Journal of Biomedical Science and Engineering
Vol.6 No.1(2013), Article ID:27046,6 pages DOI:10.4236/jbise.2013.61003
Hydrogen bonds are related to the thermal stability of 16S rRNA
![]()
Department of Clinical Laboratory Science, Graduate Course of Medical Science and Technology, School of Health Sciences, Kanazawa University, Kanazawa, Japan
Email: naka@kenroku.kanazawa-u.ac.jp
Received 29 October 2012; revised 30 November 2012; accepted 5 December 2012
Keywords: Optimal Growth Temperature; 16S Ribosomal RNA; G + C Content; Hydrogen Bonds; Base Pairs; Nucleotide Compositions
ABSTRACT
The number of base pairs in the 16S rRNA secondary structures of 51 bacterial sequences was counted, and the number of hydrogen bonds was estimated. The number of hydrogen bonds was highly correlated with the optimal growth temperature (OGT) rather than with the G + C content. Paired and unpaired nucleotides in mesophiles were compared to those in thermophiles. OGT exhibited a relationship with paired nucleotides but not with unpaired nucleotides. The total number of paired as well as unpaired nucleotides in mesophiles was very similar to that in thermophiles. However, the components in base pairs in mesophiles significantly differed from those in thermophiles. As compared with mesophiles, the number of G·C base pairs in thermophiles was high whereas that of A·U base pairs was low. In this study, we showed that hydrogen bonds are important for stabilizing 16S rRNAs at high temperatures.
1. INTRODUCTION
Bacteria can live in a wide temperature range from the freezing point of water to its boiling point. This indicates that the environment where water exists in the liquid state can be inhabited by bacteria. At their living temperature, macromolecules such as protein, DNA and RNA are stable and can perform their biological functions. DNA and RNA consist of nucleotides, sugars and phosphates. Thymine and deoxyribose in DNA are replaced by uracil and ribose in RNA. DNA is double stranded and RNA is single stranded. The method of DNA stabilization at high temperatures is different from that of RNA. The dinucleotide composition of DNA is related to the optimal growth temperature (OGT) [1,2], and mononucleotide composition i.e., G + C content of RNA is proportional to their OGT [2-6]. The uracil content of 16S rRNA has a significant inverse correlation with the OGT [7]. Hyperthermophiles have higher RNA G + C content. The G·C base pair has 3 hydrogen bonds and A·U base pair has 2 hydrogen bonds. Therefore, hydrogen bonds seem to play an important role for RNA thermal stability, however, the relationship between the number of hydrogen bonds and OGT has not reported yet.
Ribosomes are the machinery necessary to produce proteins based on the mRNA, which is a blueprint of genetic information. There are 3 types of bacterial ribosomal RNAs—5S, 16S, and 23S named according to their molecular weights. 16S rRNA is the most conservative of the 3 rRNAs, and is used to identify bacterial species on the basis of the phylogenetic tree. It is believed that their secondary structure, determined by base pairing, is more conservative than the nucleotide sequence. Three-dimensional structure of 16S rRNA of Thermus thermophilus was resolved by X-ray crystallographic studies [8,9]. The Gutell group predicted base pairs in 16S rRNA of bacteria, which are available through the web [10]. Using these data, base pairs in the 16S rRNA structures were counted and the number of hydrogen bonds was estimated. In addition to these studies, we reexamined the relationship between the G + C content of 16S rRNA and OGT.
2. MATERIALS AND METHODS
The sequences and base pairs of 16S rRNAs were retrieved from the comparative RNA web site
(http://www.rna.icmb.utexas.edu/) [10]. Base pair information of bacterial 16S rRNAs was available, but archaeal data were not available; therefore, we analyzed only bacterial data in this study. Fifty sequences were randomly selected from various species to cover a wide range of OGT (Table 1). The data for T. thermophilus was obtained from the literature [9]. The dataset included 13 sequences from thermophiles, 35 sequences from
Table 1. List of species used in this study.
