Advances in Bioscience and Biotechnology
Vol. 4 No. 7A2 (2013) , Article ID: 34154 , 9 pages DOI:10.4236/abb.2013.47A2003
Characterization of Ciprofloxacin resistant Extended Spectrum β-Lactamase (ESBL) producing Escherichia spp. from clinical waste water in Bangladesh
![]()
Department of Microbiology, University of Dhaka, Dhaka, Bangladesh
Email: #hossaina@du.ac.bd
Copyright © 2013 Nihad Adnan et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received 6 May 2013; revised 10 June 2013; accepted 20 June 2013
Keywords: Clinical Waste Water; Escherichia; Multidrug Resistance; ESBL; Ciprofloxacin
ABSTRACT
Clinical Waste Water (CWW) possesses the risks of spreading antibiotic resistant bacteria in the environment. In Bangladesh, liquid discharge is directly released into the municipal sewage system and pollutes the surroundings water bodies/rivers. Liquid samples were collected from the three CWW discharge sites connected to DMCH (Dhaka Medical College Hospital) and from one control group non-connected to DMCH. About 32, 30 and 26 times higher ciprofloxacin, cefixime and multi-drug resistant bacterial count were found in CWW over control samples. Among the isolates, two Escherichia spp. isolates, denoted 26N and 28N, were found to be resistant against fluoroquinolone (MBC of Ciprofloxacin > 1024 μg/ml), cephalosporin, glycopeptide, monobactam, penicillin, tetracycline, rifampicin, macrolides, sulfonamide and nitrofuran classes of drugs and were also ESBL positive through phenotypic assay. Plasmid curing experiment detected possible plasmid mediated resistance of fluoroquinolone, cephalosporin, tetracycline, lincosamide and nitrofuran classes of antibiotics. Phylogenetically, isolate 26N and 28N were characterized as Escherichia coli and Escherichia fergusonii. These MDR and ESBL positive bacteria are potent to disseminate resistant determinants in the surrounding environments.
1. INTRODUCTION
Antibiotics and their extensive use are pivotal in the selection of bacterial resistance towards various groups of antibiotics and spread of resistance genes [1]. This situation is initiated by the counter availability, indiscriminate and inappropriate use of antimicrobial agents [2] specially in countries like Bangladesh. An alarming situation is the progressive loss of susceptibility towards Ciprofloxacin due to its increased use [3,4] in the treatment of a broad range of clinical conditions such as urinary tract infections, upper respiratory tract infections, as a prophylaxis in neutropenic patients and in the poultry sectors [5,6].
Antibiotic resistance is particularly predominant among gram-negative bacteria [7,8], specifically within the members of Enterobacteriaceae [9]. Co-existence of fluoroquinolone and cephalosporin resistant Extended Spectrum β-Lactamase (ESBL) producing isolates are disseminating in the environment through hospital wastes [10-12]. Both chromosomal and plasmid mediated drug resistance against fluoroquinolone groups have been reported. Chromosomal resistance is due to the mutation in the DNA gyrase and topoisoerase IV [13,14], decrease in outer membrane permeability and overexpression of naturally occurring efflux [15]. Plasmid mediated quinolone resistance (PMQR) is reported to carry quinolone resistance genes: qnrA, qnrB, qnrC, qnrD, qnrS, the aminoglycoside acetyltransferase gene aac, the efflux pump genes qepA and oqxAB [16-19].
Antibiotics used in medicine are only partially metabolized [20,21] by patients and discharged into the hospital sewage system or directly into the municipal wastewater. The predicted concentrations of antibiotics in hospital wastewater are in the range of the semimaximum inhibitory concentration (MIC50) of sensitive pathogenic or beneficiary bacteria for some active substances (0.1 - 2.9 mg/l) leading to the growing antibiotic resistance of pathogenic bacteria in the environment [22].
Bacterial antibiotic resistance is transferable. If the resistant bacteria are allowed to distribute in the environment, they will most likely transfer resistance gene to other bacteria of same species or different. Dissemination of these resistant bacteria will not be restricted to a particular geographical area; drug resistance can be expected to spread steadily to all parts of the world. The scenario is plausible, but the risks cannot be estimated because of the insufficient understanding and of limited data availability.
The assessment of this pollution intensity in Bangladesh is currently at early stage and available data are limited. In our previous endeavor, high percentage of Ciprofloxacin resistant E. coli was isolated and characterized from CMCH [14]. An extended research was carried out for the largest medical college DMCH (Dhaka Medical College Hospital) located in Dhaka, Bangladesh. The disposal system of DMCH allows the liquid wastes to directly discharge into the municipal sewage system. This raises the importance of investigating the involvement of DMCH liquid wastes discharge in the development and distribution of antibiotics resistance in the environmental bacteria.
2. MATERIALS AND METHODS
2.1. Major Antibiotics Used in DMCH and Collection of Liquid Waste Samples
Prior to sampling, data of major antibiotics used by the patients of DMCH were collected within a period of September to October 2008 and September to October 2009, and the percentages of usage of different groups of antibiotics were calculated (Table 1). As Ciprofloxacin, the 2nd generation fluoroquinolone and Cefixime, the 3rd generation cephalosporin, were mostly used by the patients during the study period, bacterial resistance were analysed using these two antibiotics.
