Journal of Cancer Therapy
Vol.3 No.1(2012), Article ID:17212,8 pages DOI:10.4236/jct.2012.31003
Association of CYP2B6 Genotype with Survival and Progression Free Survival in Cyclophosphamide Treated Multiple Myeloma
![]()
1Division of Drug Research, Department of Medical and Health Sciences, Faculty of Health Sciences, Linköpings University, Linköping, Sweden; 2Department of Pharmacy, COMSATS Institute of Information Technology, Abbottabad, Pakistan; 3Department of Clinical and Experimental Medicine, Faculty of Health Sciences, Linköpings University, Linköping, Sweden; 4Department of Hematology, Karolinska University Hospital and Karolinska Institute, Huddinge, Sweden; 5Science for Life Laboratory, School of Biotechnology, Division of Gene Technology, Royal Institute of Technology, Solna, Sweden.
Email: Ingrid.jakobsen.falk@liu.se
Received November 7th, 2011; revised December 9th, 2011; accepted December 20th, 2011
Keywords: Multiple Myeloma; Cyclophosphamide; CYP2B6; Glutathion-S-Transferases (GST); Single Nucleotide Polymorphisms; Pharmacogenetics; Pyrosequencing
ABSTRACT
Objective: Cyclophosphamide is a conventional pro-drug used in Multiple Myeloma (MM) and other malignancies. The highly polymorphic CYP2B6 is suggested as a major contributor in cyclophosphamide bioactivation, and GST enzymes are involved in detoxification. Polymorphisms of these enzymes may affect enzyme expression and function as well as treatment outcome. The aim of this study was to investigate the impact of the CYP2B6 SNPs G516T, A785G and C1459T, GSTP1 SNP Ile105Val, and GSTM1 and GSTT1 null variants, on the outcome for cyclophosphamide treated MM patients, in order to find markers of value for individualised therapy. Methods: We used allele specific PCR and Pyrosequencing to investigate the impact of CYP2B6 SNPs G516T, A785G and C1459T, GSTP1 Ile105Val, and GSTM1 and GSTT1 variants, on the outcome for 26 cyclophosphamide treated multiple myeloma patients. Results and Major Conclusion: The CYP2B6 785G carriers had significantly shorter progression free survival (p = 0.048*) and overall survival (p = 0.037*) with 785G/G patients having the worst outcome compared to patients carrying the wild type. A shorter progression free survival was also indicated in patients carrying both CYP2B6 516T & 785G (p = 0.068). These results indicate a predictive role of CYP2B6 SNPs, particularly A785G, in cyclophosphamide treatment.
1. Introduction
Cyclophosphamide is an alkylating agent with anticancer activity and used in the treatment of several neoplastic disorders such as multiple myeloma, leukemias, nonhodgkin lymphoma, breast and ovarian cancer. Multiple Myeloma (MM) is characterised by clonal proliferation of abnormal plasma cells infiltrating the bone marrow, and secretion of monoclonal immunoglobulins, called Mcomponent. MM accounts for approximately 2% of all cancer deaths and 20% of deaths caused by haematological malignancies [1]. Alkylating agents, such as melphalan and cyclophosphamide, together with corticosteroids and autologous stem cell transplantation (ASCT), is an effective conventional treatment [2]. The so-called novel agents, proteasome inhibitors and IMiDs (immunomodulatory drugs) used in the treatment of MM (bortezomib, thalidomide and lenalidomide) has been established and improved the outcome of these patients. In spite of this progress, almost all patients treated with ASCT relapse due to residual disease [3]. Yet, a fraction of patients have become long-term survivors after treatment with alkylating agents and better assessment for applying current treatment modalities is required in order to understand the nature of different responses to the therapy.
Cyclophosphamide is activated by CYP450 mediated hydroxylation in the liver mainly by CYP2B6 [4]. This enzyme is polymorphic, with several single nucleotide polymorphisms (SNPs) reported to affect enzyme expression and function. The functional SNPs G516T (CYP2B6*9, exon 4, Q172H, rs3745274), A785G (CYP 2B6*4, exon 5, K262R, rs2279343) and C1459T (CYP 2B6*5, exon 9, R487C, rs3211371) are frequent in Caucasians, and the functional effects of the polymorphisms appears to be substrate specific [5-9]. Further, detoxification of drugs including cyclophosphamide is carried out by the GST enzymes including GSTM1, GSTT1 and GS TP1, by glutathione conjugation. Polymorphisms of these GST enzymes have been reported to affect enzyme function and correlate to both cancer susceptibility and treatment outcome [10-14]. In GSTM1 and GSTT1, the polymorphisms results in partial gene deletion, with the null variants completely lacking enzymatic activity [10,11]. In GSTP1, the single nucleotide polymorphism with an Isoleucine->Valine substitution in position 105 (exon 5, I105V, rs1695) results in changes in heat stability and activity [14,15].