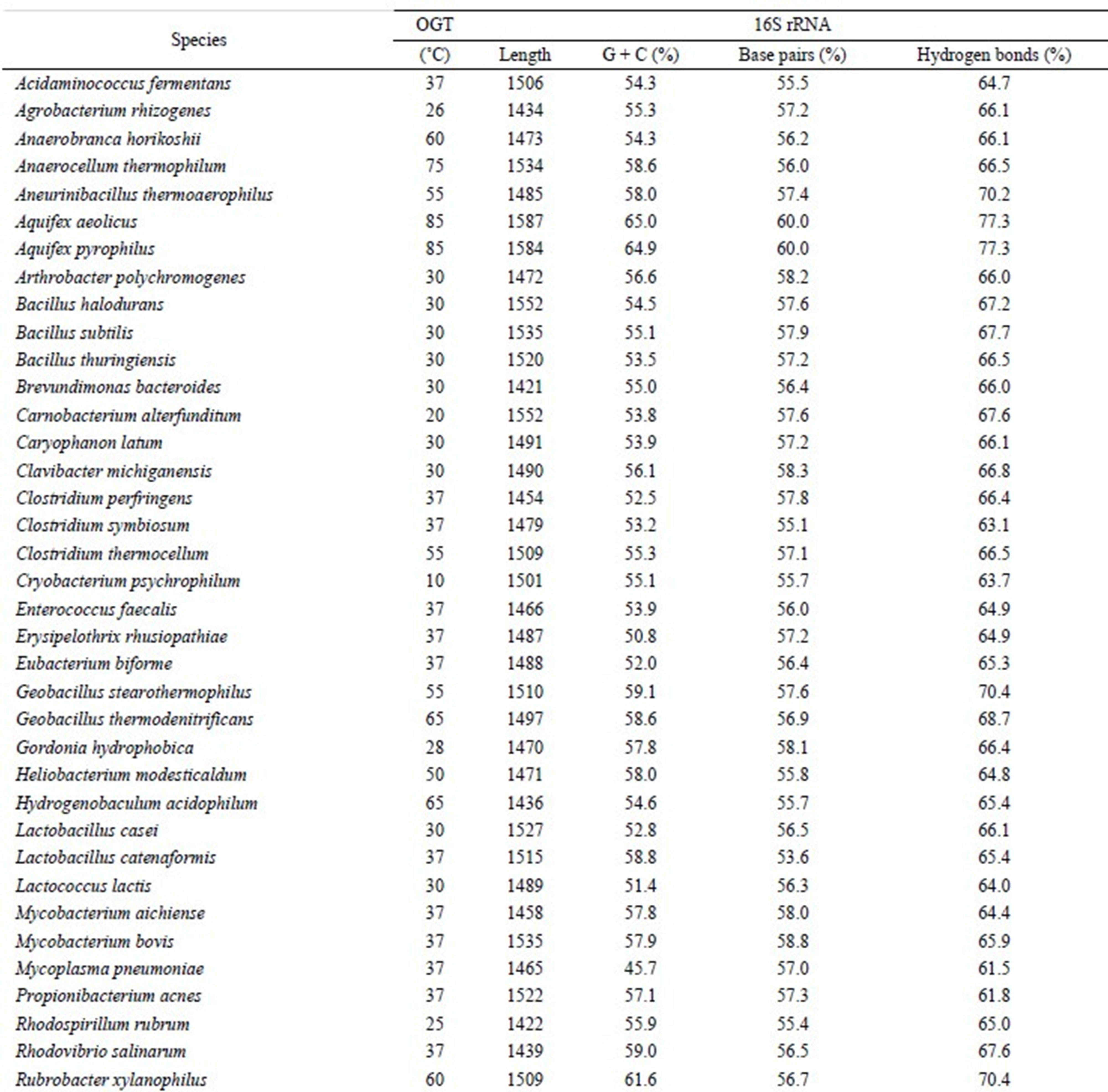
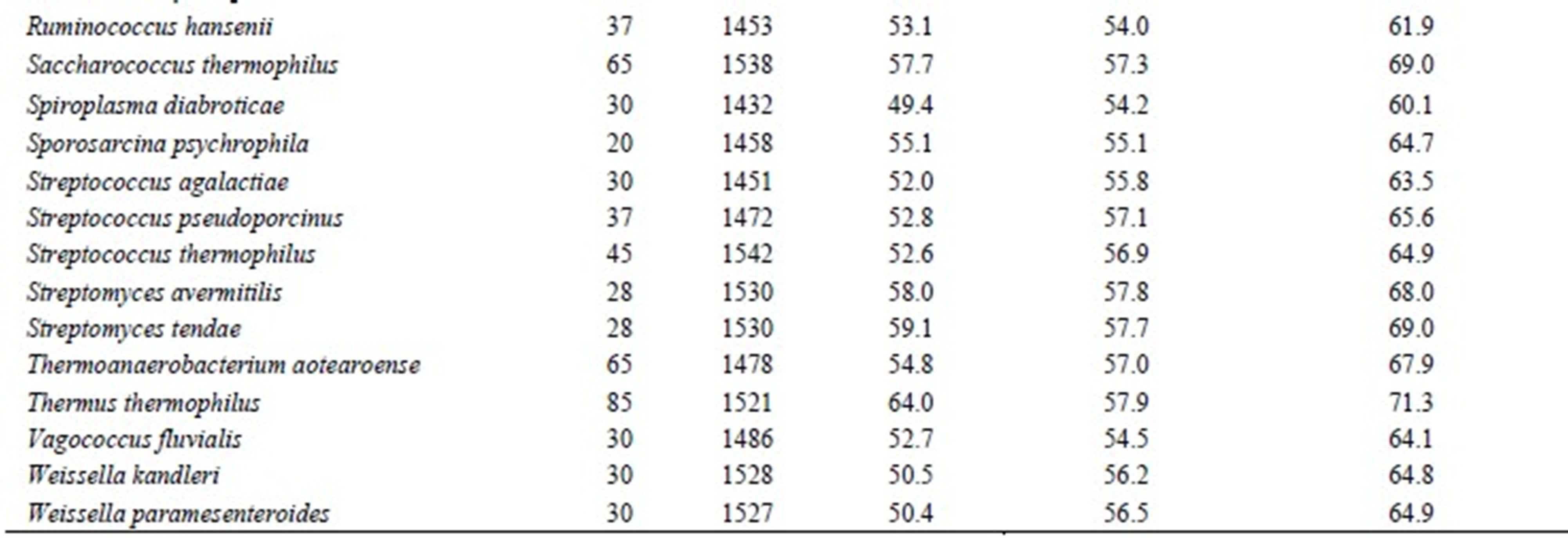
mesophiles, and 3 sequences from pyschrophiles. Thermophiles grow above 55˚C and psychrophiles grow below 20˚C. There are 4 types of nucleotides; hence, 16 types of base pairs are possible. However, an A·U base pair is identical to a U·A base pair. Therefore, 10 types of base pairs were considered; they were as follows: A·U, G·C, G·U, A·G, C·C, U·U, C·U, A·C, A·A, and G·G. Interestingly, G·C, A·U, and G·U base pairs were dominant, and the sum of other 7 base pairs equaled only approximately 5% of the total base pairing. Therefore, only the hydrogen bonds comprising G·C, A·U, and G·U base pairs were taken into account in this study. The number of