Total three groups of samples (1, 2 and 3) from DMCH liquid waste discharge and 1 control group sample (4), 400 m away from DMCH liquid discharge (Figure 1) were collected within the two months period of September-October 2008 and September-October 2009. Group 1 samples were collected from a drain where the waste falling directly from the surgical unit toilets, Group 2 and 3 samples were collected from two different drains which were directly connected to the burn unit toilets and wash rooms, respectively of the DMCH. Group 4 samples were collected directly from a safety tank of a nearby student residence called Shahidullah Hall, which had no connection to the hospital drain system. Samples were collected and transported to the laboratory using the standard me-
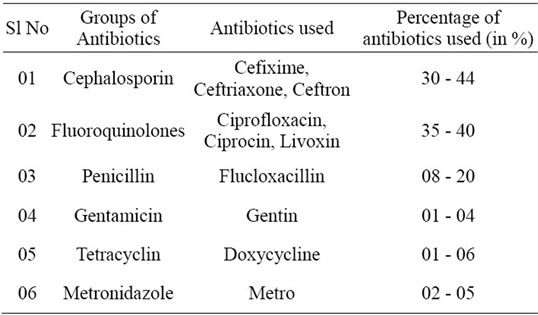
Table 1. Major antibiotics used in surgical and burn units of DMCH in September-October 2008 and September-October 2009 (a survey on 45 patients).
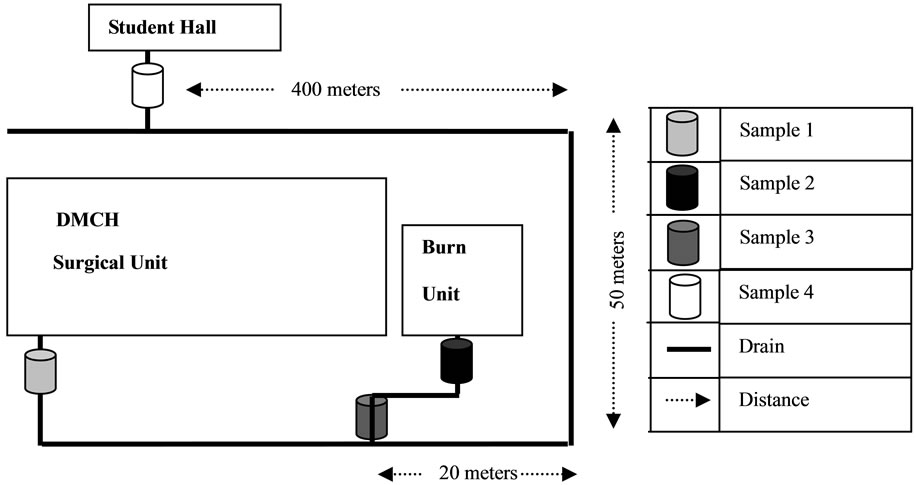
Figure 1. Dhaka Medical College Hospital (DMCH) liquid waste disposal drainage system and four sampling points. The wastes from different units of the hospital connected are indicated along with their approximate distances.
thod described previously [14].
2.2. Enumeration of Resistant Bacteria and Isolation of Ciprofloxacin Resistant Escherichia spp.
Total bacterial count (TBC), total Ciprofloxacin, Cefixime and both Ciprofloxacin and Cefixime resistant bacterial count and isolation of Ciprofloxacin resistant Escherichia spp. were done using the method described previously [14]. For total Ciprofloxacin and Cefixime resistant bacterial count, Ciprofloxacin (1 µg/ml) and Cefixime (5 µg/ml), respectively were added into the molten plate count agar (PCA) media at about 45˚C - 50˚C. From the Ciprofloxacin and Cefixime supplemented plates, 10 presumptive Escherichia spp. were selected after characteristic growth on MacConkey agar, Xylose lysine deoxycholate (XLD) agar, Eosine methylene blue (EMB) agar; and conventional biochemical tests of sugar utilization (lactose, dextrose), indole test, methyl red (MR) test, voges-proskauer (VP) test, citrate utilization test, motility Indole urease (MIU) assay, catalase and oxidase. The tests were carried out according to Bergey’s Manual of Systematic Bacteriology [23].
2.3. Antibiotic Resistance Profile of the Selected Escherichia spp.
Among the resistant isolates, two highly Ciprofloxacin presumptive Escherichia isolates (26N and 28N) were selected for detail antibiogram study by Kirby-Bauer method [24] using the commercially available standard discs (Oxoid, Basingstoke, UK) of Streptomycin (10 μg/ ml), Ciprofloxacin (5 μg/ml), Ampicillin (25 μg/ml), Rifampicin (5 μg/ml), Oxytetracyclin (30 μg/ml), Cephalosporin (30 μg/ml), Nalidixic Acid (30 μg/ml), Chloramphenicol (30 μg/ml), Nitrofurantoin (300 μg/ml), Aztreonam (30 μg/ml), Levofloxacin (5 μg/ml), Gentamycin (10 μg/ml), Ceftriaxone (30 μg/ml), Cloxacillin (5 μg/ml), Trimethoprim (5 μg/ml), Amikacin (30 μg/ml) Cefotaxime (30 μg/ml) and Ceftazidime (30 μg/ml) (Table 2). Susceptible ATCC 25922 Escherichia coli was used as a control.