Taken together, polymorphisms in these genes are of interest as potential predictors of outcome in cyclophosphamide treatment, and results from studies clarifying their impact on treatment response may improve the possibilities of individualising chemotherapy in the future. The aim of this study is to investigate the impact of the CYP2B6 SNPs as well as GSTM1, GSTT1 and GSTP1 polymorphisms on the outcome for multiple myeloma patients treated with cyclophosphamide.
2. Material and Methods
2.1. Patients
This retrospective study included samples obtained from 26 patients diagnosed with multiple myeloma between 2005 and 2009. Median age at diagnosis was 59 years (range 42 - 66 years) and mean follow-up time was 1.7 years. All patients were treated according to established treatment protocols and national guidelines with regimens containing high dose-cyclophosphamide and cortisone, followed by autologous (22 patients) or allogenic (1 patient) stem cell transplantation. 3 patients did not receive transplant.
Data on blood parameters, disease stage, treatment response, progression free survival and overall survival were registered in a clinical database. Survival times were calculated as the time from diagnosis to an event (progression, death or the latest follow-up date). Response was evaluated after transplantation according to national guidelines as follows: Complete response (CR) was defined as no detectable M-component with negative immunofixation in serum and urine, and <5% plasma cells in the bone marrow. Partial response (PR) was defined as reduction of M-component by ≥50% and urine M-component by ≥90% or <200 mg/24 h together with regress of ROTI (related organ or tissue impairment). Clinical data are summarized in Table 1.
Blood and bone marrow samples were collected at Karolinska University Hospital in Huddinge and shipped to the department of Clinical Pharmacology in Linköping,

Table 1. Patients and clinical data.
where DNA was isolated and genotyping performed. The study was approved by the local ethical committee.
2.2. DNA Isolation
DNA was isolated from peripheral blood or bone marrow cells using the QIAamp Blood Mini kit from QIAGEN (QIAGEN AB, Sweden), according to the manufacturers’ protocol. Concentration was measured and samples were diluted to10 ng/µl for the analyses. DNA was stored at –20˚C until used.
2.3. GSTM1 and GSTT1 Genotyping
Presence or partial deletion of GSTM1 and GSTT1 was analysed using a multiplex PCR method as previously published [12,13]. Briefly, 20 - 100 ng of DNA was amplified in a PCR reaction using HotStar Taq MasterMix (QIAGEN AB, Sweden), a final primer concentration of 1µM each, and a final MgCl2 concentration of 2 µM in a total volume of 20 µl. Betaglobin was co-amplified in the reactions as a control.
The PCR reactions were carried out on a Biometra TProfessional Thermocycler (Biometra GmbH, Germany) with the following temperature cycles: 1 cycle at 95˚C for 15 minutes, 30 cycles at 95˚C for 1 minute, at 57˚C for 1 minute, and at 72˚C for 1 minute, followed by a final extension cycle at 72˚C for 7 minutes. Primer sequences are presented in Table 2.

Table 2. Primers and dispensation orders for GST and CYP2B6 analysis.
Samples were run on an ethidium bromide stained 3:1 Nusieve-Agarose gel and photographed under UV-light. Band sizes were 231, 459 and 268 bp for GSTM1, GSTT1 and β-globin, respectively. Patients with absence of a GSTM1 or GSTT1 band in the presence of a β-globin band were assigned GSTM1 or T1 null genotype.
2.4. CYP2B6 and GSTP1 Genotyping
Pyrosequencing was used to determine the SNPs in CYP- 2B6 and GSTP1. Primer sequences previously published or designed using the PSQ Assay Design programme (QIAGEN AB, Sweden) was ordered from Invitrogen (Paisley, United Kingdom) and used to amplify fragments of CYP2B6 ex 4, 5 and 9 containing the SNPs G516T, A785G and C1459T, with one of the primers in each pair being biotinylated [16]. The PSQ Assay Design Programme was also used to design primers for the GSTP1 SNP 1le105Val. PCR and sequencing primers are presented in Table 2.