hydrogen bonds for G·C and A·U base pairs was estimated as 3 and 2, respectively. The number of hydrogen bonds for G·U base pair has been reported to be 1 or 2 [11]. Therefore, the number of hydrogen bonds was estimated by the following equations:
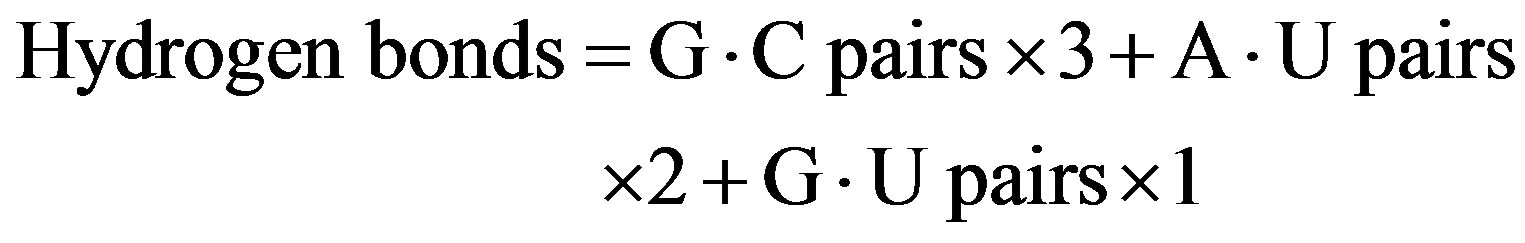
or

We also calculated the hydrogen bonds consisting of G·C and A·U base pairs as follows:
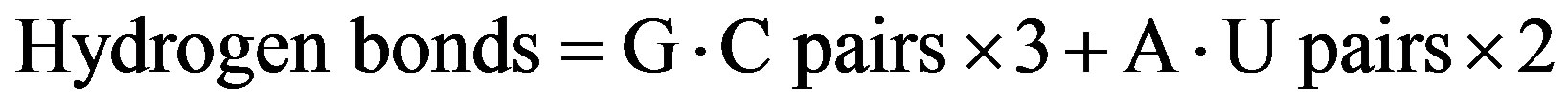
Using these equations, we calculated the number of hydrogen bonds in 3 ways. The percentage of hydrogen bonds was calculated as the number of hydrogen bonds divided by the length of 16S rRNA. The percentage of base pairs was calculated as the sum of G·C, A·U, and G·U base pairs divided by the length of 16S rRNA.