Isolate 26N and 28N that were completely resistant to 2nd generation fluoroquinolone and 3rd generation Cephalosporin, were further analyzed for the presence of Extended Spectrum β-Lactamase (ESBL) production by the method of Double Disc Synergistic Assay (DDST) [25]. In the DDST method, a lawn culture of the test organisms onto a Mueller-Hinton agar plate was performed. Augmenting disk [Amoxicillin-clavulanate] was placed at the centre of the plate and the disks containing Ceftazidime, Cefotaxime and Ceftriaxone antibiotics (30 µg each) were placed 15 mm centre to centre from the augmenting disk.
MIC (minimum inhibitory concentration) and MBC (minimum bactericidal concentration) range of the isolate 26N and 28N were analyzed by turbidimetric method [14] at an optical density of 600 nm using different concentrations of Ciprofloxacin (8 µg/ml, 32 µg/ml, 64 µg/ml, 128 µg/ml, 256 µg/ml, 512 µg/ml, 768 µg/ml, and 1024 µg/ml). 0 µg/ml Ciprofloxacin was used as a control.
2.4. Extraction of Plasmid DNA from Ciprofloxacin Resistant Escherichia spp.
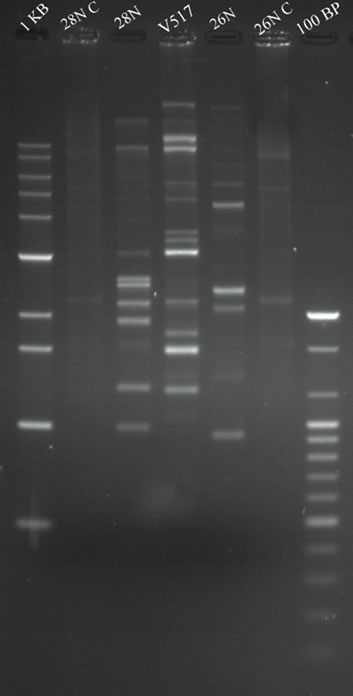
Plasmid DNA of selected presumptive Escherichia spp. 26N and 28N was extracted according to the alkaline lysis method [26]. E. coli V517 was used as a control and plasmid size marker [27]. Extracted plasmid DNA was separated by horizontal electrophoresis in 0.8% agarose gels in a Tris-acetic acid EDTA (TAE) buffer at room temperature at 70 volt (60 mA) for 2 hr. 10 µl of plasmid DNA solution was mixed with 2 µl of tracking 6X dye and was loaded into the individual well of the gel. The gel was stained with ethidium bromide for 12 minutes at room temperature and destained with distilled water for 10 minutes. DNA bands were visualized and photograph was taken using Gel Documentation with UV transilluminator (Gel Doc, BioRad, USA). The size of the unknown plasmid DNA was determined comparing with the known size (bp) plasmid markers of E. coli V517.
2.5. Curing of R-Plasmid
Plasmid curing of two ESBL positive isolates (isolate 26N and 28N) was performed by the modified method of Tomeda et al. [28]. Each test organisms was grown overnight in Luria Bertani (LB) broth. The resulting culture was diluted to 104 cells/ml and 0.5 ml was mixed with each of the 4.5 ml LB broth containing 100 µg/ml of ethidium bromide and 75 µg/ml of acridine orange respectively. Ethudium bromide and acridine orange act as curing agent. The cultures were then incubated at 37˚C in an orbital shaker at 150 rpm for 48 hours. Cured isolates were detected using patch method maintaining control plates.
