Journal of Biomedical Science and Engineering
Vol.09 No.13(2016), Article ID:72979,14 pages
10.4236/jbise.2016.913053
Metabolic Cycles: Effect of a Simultaneous Input and Output of Two Substrates
Antonio Sillero*, Víctor García-Herrero
Departamento de Bioquímica, Facultad de Medicina, Instituto de Investigaciones Biomédicas Alberto Sols (UAM/CSIC), Arzobispo Morcillo, Madrid, Spain

Copyright © 2016 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY 4.0).
http://creativecommons.org/licenses/by/4.0/



Received: October 19, 2016; Accepted: December 23, 2016; Published: December 26, 2016
ABSTRACT
The metabolic cycle firstly considered here is composed of a unique initial substrate, six enzymes, and five empty boxes to accommodate the substrates derived from the transformation of the initial substrate. This cycle was considered as a pre-Closed Metabolic Cycle (CMC). Using this model, the influence of changing the kinetic constant values of any enzyme on the substrate concentration was explored. This model was transformed into an open metabolic cycle (OMC) by the input and output of two metabolites catalyzed by two external enzymes. In this case, the relative rates of input and output of metabolites were also examined; it can be concluded that the OMC cycles form delicate and fragile structures which can be theoretically disrupted, ma- king them metabolically unfeasible.
Keywords:
Metabolic Cycles, Metabolic Regulation, Kinetic Constants, Differential Equations, Metabolites: Input and Output

1. Introduction
Several aspects of metabolic pathways (linear or cyclic) have been lately approached in our laboratory [1] . The theoretical treatment of a linear pathway was simulated assuming that an initial substrate was transformed into a final product through the successive appearance and disappearance of metabolic intermediates of the route [1] ; the addition of a new enzyme, catalyzing the transformation of the first into the last substrate converts a linear cycle into a cyclic pathway [2] . The occurrence, properties and theoretical usefulness of closed metabolic cycles (CMC) have been considered in previous publications from this laboratory [2] [3] [4] .
The vast majority of metabolic cycles in nature are open metabolic cycles (OMC), i.e. cycles with entrance and exit of metabolites at different substrate levels. The OMC cycles can be studied with different and complementary approaches, among them, by measuring the level of their components and analyzing potential changes in their concentration in different metabolic or nutritional conditions. However, these are cumbersome procedures and sometimes difficult to be implemented [5] [6] [7] . The advances in computational techniques have allowed a more comprehensive understanding of open metabolic cycles [5] [8] [9] [10] [11] [12] .
In the course of our work [2] [3] [4] [13] , we have observed a scarcity of studies on the physiological consequences of the potential transformation between linear and cyclic pathways.
Two types of complementary studies on metabolic cycles are presented in this work: a) analysis of how changes in the kinetic constants (Vmax and/or Km values) of only one of the enzymes influence the steady concentration of all substrates of the cycle; b) consequences of the transformation of a closed metabolic cycle into an open metabolic cycle caused by the simultaneous entry and exit of a metabolite at a different level; as shown below many of these changes are not metabolically feasible.
Although the practical applications of this work to specific metabolic cycles are countless, they have not been approached here; however the core of this manuscript is to facilitate its potential application to any metabolic cycle regardless the number of input and output enzymes affecting the cycle.
2. Materials and Methods
Nomenclature
A joint representation of a closed and an open metabolic cycle (CMC and OMC, respectively) is depicted in Figure 1. The CMC here considered is composed of 6 substrates, (a) to (f). The enzymes involved in the transformation of consecutive substrates are, E1a, E2b, E3c, E4d, E5e and E6f. These enzymes will be named also sub indexed with the corresponding substrate only (Ea to Ef).
In order to simplify both the presentation of the results and the calculation by Mathematica, the Km and Vmax constant values of these enzymes are simply named as, K and V, sub indexed with the corresponding substrate, 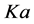 ,
, 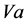 ,
, 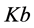 ,
, 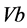 , etc. (Table 1).
, etc. (Table 1).
The Mathematica program 9 [14] was used to solve the differential equations describing the pathways. The protocol, similar to that previously described [13] is summarized in Table 1:
Part (a) contains the actual equation velocity of the 6 enzymes involved in the Closed Metabolic Cycle (v1 to v6) (Figure 1).