A 35 cycle PCR was performed on all PCR primer pairs for optimisation of MgCl2 concentration and annealing temperature, using a temperature gradient. Products were visualised on a 1.5% agarose gel stained with ethidium bromide. The final PCR reactions were carried out using HotStar Taq Mastermix (QIAGEN AB, Sweden), a final primer concentration of 0.4 µM each, a final MgCl2 concentration of 1.5 µM (CYP2B6 ex 5) or 2 µM (CYP2B6 ex 4, ex 9 and GSTP1 Ile105Val) in a total volume of 25 µl. The PCR reactions were carried out on a Biometra TProfessional Thermocycler (Biometra GmbH, Germany) with the following temperature cycles: 1 cycle at 95˚C for 15 minutes, 50 cycles at 95˚C for 30 seconds, at 50˚C (CYP2B6 ex 4) for 30 seconds or at 58˚C for 30 or 15 seconds (GSTP1 Ile105Val and CYP- 2B6 ex 5&9, respectively), and at 72˚C for 30 seconds, followed by a final extension cycle at 72˚C for 10 minutes.
For sequencing of the PCR products Pyrosequencing PSQ96MA (QIAGEN AB, Sweden) was used according to the manufacturers’ protocol. Briefly, single stranded DNA was prepared and sequencing primers in a final concentration of 0.3 µM was annealed to the templates at 80˚C for 2 min. Substrate, enzyme and nucleotides were prepared in volumes given by the PSQ 96MA SNP software and added to the samples in a predefined order, resulting in genotype specific pyrograms.
Discrepancies between the CYP2B6 and CYP2B7 pseudogene sequences within the PCR products were analysed by Pyrosequencing to detect presence of possible pseudogene co-amplification in all reactions, to ensure specificity.
2.5. Statistical Analysis
Kaplan-Meier method was used for survival curves and log-rank tests to investigate the relationship between genotypes and time to progression or overall survival. Chi2 was used to investigate agreement with Hardy-Weinberg equilibrium, as well as differences in genotype distributions between partial or complete responders.
Due to small numbers, multivariate analysis taking other clinical factors, such as del(13)(q)/–13 or t(4;14) status into account was not possible. However, distribution of these known negative prognostic factors was evaluated between genotypes.
A p-value of 0.05 was considered to be statistically significant. Softwares used were GraphPad Prism v.4 and StatCalc (EpiInfo v.3.4.3).
3. Results
3.1. GSTM1 and GSTT1 Genotyping Results
All patients were successfully genotyped for GSTM1 and GSTT1. 8 patients (31%) were found to be GSTM1 wt, while 18 (69%) had the GSTM1null genotype. No patients with the GSTT1null genotype were found in the material.
3.2. GSTP1 and CYPB6 Genotyping Results
Pyrosequencing was successfully used to analyse the Ile105Val single nucleotide polymorphism in GSTP1 as well as the SNPs G516T (ex 4), A785G (ex 5) and C1459T (ex 9) in CYP2B6.
For GSTP1 SNP Ile105Val, Pyrosequencing revealed 7 patients (27%) with the wt Ile/Ile genotype. 16 patients (61.5%) were Ile/Val, while 3 patients (11.5%) were homozygously mutated (Val/Val genotype).
In CYP2B6 ex 4, 12 patients (46%) with the wt 516G/ G genotype were found. 12 patients (46%) were heterozygous while 2 patients (8%) hade the mutated T/T genotype. Analysis of the exon 5 polymorphism A785G revealed 8 patients (31%) with the wt A/A genotype, 13 patients (50%) were heterozygously mutated, while 5 patients (19%) were homozygously mutated. For the exon 9 SNP C1459T, 22 wt patients (85%) and 4 heterozygous patients (15%) were found. No homozygously mutated T/T patients were found in the material.
8 patients (31%) were found to be wild type for both G516T and A785G, 12 (46%) were heterozygous 516G/ T 785A/G genotype. Only 2 patients (8%) were homozygously mutated (516T/T 785G/G) in both positions (*6/ *6). 4 patients (15%) carried other genotype combinations.
All genotypes were in accordance with the HardyWeinberg equation, and the analysis was found specific for CYP2B6 without any co-amplification of the CYP2- B7 pseudogene.
3.3. Correlations to Treatment Response
No significant correlations between GSTM1, GSTP1 or CYP2B6 polymorphisms and complete or partial response after transplant were found, neither for individual or combined genotypes.
3.4. GSTP1-Indication of Impact on Overall Survival
No significant correlation to progression free survival or complete response post transplant was found for the GSTP1 SNP Ile105Val. However, Kaplan-Meier analysis revealed a possible impact on overall survival. Wildtype Ile/Ile patients appear to have a longer overall survival compared to patients with at least one Val allele, although not statistically significant (p = 0.20, Figure 1). Del(13)(q)/–13 or t(4;14) status may influence this result, although no significant difference in status between genotypes were found (p > 0.05, data not shown).