2.6. PCR Amplification and Nucleotides Sequencing
DNA was extracted from the isolates 26N and 28N and the 16S rRNA gene was amplified by PCR with the primers 27F (5’-AGAGTTTGATCCTGGCTCAG-3’) and 1492R (5’-TACGGTTACCTTGTTACGACTT-3’) using the reaction conditions described previously [29]. The PCR products of the isolate 26N and 28N were purified with the Wizard PCR SV Gel and PCR Clean-Up System kit (Promega) and were sequenced in ABI sequencer (ABI prism 3130). The 16S rRNA gene sequences were
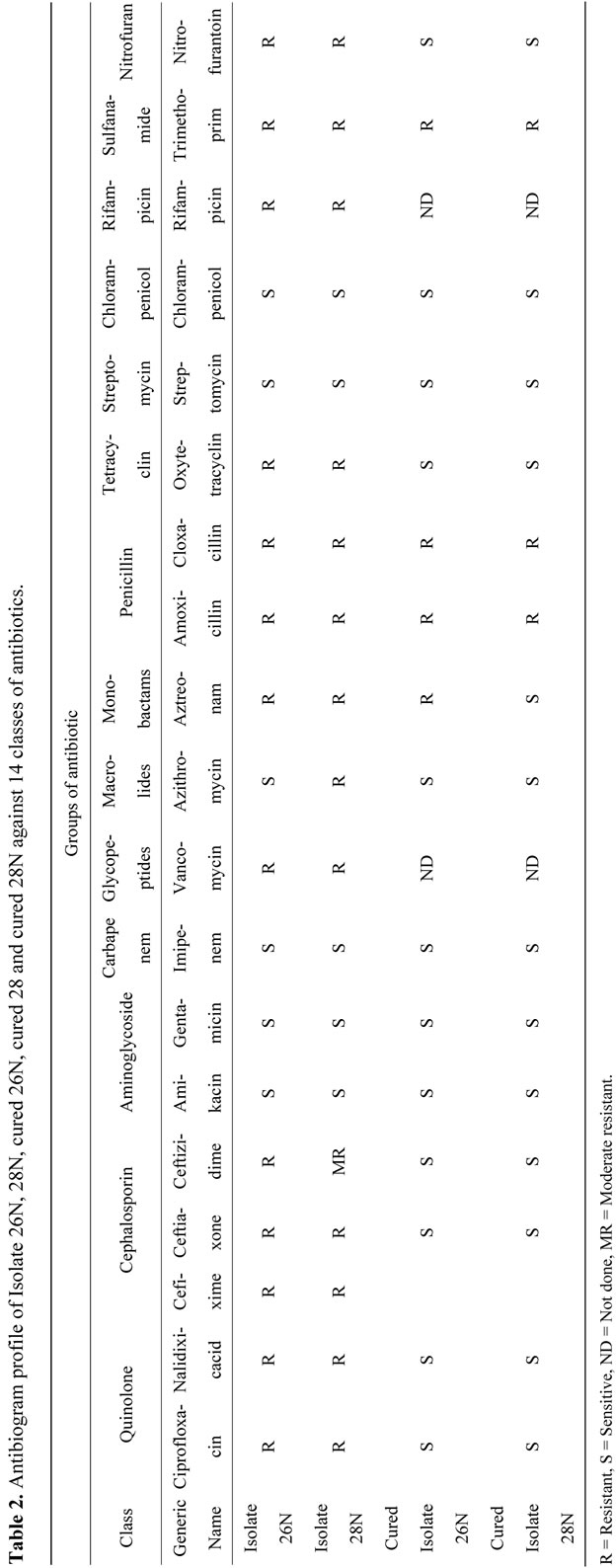
Table 2. Antibiogram profile of Isolate 26N, 28N,cured 28 and cured 28N against 14 classes of antibiotics.
compared to that of the sequences deposited in the NCBI database using the BLAST algorithm. The sequences were submitted to GenBank database under the accession numbers KC542889-KC542890.
3. RESULTS
(DMCH) Dhaka Medical College Hospital is one of the largest hospitals in Bangladesh. This is an extended study of CWW analysis in Bangladesh with respect to the frequency of specific groups of antibiotics used by the patients in DMCH; burden of R-gene pool pollution; resistance prevalence of Escherichia spp. against commonly used Ciprofloxacin and their molecular characterization.
3.1. Prevalence of Multidrug Resistant Bacteria and Isolation of Escherichia spp. 26N and 28N
The most frequently used antibiotics in the DMCH surgery and burn units were cephalosporin and fluoroquinolone groups of antibiotics during the time of this study. Among those, Cefixime within the cephalosporin and Ciprofloxacin within the fluoroquinolone groups were mostly used (Table 1).
The highest bacterial count and resistant bacterial count was observed for the burn unit samples of DMCH as compared to the surgical unit (Figure 2). Sample 2 (burn unit effluent) showed approximately 32, 30 and 26 times higher Ciprofloxacin, Cefixime and MDR bacterial count respectively as compared to the control sample 4.
From Ciprofloxacin and Cefixime supplemented plates, 10 presumptive Escherichia spp. were isolated using selective growth on MacConkey, XLD and EMB agar plates and corresponding biochemical assay. They were positive in lactose utilization, dextrose utilization, indole produc-
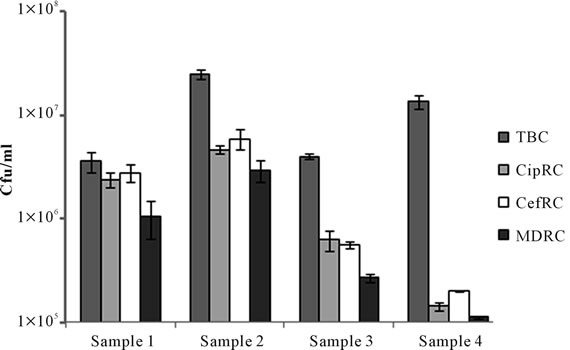
Figure 2. Total bacterial count (TBC), Ciprofloxacin resistant count (CipRC), Cefixime resistant count (CefRC) and both Ciprofloxacin and Cefixime resistant count (MDRC) of Dhaka Mediacal Colege Hospital (DMCH) sample 1 (surgical unit toilet connected drain), sample 2 (burn unit toilet connected drain), sample 3 (burn unit washroom connected drain) and sample 4 (safety tank of student dormitory).
tion, MR reaction, catalase and oxidase test and were found to be negative for VP reaction, citrate utilization and urease production. Among the ten presumptive Escherichia isolates, two isolates (isolate 26N and 28N) that were completely resistant towards Ciprofloxacin and Cefixime, were further tested to characterize their antibiotic resistance profile.