Part (b) contains the Vmax and Km values of the enzymes of the ensemble.
Part (c) contains the instructions to calculate the substrate value concentrations (profiles) along the reaction time.
Part (d) contains the instructions to Plot the substrate profiles resulting in each case from the value assigned to the respective substrates.
Figure 1. Joint representation of a pre-closed metabolic cycle (CMC) and an open metabolic cycle (OMC); the CMC is composed exclusively by the inter-conversion of 6 substrates (a) to (f) catalyzed by 6 enzymes Ea to Ef. A CMC can be converted into an OMC by the input of (c) from (xc) and exit of (e), catalyzed by enzymes E7xc and E8ew, respectively.
Table 1. Mathematica protocol to calculate the time course of substrates (Profiles) of a metabolic cycle.
3. Results
1) A preformed stable theoretical metabolic cycle is a flexible structure that can support reasonable modifications in both the amount of the starting materials and in the kinetic constant values of the concerned enzymes
Several possibilities of changes are shown below, using a kind of preformed closed metabolic cycle, composed by 6 substrates, (a) to (f) (Figure 1), six enzymes E1a, E2b, E3c, E4d, E5e, E6f and fed with only one initial substrate (a) at a concentration of 12 mM; following simulation, substrate (a) is distributed among the previously empty boxes of the cycle according to the kinetic constant values of the concerned enzymes. In Table 2, column at the left, are the names of the cycles to be explored: from DD0 to DD6L. The kinetic constants of each enzyme (Vmax and Km) are named from Va to Vf and from Ka to Kf, and the corresponding values are indicated in Table 2. The particular values for the kinetic constants Vf and Kf are also indicated in Table 2. The Mathematica protocols for the drawing of the substrate profiles are summarized in Table 1.
a) Influence of the Vmax and Km values of one enzyme (Ef) on the concentration of its own and other substrates of the cycle
Taking substrate (f) as a reference, it can be easily appreciated the grossly reciprocal influence between the Vmax and Km values of Ef on the concentration of its substrate (f) (Table 2, Figure 2). Compare the equilibrium position of substrate (f) in the standard cycle (Table 2, Figure 2, Profile of the control cycle (DD0), at the top) and after the changes introduced in: i) the Vf values(maintaining constant the Kf value) (Table 2, Figure 2, Profiles DD1 and DD2 (the left side); ii)or when to changes in the Vf values, changes in Kf values were superimposed (Table 2, Figure 2, Profiles DD3 and DD4 (right side).
Table 2. Kinetic constants values Vmax and Km (abbreviated as V and K, respectively) of the six enzymes (Ea, Eb, Ec, Ed, Ee, Ef) participating in a model Closed Metabolic Cycles (CMC). The sub-indexes of V and K correspond to the indicated enzyme (Figure 1). The kinetic constant values of the control cycle (DD0) are in the same line and are identical (=) for the enzymes Ea to Ee); the specific constants values for the Enzyme (Ef) (Vf and Kf) are indicated in the Table for the cycles DDO to DD6L.
Figure 2. Representation of 5 illustrative Closed Metabolic Cycles with the kinetic constant values consigned in Table 2. The cycle at the top corresponds to DD0 and the two at the left to DD1 and DD2; those at the right correspond to DD3 and DD4. The name of the substrates from (a) to (f) is indicated in each cycle. It is supposed that the simulation started with the only presence of (a, 12 mM), that was distributed among the previously empty boxes of the cycle according to the kinetic constant values of the enzymes. The equilibrium was settled at 300 min of reaction. Only as a reference, substrates (a) and (f) were taken as the first and the last substrates of the cycle, respectively. See the text for further details.