OGTs were retrieved from the web site http://www.dsmz.de/species/strains.htm. Monoand dinucleotide compositions of 16S rRNAs were calculated. Expected dinucleotide compositions were calculated using the mononucleotide compositions, and the ratio of observed/calculated compositions was thus obtained. The average compositions were also calculated for both thermophiles and mesophiles. The average of mesophiles was calculated, including data from three psychrophiles. The 3 psychrophiles examined did not differ significantly from the mesophiles with regard to the G + C content or the percentage of hydrogen bonds.
3. RESULTS
3.1. Hydrogen Bonds in 16S rRNA versus OGT
The plot of the percentage of hydrogen bonds consisting of G·C and A·U base pairs versus OGT expressed in degrees Celsius (˚C) showed the highest correlation (correlation coefficient, 0.65) (Figure 1). When the hydrogen bonds from the G·U base pairs were considered, the correlation coefficient was found to be 0.63 and 0.57, when the G·U base pair was assumed to contain 1 and 2 hydrogen bonds, respectively. Several G·U base pairs were observed in the 16S rRNA secondary structures; however, hydrogen bonds from G·U base pairs did not increase the correlation with the OGT. G + C content of the 16S rRNA versus the OGT is shown in Figure 2. This resulted in a correlation coefficient of 0.59, which was lower than the hydrogen bonds. The ratio of G·C base pairs in the G·C and A·U base pairs increased with OGT, and the ratio of G·C base pairs showed a high correlation with the G + C content (correlation coefficient, 0.96). This result indicates that a correlation between