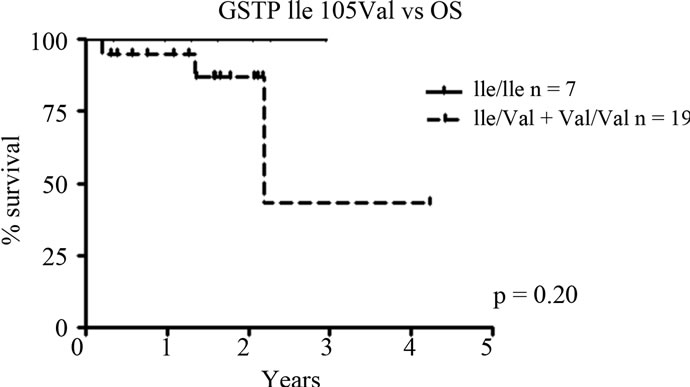
Figure 1. GSTP1 SNP Ile105Val in relation to overall survival (OS). Patients carrying the variant Val allele appear to have a shorter overall survival compared to wild type Ile/Ile patients, although not significant; p = 0.20).
3.5. CYP2B6 SNP A785G—Correlations to Progression Free and Overall Survival
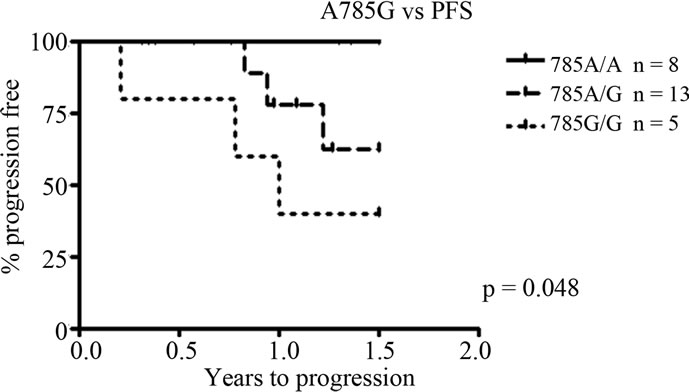
Individual and combined genotypes of CYP2B6 polymorphisms G516T, A785G and C1459T were analysed for correlations to progression free survival (PFS) and overall survival (OS). Kaplan-Meier survival analyses show a significant association between the SNP A785G and overall as well as progression free survival. The variant G allele is related to a shorter progression free survival, with homozygously mutated G/G patients having the shortest PFS followed by heterozygous individuals (p = 0.048, Figure 2(a)). This is also reflected in a difference in OS between genotypes, with homozygosity for the G allele resulting in a worse outcome (p = 0.037, Figure 2(b)). There was no difference in del(13)(q)/–13 or t(4;14) status between the genotypes (p > 0.05, data not shown). No significant correlations were found for the exon 4 and 9 SNPs G516T and C1459T.
3.6. CYP2B6*6—Possible Association to Outcome
The CYP2B6*6 allele, including both the G516T and A785G variants, was also analysed in relation to progression freeand overall survival. Due to small groups (only 2 patients were homozygously mutated in both positions, i.e. *6/*6 genotype), patients carrying two variants were grouped together in the survival analysis (*4/*9, *1/*6 or *6/*6 alleles—we cannot discriminate between*4/*9—i.e. patients with the 785 variant on one allele and the 516 variant on the other—or *1/*6 patients i.e. patients wild type for one allele and with 516 and 785 variants combined on the other allele). The results followed the same pattern as for the SNP A785G, with carriers of at least one variant in both positions having a worse outcome compared to wild type patients. For progression free survival the correlation was borderline significant (p = 0.068, Figure 3). No significant association was found for overall survival (p = 0.39). Patients carrying
 (a)
(a)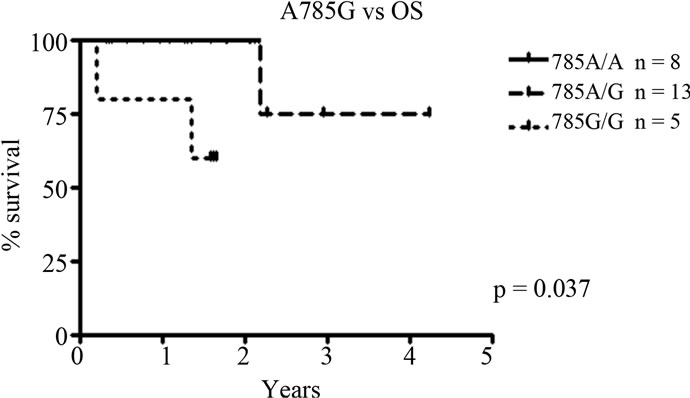 (b)
(b)
Figure 2. Correlations to progression free survival (PFS) and overall survival (OS) for CYP2B6 SNP A785G. (a) A785G vs PFS. The variant G allele is associated to a significantly shorter PFS, p = 0.048; (b) A785G vs OS. The shorter PFS related to the variant allele is also reflected in a significantly shorter OS, with a worse outcome for homozygously mutated (G/G) patients. p = 0.037.