Changes in the constant values of enzyme Ef influence not only substrate (f) but also, in varying degree the rest of substrates (Figure 2).
b) A preformed stable theoretical metabolic cycle is flexible and can support reasonable modifications in both the amount of the starting materials and in the kinetic constant values of the concerned enzymes … until a certain limit
The operator can establish this limit at will. We have here considered a cycle as not viable when one of its substrates reached, at stationary phase, a concentration above 90% to 99% the sum of the other substrates. This can be theoretically achieved by extreme changes in the Vmax or Km values of one of the enzymes of the cycle, as shown in Table 2, Figure 3: Profiles DD5 (decrease in Vf value) or Profile DD6 (increase in Kf value). For different reasons, substrate (f) accumulates in both cases. Very similar representations for DD5 and DD6 (DD5L or DD6L) are obtained, simply by making Vf = 0 (irrespective to the value assigned to Kf (results not shown)). As a matter of fact, and
Figure 3. Representation of 2 illustrative Closed Metabolic Cycles (DD5L and DD6L) with the kinetic constant values consigned in Table 2.
from a practical viewpoint, the changes in DD5, DD6, DD5L or DD6L (Figure 3) transform a closed metabolic cycle into linear cycle.
2) Transformation of a closed metabolic cycle (CMC) into an open metabolic cycle (OMC) by the simultaneous action of two additional enzymes catalyzing the entry (enzyme E7) and exit (enzyme E8) of two metabolites
The Mathematica simulation was performed following similar procedures to those described in (Table 1) (corresponding to a closed metabolic cycle composed of 6 enzymes) but with the important modification of the simultaneous addition of two external enzymes E7xc (or E7) and E8ew (or E8) (Figure 1, Table 3). These two enzymes promoted the simultaneous entry of substrate (c) by E7xc, catalyzing the transformation of one external substrate (cx) into (c), and the output of (e) by (E8ew).
These two enzymes transform the previously closed metabolic cycle (CMC), into an open metabolic cycle (OMC). The new considered kinetic constant values for the enzymes of the OMC are consigned in Table 3. Following similar procedures as those previously described [4] , Table 4 was elaborated from Table 3 under the assumption that the steady state concentration of each substrate resulted from its rate of synthesis and degradation and also from the overall rate of input and output of substrates in the cycle by enzymes E7 and E8.
For the arrangement of Table 4 the following was considered:
*If (a) reaches a steady state, a’ must be equal to 0, so
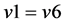 (1)
(1)
or equivalently
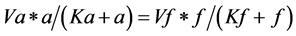 (2)
(2)
*With the same reasoning applied to (b), b' must be 0, so
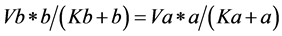 (3)
(3)
*The same applies for the substrates (d) and (f), giving the equations
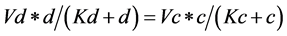 (4)
(4)
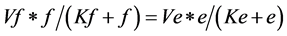 (5)
(5)
*Now, if the substrate (c) reaches a steady state, c' = 0, that is
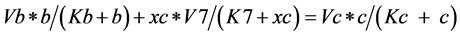 (6)
(6)
*To obtain the last formula we just have to apply the same procedure for the substrate (e), to obtain:
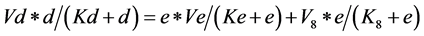 (7)
(7)
Based on the above reasoning’s, Table 4 contains: i) successive substrates to be considered at equilibrium (column at the left); ii) the corresponding formulas (column at the right); iii) the Mathematica commands used to calculate the substrate concentration in the specified conditions (panel at the bottom).
The results presented in Table 5 were obtained as follows: i) the kinetic constant values for the enzymes Ea to Ef were identical to those considered in Table 3; ii) the kinetic values for E7 and E8 (the enzymes responsible for the entry an the exit of metabolites, respectively) were as indicated in this Table 5; iii) two different concentrations were considered for the input of substrate cx, i.e. 3 and 15 mM. The resulting concentration values for the substrates (a) to (f) were calculated for each different cycle (A1 to A3) and (B1 to B3).
Cycles A1 and B1 are metabolically feasible; however, when these cycles are confronted with an increment in V7(cycles A2, B2) these cycles become not viable, probably due to the higher Vmax value (7) of enzyme E7 and the subsequent accumulation of substrate (e). But cycles A2, B2 may become feasible if the maximum velocity of enzyme E8 increases, by removing substrate (e) from the cycle (see A3 and B3).
Cycles of this type may be very sensitive to changes in the Vmax values of the enzymes V7 or V8, as exemplified in Figure 4, where the substrate profiles obtained when the V7 value of cycle A1 (see Table 5) increases from 1.2 (Panel (a)) to 1.6 (Panel (b)), are shown. This small increase in the V7 value (Panel (b)) gives rise to an unfeasible cycle, due to the abnormal steady increase of substrate (e). Panel (c) (Figure 4) shows
Table 3.Mathematica protocol to calculate the substrate profiles of a previously closed metabolic cycle with (a) 12 mM and with the simultaneous input and output of two metabolites catalyzed by V7 and V8, respectively.