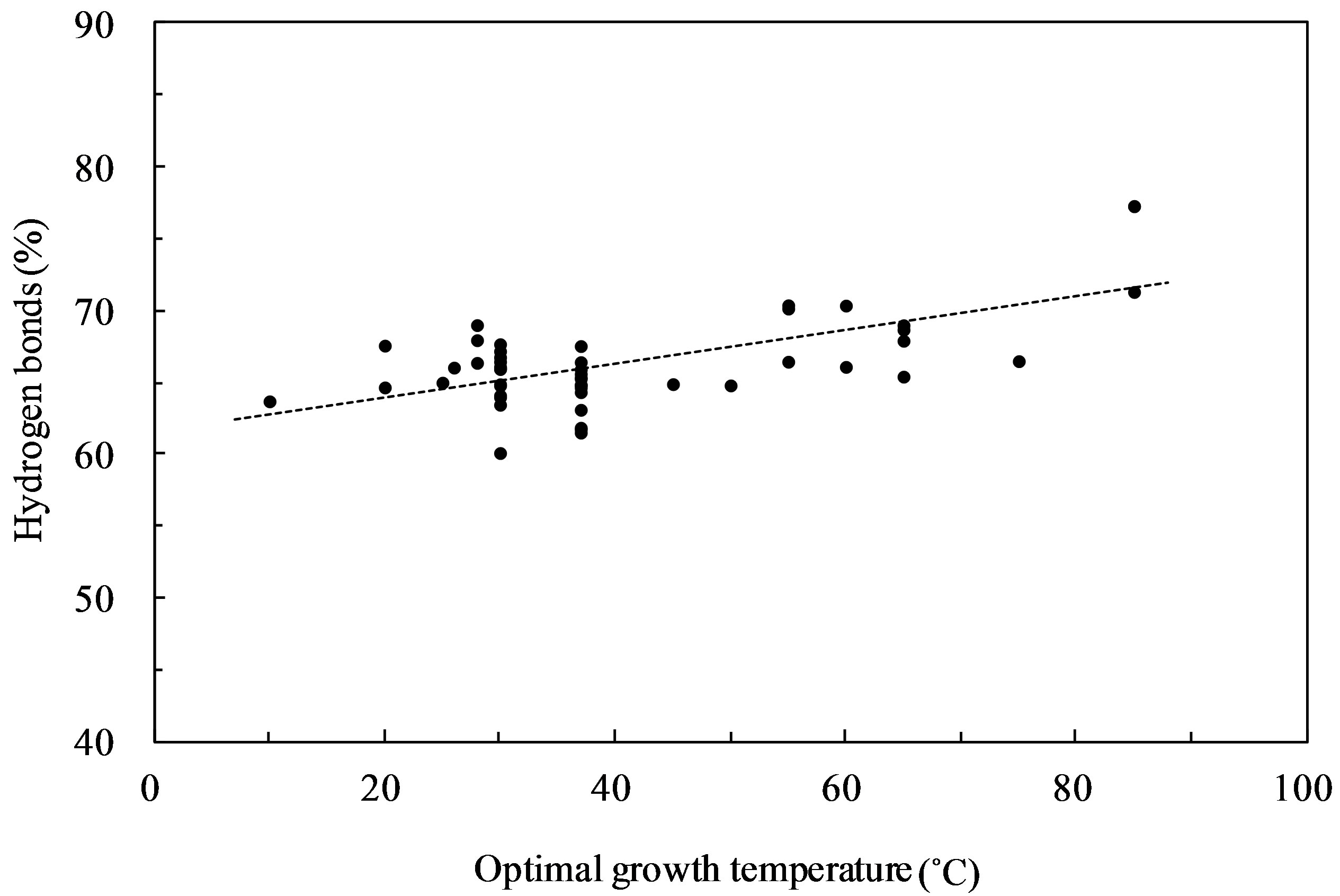
Figure 1. Hydrogen bonds (%) in 16S rRNAs against optimal growth temperature. The hydrogen bonds consist of G·C and A·U base pairs.

Figure 2. G + C contents (%) of 16S rRNAs against optimal growth temperature.
RNA G + C content and OGT is a secondary effect of the hydrogen bonds and OGT.
3.2. Nucleotide Composition of 16S rRNA
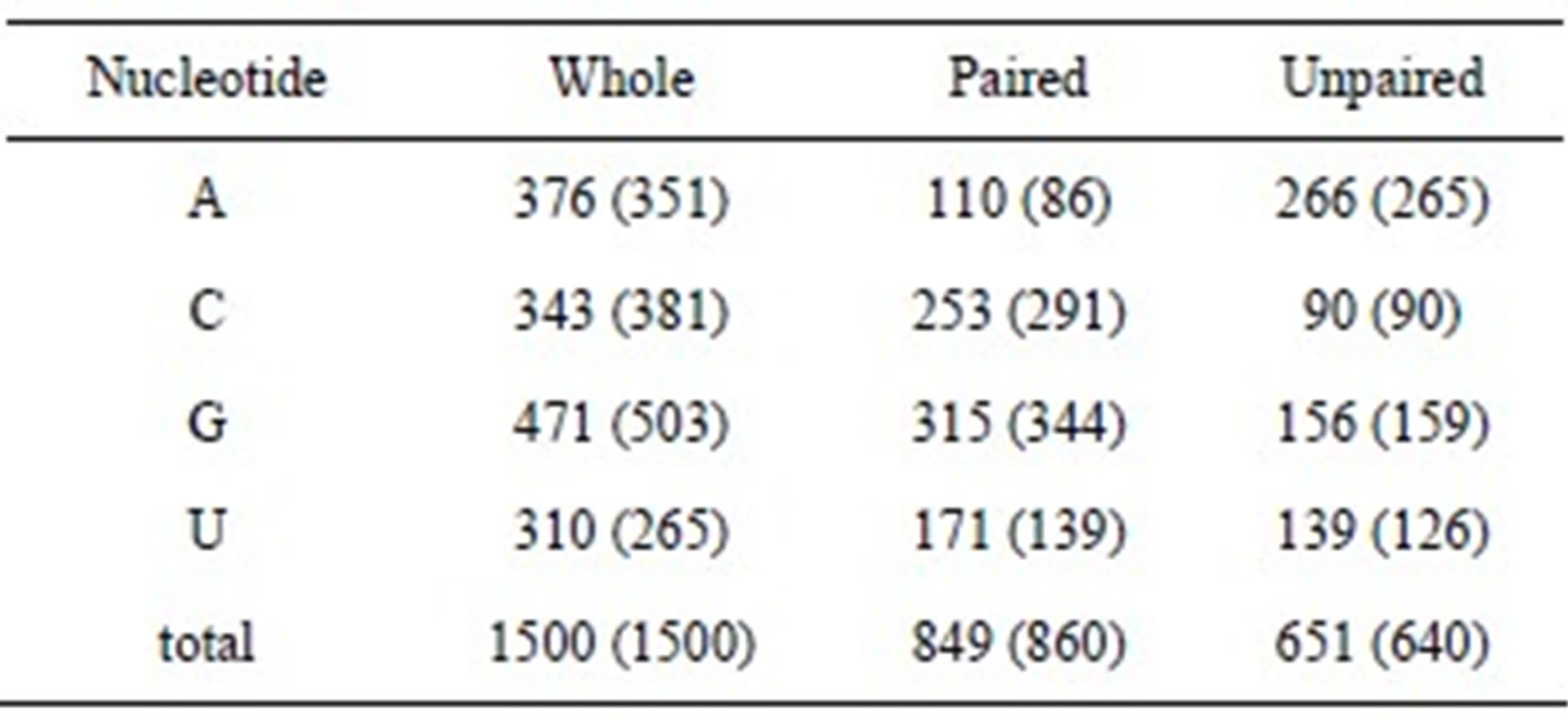
To make maximum base pairing, the guanine content should be equal to the cytosine content, and the adenine content should be equal to the uracil content. However, the guanine content was higher than that of cytosine, and the adenine content was higher than that of uracil, except in the case of Propionibacterium acnes. To show the difference in the nucleotide content, we calculated the ratios of guanine/cytosine and adenine/uracil. The average ratios of guanine/cytosine and adenine/uracil were 1.36 and 1.25, respectively. This result indicates that purines are more abundant than pyrimidines in 16S rRNA sequences. The numbers of paired and unpaired nucleotides were estimated for both thermophiles and mesophiles by using the average compositions as the length of 16S rRNA, which was assumed to be 1500 nucleotides. Table 2 shows the comparison between mesophiles and thermophiles, with the data for thermophiles represented within parentheses. The deviation of nucleotide components in the whole sequence between mesophiles and thermophiles was roughly identical with the deviation in paired nucleotides. For example, the deviation of adenine in the whole sequence was 25 and that in paired was 24. The number of unpaired nucleotides was roughly identical between mesophiles and thermophiles, with the exception of uracil. This result indicates that unpaired nucleotides are independent of the OGT. Adenine was the most abundant, and the cytosine was the least in the unpaired nucleotides. This result was consistent with the high percentage of unpaired adenine in 16S rRNA structure models [6,12]. It is reported that nearly 75% of AA dinucleotides are found in loops in rRNA sequences [10]. We found that the AA dinucleotide is the most favorable on the basis of the ratios of observed to calculated composition (see below). Total base-paired nucleotides in thermophiles were slightly higher than those in mesophiles, however, the base pair components were quite different between the 2 groups. For example, base-paired cytosine in thermophiles was 38 higher than that in mesophiles. In contrast, paired uracil was higher in mesophiles than in thermophiles. This result indicates that, in contrast to mesophiles, G·C base pairs were abundant and A·U base pairs were few in thermophiles. Thus, thermophiles increase the amount of G·C base pairs in rRNAs to adjust to high temperatures.