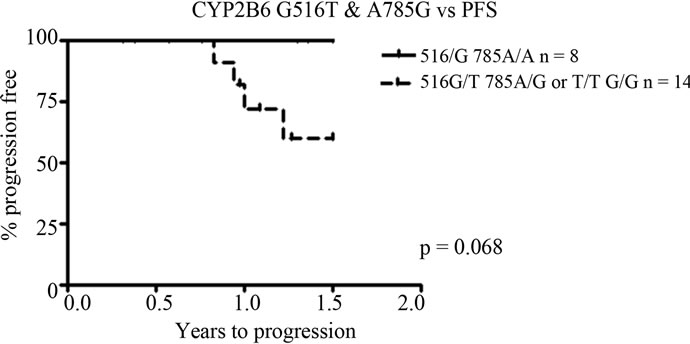
Figure 3. Presence of both G516T and A785G variants (including 2 patients with the homozygously mutated *6/*6 variant) in relation to progression free survival (PFS) and overall survival (OS). A borderline significant association between a shorter PFS and presence of at least one variant in both positions, p = 0.068.
both variants had a higher degree of del(13)(q)/–13 compared to wildtype patients, which may influence the results (p = 0.02, data not shown). There was no difference in t(4;14) status between genotypes (p > 0.05, data not shown).
4. Discussion
In the present study we investigated the impact of polymorphisms in CYP2B6 and GSTM1, GSTT1 and GSTP1 on the outcome of cyclophosphamide treatment in a small group of multiple myeloma patients. Cyclophosphamide is metabolised to its active form, 4-OH-cyclophosphamide, in the liver by CYP450 enzymes including CYP2B6 [4]. Further, GST enzymes are involved in detoxification by glutathione conjugation. Polymorphisms have been shown to affect enzyme expression and activity, and may therefore also influence treatment response and outcome for cancer patients treated with drugs metabolised by these enzymes. However, previously published studies are inconclusive.
Our results on the impact of CYP2B6 A785G on progression free and overall survival are in line with studies presenting a decreased expression and activity associated to presence of the A785G variant [7], which would result in a lower plasma concentration of the active drug and thereby a less effective treatment. This conclusion is also supported by a recently published study on breast cancer patients treated with doxorubicin and cyclophosphamide, in which the A785G variant allele was significantly associated to worse outcome [17].
Also in accordance with our results Hofmann et al. showed decreased mRNA-levels in liver samples carrying the *6 variant allele [9], and Lang et al. reported similar findings, with reduced protein levels in *6 allele carriers [5]. Tsuchiya et al. also showed a higher efavirenz plasma concentration in *6 carriers, consistent with decreased enzyme activity [7]. However, it is not clear that reduced protein levels are associated with decreased cyclophosphamide bioactivation; any effects of the polymorphisms may rather be due to functional changes of the enzyme. Other enzymatic pathways and polymorphic variations, such as those of CYP2C19, are also suggested as contributors to the variability [18]. In contrast, presence of the *6 allele (516T 785G) has been shown to result in increased cyclophosphamide activation in liver microsome preparations and in patients with haematological malignancies [19] which is in agreement with the report by Nakajima et al. showing a higher clearance and shorter t½ of cyclophosphamide in 103 Japanese patients with malignant lymphoma or breast cancer carrying the *6 variant allele [8]. Kirchheiner et al. also showed an increased clearance of bupropion in *4 (A785G) carriers but not in *6 [6]. Possible explanations for discrepancies between studies may include differences in patient groups, type of disease and different concomitant chemotherapy.