Table 4. A general Mathematica procedure to calculate the concentration of the substrates of an Open Metabolic Cycle (OMC), composed of 6 substrates, and with the input of an external substrate (cx), (3 or 15 mM) and the output of cyclic substrate (e) catalyzed by E7 and E8, respectively. Note as the cycle was started with only (a) 12 mM.
Table 5. Influence of changes in the Vmax and Km of the enzymes catalyzing the input (v7) or output (v8) of substrates into a closed metabolic cycle, at two different concentration of the input substrate (cx).
how the increase of substrate (e) was reversed by increasing the Vmax of the output enzyme (E8) from a value of V8 = 1 to a value of V8 = 7.
4. Discussion
By definition a true open metabolic cycle contains at least one pathway for the entry and another one for the exit of metabolites. The occurrence of this type of cycles is well documented in several instances: Krebs/glyoxylate: [7] [12] [15] [16] [17] ; urea: [5] [17]
Figure 4. Influence of small changes in the rate of the maxima velocities of the enzymes catalyzing the input (E7) and output (E8) of substrates in an Open Metabolic Cycle.
[10] ; pentose and Calvin cycles: [18] [19] .
However, contemplating general maps of biochemical pathways it seemed to us that groups of substrates are so interconnected among them and with distant metabolites, to the point of making difficult to delimitate whether they are part of metabolic cycles. Particular examples of this type of networks can be visualized at the level of the synthesis and degradation of lipids, purine or pyrimidine nucleoside/nucleotides, glycogen and monosaccharide’s, etc.
There is scarce information on how these cycles have emerged or even how life started: the prebiotic chemistry of life [20] [21] and how the early chemical evolution took place [21] [22] through a long evolutionary robust response towards environmental fluctuations [23] ; the possibility that the diversity of metabolism aroused by the evolution of few metabolic modules was explored in [24] . A plethora of metabolic pathways have been reported in the Central Nervous System [25] . Particular interest has been also paid to the evolution of the primitive cells and the role and existence of the Last Universal Common Ancestor (LUCA) [19] [20] [21] [22] [26] .
Several examples of metabolic cycles are shown in this work, each one composed of 6 enzymes with distinct kinetic constant values. We started considering a kind of preformed closed metabolic cycle (CMC) with 5 empty boxes ready to accommodate 5 substrates and a unique starting substrate located in one of the boxes at a predetermined concentration. As soon as the cycle is initiated the unique substrate is distributed among the previously empty boxes as if the cycle were actually an irreversible pathway. After few min, and as consequence of the repetition of the cyclic pathway, the substrates of the cycle reach equilibrium at a fixed concentration and the derivatives of the profiles are zero.
5. Concluding Remarks
Simulation of metabolic cycles uncovers some interesting peculiarities practically impossible to be tested in vivo: a) the metabolic cycles are interrelated structures in which changes in the kinetic constant values in one of its enzymes may modify all the substrate concentrations; b) by the same token modifications of only one of the substrates is followed by its re-equilibration with the rest of substrates of the cycle; c) possible inborn errors of metabolism mainly because of the absence of one enzyme, disrupts the cycle, totally or partially; in the proper open metabolic cycles (OMC) it is critical the ratio between the inflow and outflow of material; inadequate balance between these two processes, may have fatal consequences for a correct cyclic performance; d) finally the therapeutic effect of many drugs could be due to their effect on the Km values of some enzymes, some of them corresponding to linear or cyclic pathways.
Acknowledgements
We thank Javier Pérez for very helpful drawing assistance and Dr. Maria Antonia Günther Sillero for helpful comments on the manuscript. This investigation was supported by Grant BU2009-08877.
Conflict of Interest Statement
None declared.