The relationship between G+C content and OGT can be expressed as shown in Eq.1 from our previous study [2];
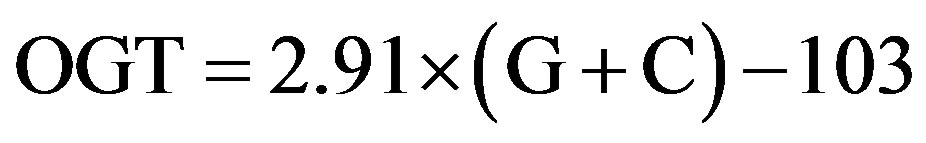 (1)
(1)
where OGT is estimated in degrees Celsius (˚C), and G + C refers to the percentage of guanine and cytosine content in 16S rRNA. In this study, the relationship obtained by least-square regression analysis was slightly different
Table 2. Comparisons of paired and unpaired nucleotides of 16S rRNA between mesophiles and thermophiles. Data for thermophiles are indicated within parentheses.
from Eq.1 and was expressed as Eq.2:
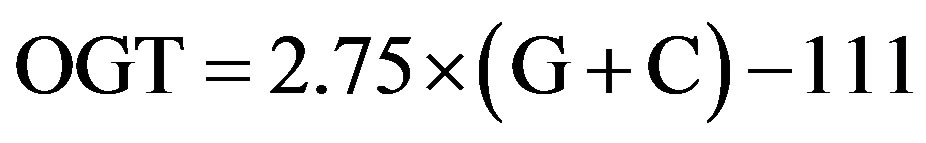 (2)
(2)
Favorable and unfavorable dinucleotides were estimated in terms of ratios of the observed to calculated compositions. The AA and UG dinucleotides showed average ratios greater than 1.1 and were considered favorable, and AU and UC had average ratios less than 0.9 and were considered unfavorable. The other 12 dinucleotides showed average ratios in the range of 0.9 - 1.1.
4. DISCUSSION
The number of hydrogen bonds was estimated by the secondary structures of the paired bases. Therefore, the accuracy of base pair is very important. Gutell et al. evaluated the 16S and 23S rRNA structure models against the crystal structures, and they reported approximately 97% - 98% of the base pairings predicted were indeed corresponding to their experimental data [13].
A simple way to attain thermal stability of nucleic acids is to increase the number of hydrogen bonds, i.e., the G + C content. This is indeed observed for the 16S rRNAs of thermophiles, in which the G + C content is increased with OGT. If the same strategy for thermal stability as observed in RNA was applicable to DNA as well, the increased G + C content would have significant effects on the amino acid composition. For example, amino acids encoded by G + C rich codons such as Ala, Arg, Gly, and Pro would be abundant, whereas those encoded by G + C poor codons such as Lys, Ile, Tyr, and Phe would be less represented in thermophilic proteins. We found no correlation between the G + C content of DNA and OGT. Instead, the dinucleotide composition of DNA was found to be correlated with OGT [2]. rRNAs do not encode proteins; therefore, G + C content of rRNA seemed to have no restrictions. In fact, the G + C content of RNA depend on the G + C content of genomic DNA in mesophiles [14], whereas no correlation was observed between the G + C contents of RNA and genomic DNA in thermophiles [2]. As shown in Figure 2, the G + C contents from mesophiles showed greater deviations from the regression line than the plots in hydrogen bonds (Figure 1). This result suggests that mesophiles have more freedom to have various G + C content in 16S rRNA than hydrogen bonds.
Hydrogen bonds of G·U base pairs did not increase the correlation to OGT. The hydrogen bond of a G·U base pair was observed in the syn conformation of guanine, on the edge of the bulge loop in the hairpin loop structure [11]. If G·U base pairs are located toward the center of the secondary structure, which forms the helical structure, it is difficult for guanine to assume the syn conformation; hence, it might be difficult to form hydrogen bonds between the G·U base pairs. It is reported that G·U base pairs are found in different conformations in different chemical and structural environments, and the RNA double helix can be more easily altered at sites of G·U base pairs [15]. Future studies are needed to examine the relationship between hydrogen bonds of G·U base pairs and thermal stability.