3.2. Analysis of Antibiotic Resistance Pattern and Determination of ESBL Production
Isolate 26N and 28N were resistant against wide groups of antibiotics (Table 2). These two isolates showed common resistance against fluoroquinolones, cephalosporins, monobactams, penicillin, tetracycline, glycopeptides, rifampicin, sulfanamide and nitrofuran groups of antibiotics except macrolides. Isolate 26N were sensitive to Azithromycin whereas isolate 28N were resistant. Both of them were sensitive against aminoglycosides, carbapenem, chlorampenicol and streptomycin classes of antibiotics (Table 2). Isolate 26N and 28N were found to be pure ESBL producers through the DDST method showing characteristics zone of enhancement towards the augmenting disc. MBC of the isolate 26N and 28N couldn’t be attained even at 1024 µg/ml of Ciprofloxacin (data not shown).
3.3. Plasmid Profile Analysis before and after Curing Experiment
Isolate 26N and 28N contained multiple plasmids of variable molecular weight range from <2.1 kbp to upto 53.7 kbp (Figure 3(a)). Both the isolates contain multiple size ranged plasmids of low and high molecular weight. As isolate 26N and 28N contained several small plasmids, plasmid curing experiments were carried out to investigate the plasmid mediated resistant property. After plasmid curing of the isolate 26N and 28N with curing agents, it was observed that most of the small plasmids were successfully cured. After curing, the isolates showed sensitivity against Ciprofloxacin, Cefixime, Ampicillin and Nalidixic acid with whom they were resistant before (Figure 3(b)).
3.4. Sequencing and Phylogenetic Analysis
The 16S rRNA gene sequence analysis of the isolate 26N and 28N showed close similarity to to Escherichia coli and Escherichia fergusonii respectively with a maximum similarity of 99% to 100% (Figure 4). Phylogenetically, they were well aligned within the Shigella-Escherichia cluster showing homology to Escherichia spp. (Figure 4).
4. DISCUSSION
Use of common antibiotics to control infectious diseases
 (a)
(a)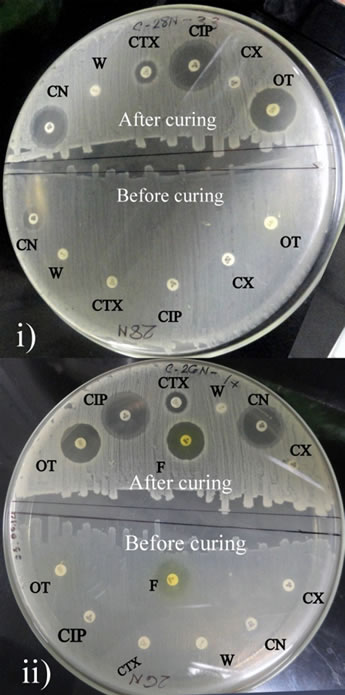 (b)
(b)
Figure 3. (a) Agarose gel electrophoresis (0.8% gel) of plasmid DNA showing the multiple plasmid patterns of E. coli isolates 26N and 28N before curing (Lane 5, 3) and after curing (Lane 6, 2) experiment. Lane 1 and 7 shows 1 kb and 100 bp markers (Bioneer, USA). E. coli V517 (Lane 4) was used as reference plasmid marker. (b) Antibiogram (plate diameter 200 mm) of 26N ii) and 28N i) showing difference in resistance pattern before and after curing of plasmids. CIP-Ciprofloxacin; OT-Oxitetracicline; F-Nitrofurantoin; W-Trimethoprim; CN-Gentamicin; CX-Cloxacillin and CTX-Cefotaxime.
is gradually becoming ineffective due to the development of antibiotic resistance within the microorganisms and its’ horizontal/vertical transfer in the microbial population [30]. Developing countries like Bangladesh is suffering from this crisis at exceeding level due to indiscriminate use of antibiotics by the clinical practitioners as well as by the community people without any prescriptions [31]. A considerable amount of antibiotics from the users are excreted out of the body through urine and other means [32]. As a consequence, the treatment of disease with antibiotic not only kills the pathogen, but also favors the development of resistance in the environment [33].
A practitioner of surgical and burn unit relies on broad spectrum antibiotics to fight against unwanted infections. Data collected from these units of DMCH showed that broad spectrum antibiotic Ciprofloxacin and Cefixime were prescribed at the highest frequency during our study period. To support the hypothesis that these hospital units discharge a large amount of resistant bacteria than the non-hospital connected waste, surgical and burn unit waste were chosen. Sampling site 1, 2, and 3 were directly connected to surgical unit toilets, burn unit toilets and burn unit wash rooms. A control sample 4 was collected from the outflow of nearby student hall that were not directly connected to the hospital water outflow (Figure 1). The results clearly showed that hospital waste or hospital connected sample 1, 2 and 3 contained enormously high numbers of resistant bacteria compared to hospital non-connected control sample 4 (Figure 2).