Cite this paper
Sillero, A. and Gar- cía-Herrero, V. (2016) Metabolic Cycles: Ef- fect of a Simultaneous Input and Output of Two Substrates. J. Biomedical Science and Engineering, 9, 624-637. http://dx.doi.org/10.4236/jbise.2016.913053
References
- 1. López-Canovas, F., Gomes, P.J. and Sillero, A. (2013) Mathematica Program: Its Use to Simulate Metabolic Irreversible Pathways and Inhibition of the First Enzyme of a Pathway by Its End Product as Visualized with the Reservoir Model. Computers in Biology and Medicine, 43, 853-864.
https://doi.org/10.1016/j.compbiomed.2013.04.003 - 2. García-Herrero, V., López-Cánovas, F.J. and Sillero, A. (2014) A Model Metabolic Cycle Simulated with the Mathematica Program. Journal of Biomedical Science and Engineering, 7, 286-295.
https://doi.org/10.4236/jbise.2014.75031 - 3. García-Herrero, V. and Sillero, A. (2015) Pedagogical View of Model Metabolic Cycles. Biochemistry and Molecular Biology Education, 43, 468-475.
https://doi.org/10.1002/bmb.20920 - 4. Sillero , A. and García-Herrero, V. (2015) Theoretical Evaluation of Both Unknown Substrate Concentrations and Enzyme Kinetics Constants of Metabolic Cycles. Journal of Biomedical Science and Engineering, 8, 479-489.
https://doi.org/10.4236/jbise.2015.88045 - 5. Alberty, R.A. (2011) Enzyme Kinetics. Rapid-Equilibrium Applications of Mathematica. Methods of Biochemical Analysis, 53, 1-445.
https://doi.org/10.1002/9780470940020 - 6. Morris Jr., S.M. (2002) Regulation of Enzymes of the Urea Cycle and Arginine Metabolism. Annual Review of Nutrition, 22, 87-105.
https://doi.org/10.1146/annurev.nutr.22.110801.140547 - 7. Albe, K.R. and Wright, B.E. (1992) Systems Analysis of the Tricarboxylic Acid Cycle in Dictyostelium Discoideum. II. Control Analysis. The Journal of Biological Chemistry, 267, 3106-3114.
- 8. Korla, K. and Mitra, C.K. (2014) Modelling the Krebs Cycle and Oxidative Phosphorylation. Journal of Biomolecular Structure and Dynamics, 32, 242-256.
https://doi.org/10.1080/07391102.2012.762723 - 9. Chalhoub, E., Hanson, R.W. and Belovich, J.M. (2007) A Computer Model of Gluconeogenesis and Lipid Metabolism in the Perfused Liver. American Journal of Physiology. Endocrinology and Metabolism, 293, E1676-E1686.
https://doi.org/10.1152/ajpendo.00161.2007 - 10. Maher, A.D., Kuchel, P.W., Ortega, F., de Atauri, P., Centelles, J. and Cascante, M. (2003) Mathematical Modelling of the Urea Cycle. A Numerical Investigation into Substrate Channelling. European Journal of Biochemistry, 270, 3953-3961.
https://doi.org/10.1046/j.1432-1033.2003.03783.x - 11. Bachmann, C. and Colombo, J.P. (1981) Computer Simulation of the Urea Cycle: Trials for an Appropriate Model. Enzyme, 26, 259-264.
- 12. Wu, F., Yang, F., Vinnakota, K.C. and Beard, D.A. (2007) Computer Modeling of Mitochondrial Tricarboxylic Acid Cycle, Oxidative Phosphorylation, Metabolite Transport, and Electrophysiology. The Journal of Biological Chemistry, 282, 24525-24537.
https://doi.org/10.1074/jbc.M701024200 - 13. Sillero , A. and García-Herrero, V. (2016) Closed and Open Metabolic Cycles: Transition Time. Journal of Biomedical Science and Engineering, 9, 127-140.
https://doi.org/10.4236/jbise.2016.92009 - 14. Wolfram, S. (2013) Wolfram Mathematica Tutorial Collection.
http://www.wolfram.com/learningcenter/tutorialcolection - 15. Oliveira, J.S., Bailey, C.G., Jones-Oliveira, J.B., Dixon, D.A., Gull, D.W. and Chandler, M.L. (2003) A Computational Model for the Identification of Biochemical Pathways in the Krebs Cycle. Journal of Computational Biology, 10, 57-82.