REFERENCES
- Kawashima, T., Amano, N., Koike, H., Makino, S., Higuchi, S., Kawashima-Ohya, Y., Watanabe, K., Yamazaki, M., Kanehori, K., Kawamoto, T., Nunoshiba, T., Yamamoto, Y., Aramaki, H., Makino, K. and Suzuki, M. (2000) Archaeal adaptation to higher temperatures revealed by genomic sequence of Thermoplasma volcanium. Proceedings of the National Academy of Sciences of the United States of America, 97, 14257-14262. doi:10.1073/pnas.97.26.14257
- Nakashima, H., Fukuchi, S. and Nishikawa, K. (2003) Compositional changes in RNA, DNA and proteins for bacterial adaptation to higher and lower temperatures. The Journal of Biochemistry, 133, 507-513. doi:10.1093/jb/mvg067
- Galtier, N. and Lobry, J.R. (1997) Relationships between genomic G + C content, RNA secondary structures, and optimal growth temperature in prokaryotes. Journal of Molecular Evolution, 44, 632-636. doi:10.1007/PL00006186
- Galtier, N., Tourasse, N. and Gouy, M. (1999) A nonhyperthermophilic common ancestor to extant life forms. Science, 283, 220-221. doi:10.1126/science.283.5399.220
- Hurst, L.D. and Merchant, A.R. (2001) High guaninecytosine content is not an adaptation to high temperature: A comparative analysis amongst prokaryotes. Proceeding of the Royal Society B. Biological Sciences, 268, 493-497.
- Wang, H.-C. and Hickey, D.A. (2002) Evidence for strong selective constraint acting on the nucleotide composition of 16S ribosomal RNA genes. Nucleic Acids Research, 30, 2501-2507. doi:10.1093/nar/30.11.2501
- Khachane, A.N., Timmis, K.N. and Martins dos Santos, V.A.P. (2005) Uracil content of 16S rRNA of thermophilic and psychrophilic prokaryotes correlates inversely with their optimal growth temperatures. Nucleic Acids Research, 33, 4016-4022. doi:10.1093/nar/gki714
- Yusupov, M.M., Yusupova, G.Z., Baucom, A., Lieberman, K., Earnest, T.N., Cate, J.H.D. and Noller, H.F. (2001) Crystal structure of the ribosome at 5.5A resolution. Science, 292, 883-896. doi:10.1126/science.1060089
- Brodersen, D.E., Clemons Jr., W.M., Carter, A.P., Wimberly, B.T. and Ramakrishnan, V. (2002) Crystal structure of the 30S ribosomal subunit from Thermus thermophilus: Structure of the proteins and their interactions with 16S RNA. Journal of Molecular Biology, 316, 725- 768. doi:10.1006/jmbi.2001.5359
- Cannone, J.J., Subramanian, S., Schnare, M.N., Collett, J.R., D’Souza, L.M., Du, Y., Feng, B., Lin, N., Madabusi, L.V., Müller, K.M., Pande, N., Shang, Z., Yu, N. and Gutell, R.R. (2002) The comparative RNA web (CRW) site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics, 3, 1-31. doi:10.1186/1471-2105-3-1
- Cheong, C., Varani, G. and Tinoco Jr., I. (1990) Solution structure of an unusually stable RNA hairpin, 5’GGAC(UUCG)GUCC. Nature, 346, 680-682. doi:10.1038/346680a0
- Gutell, R.R., J.J. Cannone, J.J., Shang, Z., Du, Y. and Serra, M.J. (2000) A story: Unpaired adenosine bases in ribosomal RNAs. Journal of Molecular Biology, 304, 335-354. doi:10.1006/jmbi.2000.4172
- Gutell, R.R., Lee, J.C. and Cannone, J.J. (2002) The accuracy of ribosomal RNA comparative structure models. Current Opinion in Structural Biology, 12, 301-310. doi:10.1016/S0959-440X(02)00339-1
- Muto, A. and Osawa, S. (1987) The guanine and cytosine content of genomic DNA and bacterial evolution. Proceedings of the National Academy of Sciences of the United States of America, 84, 166-169. doi:10.1073/pnas.84.1.166
- Varani, G. and McClain, W.H. (2000) The G·U wobble base pair. EMBO Reports, 1, 18-23. doi:10.1093/embo-reports/kvd001

