rodríguez et al. / Agricultural Sciences 1 (2010) 95-101 Copyright © 2010 SciRes. Openly accessible at http://www.scirp.org/journal/AS/ 101 101 [22] Lemos, E.G.M., Silva, C.L. and Zcaidad, H.A. (2002) Identification of sex in Carica papaya L. using RAPD markers. Euphytica, 127, 179-184. [23] Urasaki, N., Tokumoto, M., Tanora, K., Kayano, T., Tamaka, H., Oku, H. and Terauchi, P. (2002) A male and hermaphrodite specific RAPD markers for papaya. Theoretical and Applied Genetics, 104, 281-285. [24] da Silva, F.F., Pereira, G.M., Ferrerira-Campos, W., Damasceo-Junior, P.C., Santana-Pereira, T.M., Souza- Filho, G.A., Cancela-Ramos, H.G., Pio-Viana, A. and Ferregeti, G.A. (2007b) DNA marker-assisted sex con- version in elite papaya genotype (Carica papaya L.). Crop Breeding and Applied Biotechnology, 7, 52-58. [25] Niroshini, E., Everard, J.M., Karunahayane, E.M. and Tirimanne, M.C.S. (2008) Detection of sequence charac- terized amplified region (SCAR) markers linked to sex expression in Carica papaya L. Journal of National Sci- ence Foundation of Sri Lanka, 36, 145-150. [26] Sondur, S.N., Manshard, R.M. and Stiles, J.I. (1996) A genetic linkage map of papaya based on RAPD markers. Theoretical and Applied Genetics, 93, 542-553. [27] Narvel, J.M., Chu, W.C., Fehr, W.R., Cregan, P.B. and Shoemaker, R.C. (2000) Development of multiplex sets of simple sequence repeat DNA markers covering the soybean genome. Molecular Breeding, 6, 175-183. [28] Sánchez-Pérez, R., Dicenta, F. and Martínez-Gómez, P. (2004) Identification of S-alleles in almond using multi- plex PCR. Euphytica, 138, 263-269. [29] Hayden, M.J., Nguyen, T.M., Waterman, A. and Chal- mers, K.J. (2008) Multiplex-ready PCR: A new method for multiplexed SSR and SNP genotyping. BMC Genom- ics, 9, 80 [30] Doyle, J.J. and Doyle, J.L. (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phy- tochemestry Bulletin, 19, 11-15. [31] Tamura, K., Dudley, J., Nei, M. and Kumar, S. (2007) MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution, 24, 1596-1599. [32] Nei, M. and Li, W.H. (1979) Mathematical model for studying genetic variation in terms of restriction. Pro- ceedings of the National Academy of Science of USA, 76, 5269-5273. [33] Dax, E., Livneh, O., Edelbaum, O., Kedar, N., Gavish, N., Karchi, H., Milo, J., Sela, I. and Rabinowit, H.D. (1993) A random amplified polymorphic DNA (RAPD) mo- lecular marker for the Tm-2agene in tomato. Euphytica, 1-2, 159-163. [34] Gérard, P.R., Fernández-Manjarrés, J.F., Bertolino, P., Dufour, J., Raquin, C. and Frascaria-Lacostem, F. (2006) New insights in the recognition of the European ash spe- cies Fraxinus excelsior and Fraxinus augustifolia as useful tools for forest management. Annals of Forest Science, 63, 733-738. [35] Yamada, M.M., Faleiro, F.G., Lopes, U.V., Bahia, R.C., Pires, J.L., Gomes, L.M.C. and Melo, G.R.P. (2001) Genetic variability in cultivated cacao populations in Bahia, Brazil, detected by isozymes and RAPD markers. Crop Breeding and Applied Biotechnology, 1, 377-384. [36] Teixeira-Cabral, T.A., Sakiyama, N.S., Zambolim, L., Pereira, A.A., Gonçalves-Barros, E. and Sakiyama, C.C.H. (2002) Reproducibility of the RAPD marker and its efficiency in coffee tree genotype grouping analysis. Crop Breeding and Applied Biotechnology, 2, 121-129. [37] Wang, J.C., Hu, J., Liu, N.N., Xu, H.M. and Zhang, S. (2006) Investigation of combining plant genotypic values and molecular marker information for constructing core subsets. Journal of Integrated Plant Biology, 48, 1371- 1378. [38] Lachenaud, P. and Zhang, D. (2008) Genetic diversity and population structure in wild stands of cacao trees (Theobroma cacao L.) in French Guiana. Annals of For- est Science, 65, 310. [39] Magdalita, P.M., Villegas, V.M., Pimentel, R.B. and Bayot, R.G. (1988) Reaction of papaya and related spe- cies to ringspot virus Phillipp. Journal of Crop Science, 3, 232-234. [40] Persley, D.M. and Thomas, J.E. (1994) Screening for PRSV resistance. Annual Meeting of Control of Ringspot in Papaya, Hawaii, 41-44. [41] Chavarriaga-Aguirre, P., Maya, M.M., Tohme, J., Duque, M.C., Iglesias, C., Bonierbale, M.W., Kresovich, S. and Kochert, G. (1999) Using microsatellites, isozymes and AFLPs to evaluate genetic diversity and redundancy in the cassava core collection and to assess the usefulness of DNA-based markers to maintain germplasm collections. Molecular Breeding, 5, 263-273. [42] Bortolini, F., Dall’Agnol, M. and Schifino-Wittmann, M.T. (2006) Molecular characterization of the USDA white clover (Trifolium repens L.) core collection by RAPD markers. Genetic Resources and Crop Evolution, 53, 1081-1087. [43] Pérez, J.O., Dambier, D., Ollitrault, P., D’Eeckenbrugge, G.O., Brotiers, P., Froelicher, Y. and Risterucci, A.M. (2006) Microsatellite markers in Carica papaya L.: Isolation, characterization and transferability to Vascon- cellea species. Molecular Ecology Research, 6, 212-217. [44] Eustice, M., Yu, Q., WanLai, C., Thimmapuran, J., Liu, L., Afan, M., Presting, G. and Ming, R. (2008) Develop- ment and application of microsatellite markers for ge- nomics analysis in papaya. Tree Genetic and Genomic, 4, 333-341. |