https://doi.org/10.1089/106652703763255679 - 16. Nazaret, C., Heiske, M., Thurley, K. and Mazat, J.P. (2009) Mitochondrial Energetic Metabolism: A Simplified Model of TCA Cycle with ATP Production. Journal of Theoretical Biology, 258, 455-464.
https://doi.org/10.1016/j.jtbi.2008.09.037 - 17. Amador-Noguez, D., Feng, X.J., Fan, J., Roquet, N., Rabitz, H. and Rabinowitz, J.D. (2010) Systems-Level Metabolic Flux Profiling Elucidates a Complete, Bifurcated Tricarboxylic Acid Cycle in Clostridium acetobutylicum. Journal of Bacteriology, 192, 4452-4461.
https://doi.org/10.1128/JB.00490-10 - 18. Melendez-Hevia, E. (1990) The Game of the Pentose Phosphate Cycle: A Mathematical Approach to Study the Optimization in Design of Metabolic Pathways during Evolution. Biomed Biochim Acta, 49, 903-916.
- 19. Ranea, J.A., Sillero , A., Thornton, J. and Orengo, C. (2006) Protein Superfamily Evolution and the Last Universal Common Ancestor (LUCA). Journal of Molecular Evolution, 63, 513-525.
https://doi.org/10.1007/s00239-005-0289-7 - 20. Lazcano, A. and Miller, S.L. (1996) The Origin and Early Evolution of Life: Prebiotic Chemistry, the Pre-RNA World, and Time. Cell, 85, 793-798.
https://doi.org/10.1016/s0092-8674(00)81263-5 - 21. Wachtershauser, G. (1990) Evolution of the First Metabolic Cycles. Proceedings of the National Academy of Sciences, 87, 200-204.
https://doi.org/10.1073/pnas.87.1.200 - 22. De Duve, C. (1987) Selection by Differential Molecular Survival: A Possible Mechanism of Early Chemical Evolution. Proceedings of the National Academy of Sciences, 84, 8253-8256.
https://doi.org/10.1073/pnas.84.23.8253 - 23. Jeong, H., Tombor, B., Albert, R., Oltvai, Z.N. and Barabasi, A.L. (2000) The Large-Scale Organization of Metabolic Networks. Nature, 407, 651-654.
https://doi.org/10.1038/35036627 - 24. Marakushev, S.A. and Belonogova, O.V. (2013) The Divergence and Natural Selection of Autocatalytic Primordial Metabolic Systems. Origins of Life and Evolution of Biospheres, 43, 263-281.
https://doi.org/10.1007/s11084-013-9340-7 - 25. Siegel, G.J., Sgranoff, B.W., Albers, R.W. and Molinoff, P.B. (1994) Basic Neurochemistry. 5th Edition, Raven Press, New York.
- 26. Woese, C.R. (2002) On the Evolution of Cells. Proceedings of the National Academy of Sciences, 99, 8742-8747.
https://doi.org/10.1073/pnas.132266999
Abbreviation Note List
OMC: Open metabolic cycle.
CMC: Closed metabolic cycle.

























DD0, DD1, DD2, DD3, DD4, DD5, DD5L, DD6L, Name of the indicated cycles, with the constant Values specified in (Table 2).
E1a or Ea: enzyme converting substrate a into substrate b; E2b or Eb: enzyme converting substrate b into substrate c; E3c or Ec: enzyme converting substrate c into substrate d; E4d or Ed: enzyme converting substrate d into substrate e; E5e or Ee: enzyme converting substrate e into substrate f; E6f or Ef: enzyme converting substrate f into substrate a. E7xc: enzyme converting substrate xc into substrate c; E8e: enzyme converting substrate e into substrate ew (Figure 1).
Submit or recommend next manuscript to SCIRP and we will provide best service for you:
Accepting pre-submission inquiries through Email, Facebook, LinkedIn, Twitter, etc.
A wide selection of journals (inclusive of 9 subjects, more than 200 journals)
Providing 24-hour high-quality service
User-friendly online submission system
Fair and swift peer-review system
Efficient typesetting and proofreading procedure
Display of the result of downloads and visits, as well as the number of cited articles
Maximum dissemination of your research work
Submit your manuscript at: http://papersubmission.scirp.org/
Or contact jbise@scirp.org