Bacteriological analysis showed that DMCH surgical unit outflow (sample 1) contained the highest percentage of Ciprofloxacin, Cefixime and MDR bacteria as compared to the non-hospital connected sewage outflow (sample 4) (Figure 2). This may be due to the enormous amount of antibiotic usage and excreted from the hospital units. The burn unit sample connected to the wash room (sample 3) perhaps undergone more dilution and therefore showed the lowest percentage of antibiotic resistant bacteria among the three sampling locations. Also the MDR count in all three samples were considerably high as compared to the control sample (Figure 2), indi-
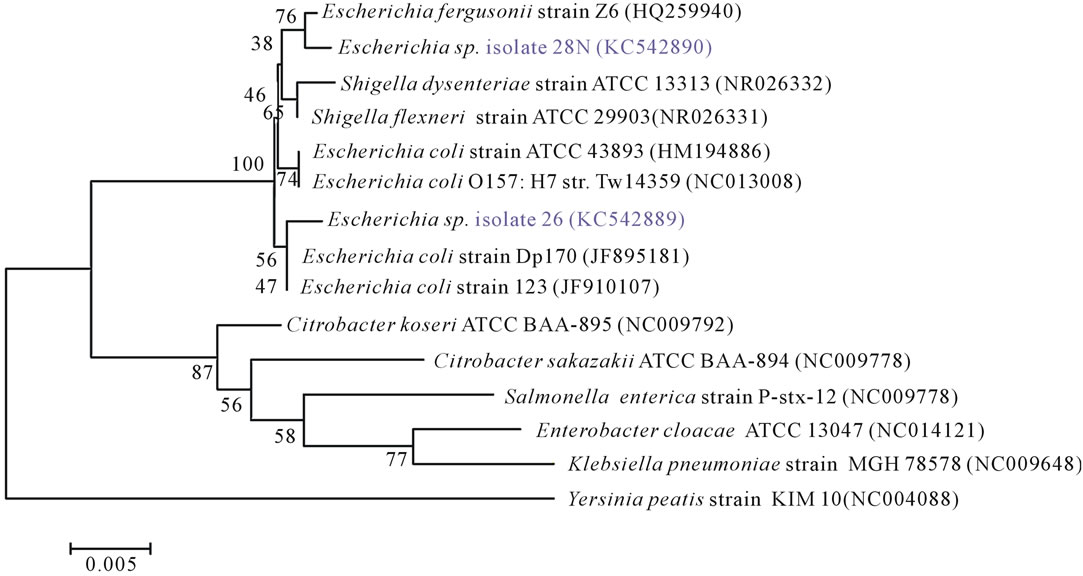
Figure 4. Phylogenetic tree of 16S rRNA gene sequences of Escherichia spp. isolates and close relative NCBI reference organisms retrieved from database with accession numbers. The tree was generated in program MEGA 5 using the Neighbour-Joining algorithm with the Yersinia sequence serving as outgroup. Bootstrap values (n = 1000 replicates) are shown at branch nodes and the scale bar represents the number of changes per nucleotide position.
cating the severity of the resistance situation in hospital wastewater effluents.
Escherichia coli is one of the most predominant organisms that is excreted out of the body through stools and sometimes urine and also it is an indicator organism for other water-borne pathogens [34]. Here, in this study Escherichia spp. was selected as a representative model organism, because MDR E. coli were randomly selected for antibiotic resistant profile analysis. Isolate 26N and 28N, phylogenetically closely related to Escherichia coli and Escherichia fergusonii respectively, showed wide range of resistance against 9 classes of antibiotics among 14 classes tested (Table 2). Their MIC and MBC against Ciprofloxacin and Cefixime were very high and they were both ESBL producing organisms.
Plasmid mediated Ciprofloxacin and Cefixime resistance is a common scenario [35] now a day, but usually the resistance level is around 32 µg/ml [36]. But the isolates of the present study are highly resistant at a level of >1024 µg/ml and carries plasmid mediated resistant determinants. If this level of resistance is present in the nature and disseminating through transferrable plasmids, soon highly effective fluoroquinolones will lose their implication in medical use. This study also shows the presence of ESBL in highly quinolone resistant Escherichia spp. from the hospital wastewater indicating that patients are spreading multi-drug resistant ESBL positive isolates in the environment, through hospital wastes.
Plasmid extraction of the cured isolates of Escherichia coli 26N and Escherichia fergusonii 28N showed that they lost most of their plasmids after curing experiments and became sensitive towards Ciprofloxacin, Cefixime and others (Figure 3). Previous evidence of interspecies O antigen gene cluster transfer between Shigella boydii 15 and Escherichia fergusonii was reported from Bangladesh [37]. Also lateral transfer of plasmid mediated ampicillin resistance and ESBL gene was observed in E. fergusonii isolates [38]. Recently plasmid mediated quinolone resistance determinants qnr was found in clinical isolates of E. coli from Norway [39] and Egypt [40] and our unpublished data also supported the view for Escherichia spp 26N and 28N. Therefore, isolate 26N and 28N are potent to transfer plasmids harbored resistance genes within other bacteria imparting antibiotic gene pollution.