In this pilot study we show that individuals carrying the CYP2B6 785G allele have a shorter progression free survival and a shorter overall survival, with homozygously mutated patients having the worse outcome. A similar tendency was seen in carriers of the *6 allele, although significance could not be determined due to small groups. This may indicate that the effect on survival is caused by the A785G SNP rather than a combination of G516T and A785G, since no individual effect of G516T was seen.
No difference in del(13)(q)/–13 or t(4;14) status, a previously reported negative predictor in MM, was seen between A785G genotypes. This indicates that the negative effect of the 785G allele seen on progression free and overall survival are less likely to be a result of del(13)(q)/–13, and it is generally observed that the negative impact on prognosis in MM is not limited to del(13)(q)/–13, but is due to association with t(4;14) and/or del(17)(p) [20].
No effect of the exon 9 SNP C1459T was seen in our material, contradictory to a recent study identifying this as a loss of function variant with effect on cyclophosphamide bioactivation [18]; however, both that study and ours are on a small material with low frequency of the variant T allele.
The GSTP1 polymorphism Ile105Val is located in the substrate binding region of the enzyme, and results in altered heat stability and enzymatic properties with substrate specific differences [14,15]. We found that patients with the wild type Ile/Ile genotype may have a tendency to improved overall survival compared to carriers of at least one Val allele. Previously, Dasgupta et al. reported a longer progression free survival and a trend to improved overall survival for the Val allele in myeloma patients treated with standard dose chemotherapy, but no impact in patients with high-dose treatment [21]. Better outcome with the Val allele has also been reported by others; Sweeney et al. described an improved survival in breast cancer patients with Val/Val genotype compared to the wildtype Ile/Ile, and the same findings were reported in ovarian cancer patients with carriers of the Val allele having lower risk of progression as well as longer overall survival [22,23].
In contrast, Khrunin et al. showed an increased progression free survival in ovarian cancer patients carrying the Ile allele when treated with cisplatin-cyclophosphamide regimens [24], similar to our results. Also, Maggini et al. reported a significantly better response to chemotherapy and a tendency to better overall survival in myeloma patients with the wild type genotype, when treated with DAV (dexamethasone-doxorubicin-vincristine), with cyclophosphamide and G-CSF (granulocyte colony stimulating factor) or melphalan as a conditioning regimen [25].
Although generally a decreased enzymatic activity (and heat stability) in the Val-variant is described, substrate specific differences appear likely since the polymorphism is positioned in the substrate binding region, changing size and shape of the pocket. Interactions between substrate, substrate binding pocket and nearby residues may increase or decrease binding efficacy and conjugation depending on chemical properties of the substrate and side chain-interaction variations [15]. If the Ile/Ile genotype have a decreased ability to bind and conjugate cyclophosphamide metabolites compared to the Val-variants, and thereby give a prolonged effect of the active drug, this would explain the possible advantage for our patients. This theory is supported by an article by Ishimoto and Ali-Osman, showing differential cytoprotective activity of GSTP1 alleles against different anticancer agents, with the protection against hydroperoxyifosfamide (a drug structurally and in its action similar to cyclophosphamide) being lower for the Ile allele compared to the Val allele [14].
In conclusion, we have shown that polymorphisms in CYP2B6, especially the SNP A785G, as well as the GSTP1 SNP Ile105Val, are indicated to influence the outcome in cyclophosphamide treatment in myeloma patients with possible effects on progression free survival and overall survival. These results indicate that patients carrying the CYP2B6 785A/A genotype might benefit from the traditional cyclophosphamide treatment while patients carrying the variant might profit from the use of novel agents such as proteasome inhibitors and IMiDs. Due to the small number of patients and short follow up time of this pilot study, and the inconclusiveness of previously published studies regarding these polymorphisms in different settings, further studies on a larger material of cyclophosphamide treated patients are needed. Access to a larger material would enable the investigation of the combined impact of polymorphisms in activating (CYP- 2B6) as well as detoxifying (GSTs) enzymes. Inclusion of details on other clinically relevant prognostic markers, treatment duration, toxicity data and data on pharmacokinetics could also be useful to further elucidate the impact of genetic variation in CYP2B6 and GST enzymes. Confirmation of a predictive role of SNPs in these enzymes could contribute to future individualised chemotherapy in myeloma patients and thereby a better treatment outcome and long term health improvement.
5. Acknowledgements
This work was supported by grants from the Swedish Cancer Society, Swedish Research Council, Cancer Society in Stockholm, Karolinska Institutet, and the County Council in Östergötland. We also thank Dr. Kourosh Lotfi at the Clinical Pharmacology and Division of Haematology, Linköping University Hospital, for valuable input.