5. CONCLUSION
DMCH hospital wastes pollutes environment with large number of multi-drug resistant bacteria; high level of Ciprofloxacin (>1024 µg/ml) resistant Escherichia spp. are present within the waste; these strains are found to be MDR-ESBLs and are potent to harbor multi-drug resistant genes within transferrable plasmids. The current investigation is one of the earliest reports from Bangladesh with respect to plasmid harbored high level of Ciprofloxacin resistance in Escherichia spp. especially in Escherichia fergusonii. Further studies will be carried out to identify the genes responsible for this high level of ciprofloxacin resistant and typing of the ESBL.
6. ACKNOWLEDGEMENTS
The study was supported by a grant from UGC (University Grant Commission) and Ministry of Science and Technology (Grant No: 3-2605- 3993-5901/551) and Ministry of Education, Bangladesh.
![]()
![]()
REFERENCES
- Levy, S.B. (2002) The 2000 Garrod lecture. Factors impacting on the problem of antibiotic resistance. Journal of Antimicrobial Chemotherapy, 49, 25-30. doi:10.1093/jac/49.1.25
- Okeke, I.N., Lamikanra, A. and Edelman, R. (1999) Socioeconomic and behavioral factors leading to acquired bacterial resistance to antibiotics in developing countries. Emerging Infectious Diseases, 5, 18-27. doi:10.3201/eid0501.990103
- Sahm, D.F., et al. (2001) Need for annual surveillance of antimicrobial resistance in Streptococcus pneumoniae in the United States: 2-year longitudinal analysis. Antimicrobial Agents and Chemotherapy, 45, 1037-1042. doi:10.1128/AAC.45.4.1037-1042.2001
- Lockhart, S.R., et al. (2007) Antimicrobial resistance among Gram-negative bacilli causing infections in intensive care unit patients in the United States between 1993 and 2004. Journal of Clinical Microbiology, 45, 3352- 3359. doi:10.1128/JCM.01284-07
- Barry, A.L., et al. (1990) Prevalence of fluoroquinoloneresistant bacterial isolates in four medical centers during the first quarter of 1990. European Journal of Clinical Microbiology & Infectious, 9, 906-908. doi:10.1007/BF01967511
- Bazile-Pham-Khac, S., et al. (1996) Resistance to fluoroquinolones in Escherichia coli isolated from poultry. Antimicrobial Agents and Chemotherapy, 40, 1504-1507.
- Thomson, C.J. (1999) The global epidemiology of resistance to ciprofloxacin and the changing nature of antibiotic resistance: A 10-year perspective. Journal of Antimicrobial Chemotherapy, 43, 31-40. doi:10.1093/jac/43.suppl_1.31
- Toltzis, P. (2004) Antibiotic-resistant gram-negative bacteria in hospitalized children. Clinics in Laboratory Medicine, 24, 363-380. doi:10.1016/j.cll.2004.03.001
- Schlackow, I., et al. (2012) Increasing incidence of Escherichia coli bacteraemia is driven by an increase in antibiotic-resistant isolates: Electronic database study in Oxfordshire 1999-2011. Journal of Antimicrobial Chemotherapy, 67, 1514-1524. doi:10.1093/jac/dks082
- Diwan, V., et al. (2012) Identification of extended-spectrum beta-lactamase and quinolone resistance genes in Escherichia coli isolated from hospital wastewater from central India. Journal of Antimicrobial Chemotherapy, 67, 857-859. doi:10.1093/jac/dkr564
- Liu, B.T., et al. (2012) Plasmid-mediated quinolone resistance determinant qepA1 and extended-spectrum betalactamase gene blaCTX-M-14 co-located on the same plasmid in two Escherichia coli strains from China. Journal of Medical Microbiology, 61, 603-605. doi:10.1099/jmm.0.039347-0
- Tausova, D., et al. (2012) Escherichia coli with extendedspectrum beta-lactamase and plasmid-mediated quinolone resistance genes in great cormorants and mallards in Central Europe. Journal of Antimicrobial Chemotherapy, 67, 1103-1107. doi:10.1093/jac/dks017
- Drlica, K. and Zhao, X. (1997) DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiology and Molecular Biology Reviews, 61, 377-392.
- Akter, F., et al. (2012) Ciprofloxacin-resistant Escherichia coli in hospital wastewater of Bangladesh and prediction of its mechanism of resistance. World Journal of Microbiology and Biotechnology, 28, 827-834. doi:10.1007/s11274-011-0875-3
- Poole, K. (2005) Efflux-mediated antimicrobial resistance. Journal of Antimicrobial Chemotherapy, 56, 20-51. doi:10.1093/jac/dki171
- Cavaco, L.M., et al. (2007) First detection of plasmidmediated quinolone resistance (qnrA and qnrS) in Escherichia coli strains isolated from humans in Scandinavia. Journal of Antimicrobial Chemotherapy, 59, 804- 805. doi:10.1093/jac/dkl554
- Tran, J.H. and Jacoby, G.A. (2002) Mechanism of plasmid-mediated quinolone resistance. Proceedings of the National Academy of Sciences of the United States of America, 99, 5638-5642. doi:10.1073/pnas.082092899
- Yamane, K., et al. (2007) New plasmid-mediated fluoroquinolone efflux pump, QepA, found in an Escherichia coli clinical isolate. Antimicrobial Agents and Chemotherapy, 51, 3354-3360. doi:10.1128/AAC.00339-07
- Tamang, M.D., et al. (2008) Plasmid-mediated quinolone resistance determinants qnrA, qnrB, and qnrS among clinical isolates of Enterobacteriaceae in a Korean hospital. Antimicrobial Agents and Chemotherapy, 52, 4159-4162. doi:10.1128/AAC.01633-07
- Golet, E.M., et al. (2003) Environmental exposure assessment of fluoroquinolone antibacterial agents from sewage to soil. Environmental Science & Technology, 37, 3243-3249. doi:10.1021/es0264448
- Martinez, J.L. (2009) Environmental pollution by antibiotics and by antibiotic resistance determinants. Environmental Pollution, 157, 2893-2902. doi:10.1016/j.envpol.2009.05.051
- Kummerer, K. (2001) Drugs in the environment: Emission of drugs, diagnostic aids and disinfectants into wastewater by hospitals in relation to other sources—A review. Chemosphere, 45, 957-969. doi:10.1016/S0045-6535(01)00144-8
- Brenner, D.J., et al. (2005) Bergey’s manual of systematic bacteriology. Springer.
- Bauer, A.W., et al. (1966) Antibiotic susceptibility testing by a standardized single disk method. American Journal of Clinical Pathology, 45, 493-496.
- Jarlier, V., et al. (1988) Extended broad-spectrum betalactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: Hospital prevalence and susceptibility patterns. Reviews of Infectious Diseases, 10, 867-878. doi:10.1093/clinids/10.4.867
- Birnboim, H.C. and Doly, J. (1979) A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Research, 7, 1513-1523. doi:10.1093/nar/7.6.1513
- Macrina, F.L., et al. (1978) A multiple plasmid-containing Escherichia coli strain: Convenient source of size reference plasmid molecules. Plasmid, 1, 417-420. doi:10.1016/0147-619X(78)90056-2
- Tomoeda, M., et al. (1968) Effective elimination of drug resistance and sex factors in Escherichia coli by sodium dodecyl sulfate. Journal of Bacteriology, 95, 1078-1089.
- Brosius, J., et al. (1978) Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America, 75, 4801-4805. doi:10.1073/pnas.75.10.4801
- Lawrence, J.G. (2005) Horizontal and vertical gene transfer: the life history of pathogens. Contributions to Microbiology, 12, 255-271. doi:10.1159/000081699
- Hossain, M.A., et al. (1998) Increasing frequency of mecillinam-resistant shigella isolates in urban Dhaka and rural Matlab, Bangladesh: A 6-year observation. Journal of Antimicrobial Chemotherapy, 42, 99-102. doi:10.1093/jac/42.1.99
- Zuccato, E., et al. (2010) Source, occurrence and fate of antibiotics in the Italian aquatic environment. Journal of Hazardous Materials, 179, 1042-1048. doi:10.1016/j.jhazmat.2010.03.110
- Ding, C. and He, J. (2010) Effect of antibiotics in the environment on microbial populations. Applied Microbiology and Biotechnology, 87, 925-941. doi:10.1007/s00253-010-2649-5
- Cabral, J.P. (2010) Water microbiology. Bacterial pathogens and water. International Journal of Environmental Research and Public Health, 7, 3657-3703. doi:10.3390/ijerph7103657
- Rahman, M., et al. (2007) Increasing spectrum in antimicrobial resistance of Shigella isolates in Bangladesh: Resistance to azithromycin and ceftriaxone and decreased susceptibility to ciprofloxacin. Journal of Health, Population and Nutrition, 25, 158-167.
- Chaudhry, U., et al. (2002) Mutation patterns in gyrA and parC genes of ciprofloxacin resistant isolates of Neisseria gonorrhoeae from India. Sexually Transmitted Infections, 78, 440-444. doi:10.1136/sti.78.6.440
- Azmuda, N., et al. (2012) Evidence of interspecies O antigen gene cluster transfer between Shigella boydii 15 and Escherichia fergusonii. APMIS, 120, 959-966. doi:10.1111/j.1600-0463.2012.02926.x
- Rayamajhi, N., et al. (2011) Plasmid typing and resistance profiling of Escherichia fergusonii and other Enterobacteriaceae isolates from South Korean farm animals. Applied and Environmental Microbiology, 77, 3163-3166. doi:10.1128/AEM.02188-10
- Karah, N., et al. (2010) Plasmid-mediated quinolone resistance determinants qnr and aac(6’)-Ib-cr in Escherichia coli and Klebsiella spp. from Norway and Sweden. Diagnostic Microbiology and Infectious Disease, 66, 425-431. doi:10.1016/j.diagmicrobio.2009.12.004
- Hassan, W.M., Hashim, A. and Domany, R. (2012) Plasmid mediated quinolone resistance determinants qnr, aac (6’)-Ib-cr, and qep in ESBL-producing Escherichia coli clinical isolates from Egypt. Indian Journal of Medical Microbiology, 30, 442-447. doi:10.4103/0255-0857.103766
NOTES
*Current address: Department of Biomolecular Engineering, Tokyo Institute of Technology, Tokyo, Japan.
#Corresponding author.