REFERENCES
- W. M. Kuehl and P. L. Bergsagel, “Multiple Myeloma: Evolving Genetic Events and Host Interactions,” Nature Reviews Cancer, Vol. 2, No. 3, 2002, pp. 175-187. doi:10.1038/nrc746
- M. C. Minnema, E. van der Spek, N. W. C. J van de Donk and H. M. Lokhorst, “New developments in the treatment of patients with multiple myeloma,” The Netherlands Journal of Medicine, Vol. 68, No. 1, 2010, pp. 24-32.
- H. Kaufman, E. Urbauer, J. Ackermann, H. Huber and J. Drach, “Advances in the biology and therapeutic management of multiple myeloma,” Annals of Hematology, Vol. 80, No. 8, 2001, pp. 445-451. doi:10.1007/s002770100348
- T. K. H. Chang, G. F. Weber, C. L. Crespi and D. J. Waxman, “Differential Activation of Cyclophosphamide and Ifosphamide by Cytochromes P-450 2B and 3A in Human Liver Microsomes,” Cancer Research, Vol. 53, No. 23, 1993, pp. 5629-5637.
- T. Lang, K. Klein, J. Fischer, A. K. Nüssler, P. Neuhaus, U. Hofmann, et al., “Extensive Genetic Polymorphism in the human CYP2B6 gene with impact on expression and function in human liver,” Pharmacogenetics, Vol. 11, No. 7, 2001, pp. 399-415. doi:10.1097/00008571-200107000-00004
- J. Kirchheiner, C. Klein, I. Meineke, J. Sasse, U. M. Zanger, T. E. Mürdter, et al., ”Bupropion and 4-OH-bupropion pharmacokinetics in relation to genetic polymorphisms in CYP2B6,” Pharmacogenetics, Vol. 13, No. 10, 2003, pp. 619-626. doi:10.1097/00008571-200310000-00005
- K. Tsuchiya, H. Gatanaga, N. Tachikawa, K. Teruya, Y. Kikuchi, M. Yoshino, et al., “Homozygous CYP2B6 *6 (Q172H and K262R) Correlates with High Plasma Efavirenz Concentrations in HIV-1 Patients Treated with Standard Efavirenz-Containing Regimens,” Biochemical and Biophysical Research Communications, Vol. 319, No. 4, 2004, pp. 1322-1326. doi:10.1016/j.bbrc.2004.05.116
- M. Nakajima, S. Komagata, Y. Fujiki, Y. Kanada, H. Ebi, K. Itoh, et al., “Genetic polymorphisms of CYP2B6 affect the pharmacokinetics/pharmacodynamics of cyclophosphamide in Japanese cancer patients,” Pharmacogenetics and Genomics, Vol. 17, No. 6, 2007, pp. 431-445. doi:10.1097/FPC.0b013e328045c4fb
- M. H. Hofmann, J. K. Blievernicht, K. Klein, T. Saussele, E. Schaeffeler, M. Schwab, et al., “Aberrant Splicing Caused by Single Nucleotide Polymorphism c.516G>T (Q172H), a Marker of CYP2B6*6, Is Responsible for Decreased Expression and Activity of CYP2B6 in Liver,” The Journal of Pharmacology and Experimental Therapeutics, Vol. 325, No. 1, 2008, pp. 284-292. doi:10.1124/jpet.107.133306
- J. Seidegård, W. R. Vorachek, R. W. Pero and W. R. Pearson, “Hereditary differences in the expression of the human glutathione transferase Active on trans-stilbene oxide are due to a gene deletion,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 85, No. 19, 1988, pp. 7293-7297. doi:10.1073/pnas.85.19.7293
- S. Pemble, R. Klaus, K. R. Schroeder, S. R. Spencer, D. J. Meyer, E. Hallier, et al., “Human glutathione S-transferase Theta (GSTT1): cDNA Cloning and the Characterization of a Genetic Polymorphism,” The Biochemical Journal, Vol. 300, No. 1, 1994, pp. 271-276.
- D. A. Bell, J. A. Taylor, D. F. Paulson, C. N. Robertson, J. L. Mohler and G. W. Lucier, “Genetic Risk and Carcinogen Exposure: A Common Defect of the CarcinogenMetabolism Gene Glutathione S-Transferase M1 (GSTM1) That Increases Susceptibility to Bladder Cancer,” Journal of the National Cancer Institute, Vol. 85, No. 14, 1993, pp. 1159-1164. doi:10.1093/jnci/85.14.1159
- A. Ahmadi, P. Jönsson, U. Flodin and P. Söderkvist, ”Interaction between smoking and glutathione S-transferase polymorphisms in solvent-induced chronic toxic encephalopathy,” Toxicology and Industrial Health, Vol. 18, No. 6, 2002, pp. 289-296. doi:10.1191/0748233702th152oa
- T. M. Ishimoto and F. Ali-Osman, “Allelic variants of the human glutathione S-transferase P1 gene confer differential cytoprotection against anticancer agents in Escherichia coli,” Pharmacogenetics, Vol. 12, No. 7, 2002, pp. 543-553. doi:10.1097/00008571-200210000-00006
- A.-S. Johansson, G. Stenberg, M. Widersten and B. Mannervik, “Structure-Activity Relationships and Thermal Stability of Human Glutathione Transferase P1-1 Governed by the H-Site Residue 105,” Journal of Molecular Biology, Vol. 278, No. 3, 1998, pp. 687-698. doi:10.1006/jmbi.1998.1708
- M. Rohrbacher, A. Kirchhof, G. Geisslinger and J. Lötsch, “Pyrosequencing™-based screening for genetic polymorphisms in Cytochrome P450 2B6 of potential clinical relevance,” Pharmacogenomics, Vol. 7, No. 7, 2006, pp. 995-1002. doi:10.2217/14622416.7.7.995
- J. Bray, J. Sludden, M. J. Griffin, M. Cole, M. Verril, D. Jamieson, et al., “Influence of pharmacogenetics on response and toxicity in breast cancer patients treated with doxorubicin and cyclophosphamide,” British Journal of Cancer, Vol. 102, No. 6, 2010, pp. 1003-1009. doi:10.1038/sj.bjc.6605587
- N. A. Hesby, C.-Y. Hui, M. A. Goldthorpe, J. K. Colle, M. C. Soh, P. J. Gow, et al., “The combined impact of CYP2C19 and CYP2B6 pharmacogenetics on cyclophosphamide bioactivation,” British Journal of Clinical Pharmacology, Vol. 70, No. 6, pp. 844-853. doi:10.1111/j.1365-2125.2010.03789.x
- H. Xie, L. Griskevicius, L. Ståhle, Z. Hassan, Ü. Yasara, A. Rane, et al., “Pharmacogenetics of cyclophosphamide in patients with hematological malignancies,” European Journal of Pharmaceutical Sciences, Vol. 27, No. 1, 2006, pp. 54-61.
- H. Avet-Loiseau, M. Attal, P. Moreau, C. Charbonnel, F. Garban, C. Hulin, et al., “Genetic abnormalities and survival in multiple myeloma: the experience of the Intergroupe Francophone du Myélome,” Blood, Vol. 109, No. 8, 2007, pp. 3489-3495. doi:10.1182/blood-2006-08-040410
- R. K. Dasgupta, P. J. Adamson, F. E. Davies, S. Rollinson, P. L. Roddam, A. J. Ashcroft, et al., “Polymorphic variation in GSTP1 modulates outcome following therapy for multiple myeloma,” Blood, Vol. 102, No. 7, 2003, pp. 2345-2350. doi:10.1182/blood-2003-02-0444
- C. Sweeney, G. Y. McClure, M. Y. Fares, A. Stone, B. F. Coles, P. A. Thompson, et al., “Association between Survival after Treatment for Breast Cancer and Glutathione S-Transferase P1 Ile105Val Polymorphism,” Cancer Research, Vol. 60, No. 20, 2000, pp. 5621-5624.
- A. Beeghly, D. Katsaros, H. Chen, S. Fracchioli, Y. Zhang, M. Massobrio, et al., “Glutathione S-transferase polymorphisms and ovarian cancer treatment and Survival,” Gynecologic Oncology, Vol. 100, No. 2, 2006, pp. 330- 337. doi:10.1016/j.ygyno.2005.08.035
- A. V. Khrunin, A. Moisseev, V. Gorbunova and S. Limborska, “Genetic polymorphisms and the efficacy and toxicity of cisplatin-based chemotherapy in ovarian cancer Patients,” Pharmacogenomics Journal, Vol. 10, No. 1, 2010, pp. 54-61. doi:10.1038/tpj.2009.45
- V. Maggini, G. Buda, S. Galimberti, E. Conidi, N. Giuliani, F. Morabito, et al., “Response to chemotherapy and tandem autologous transplantation of multiple myeloma patients and GSTP1 and TYMS polymorphisms,” Leukemia Research, Vol. 32, No. 1, 2008, pp. 49-53. doi:10.1016/j.leukres.2007.03.029

