Advances in Bioscience and Biotechnology
Vol.1 No.4(2010), Article ID:2854,5 pages DOI:10.4236/abb.2010.14036
Nickel uptake and intracellular localization in Cupriavidus pauculus KPS 201, native to ultramafic ecosystem
![]()
1Department of Botany, Serampore College, Serampore, Hooghly, West Bengal, India;
2Microbiology Laboratory, Department of Botany, University of Calcutta, Kolkata, West Bengal, India.
Email: arundhatipalcu@gmail.com, akpbot@caluniv.ac.in
Received 14 July 2010; revised 21 July 2010; accepted 25 July 2010.
Keywords: Cupriavidus Pauculus; Inducible Ni Resistance; Intracellular Uptake; Periplasmic Proteins; Ultramafic Soil
ABSTRACT
The nickel-resistant bacterium, Cupriavidus pauculus KPS 201 was isolated from the rhizosphere of Rinorea bengalensis (Wall.) O. K. endemic to metal-percolated ultramafic ecosystem of Andaman, India. This study investigates nature of Ni resistance, growth associated uptake and localization of Ni in cellular compartments of KPS 201. Growth kinetics of C. pauculus KPS 201 exhibited a typical inducible Ni resistance in Ni-supplemented (1.0-10.0 mM) Tris-minimal medium. The Ni-induced cells showed a high degree of Ni resistance and accumulated a maximum of 29.3 µM Ni/g protein after 48 h of growth in 5 mM Ni. The accumulated Ni was preferentially retained (90.6%) in the periplasm and was associated with the expression of two periplasmic proteins (74 and 66 kDa) under Ni-induced condition. Inducible nickel resistance in C. pauculus KPS 201 may possibly be due to extracytoplasmic binding and accumulation coupled with expression of specific periplasmic proteins. These findings will provide an insight in understanding metal-microbe interaction in geogenous environments and their exploitation in bioremediation of heavy metal pollutants.
1. INTRODUCTION
Ultramafic ecosystem comprises nutritionally poor, metal enriched soils characterized by high concentrations of nickel in addition to chromium and cobalt [1]. Majority of the endemic metallophytes inhabiting such ecosystem are Ni-hyperaccumulators which provide a niche for Ni-resistant bacteria in their rhizosphere [2]. These Ni-resistant microorganisms bear strong homologies with those isolated from anthropogenically Ni-polluted ecosystems [3]. Although Ni is an essential micronutrient, high levels of Ni along with Cr and Co favor inhibition of microbial population, growth and activity. However, the indigenous Ni-metallophiles from such Ni-percolated habitat has adopted diverse strategies to overcome Ni toxicity. Ni detoxification in bacteria takes place either by avoiding entry of metal into the cell or via intracellular sequestration and compartmentalization of Ni in different cell sectors. Resistance to Ni in Cupriavidus metallidurans CH 34 (formerly Ralstonia metallidurans, Alcaligenes eutrophus) isolated from a zinc-decantation tank in Belgium, is inducible and due to energy dependent efflux driven by chemiosmotic protonantiporter system [4]. Bioaccumulation of Ni by growing or resting cells was found linked to cellular metabolism. Pre-induced cells of Burkholderia 32W-2, native to serpentines of New Caledonia, showed nickel accumulation during growth [3]. Nickel transport across the cell membrane occurs in Escherichia coli [5] and Pseudomonas aeruginosa [6] under Ni induction and was found to encode a periplasmic Ni-binding protein.
We have screened Ni resistance in microorganisms underneath Ni-hyperaccumulators, Rinorea bengalensis and Dichapetelum gelonioides ssp. andamanicum, endemic to Indian ultramafics located in Andaman Islands. One of the 123 microbial isolates, the bacterium Cupriavidus pauculus KPS 201 (MTCC 6280) showed high resistance to Ni (MIC 29 mM Ni in nutrient broth) and resting cells pre-grown in nutrient broth accumulated 224 µM Ni/g protein in 60 min from aqueous Ni(II) solution in HEPES buffer [7]. Our present study investigates the nature of Ni resistance, growth associated uptake and possible localization of accumulated Ni in C. pauculus KPS 201 cells.
2. MATERIALS AND METHODS
2.1. Organism and Culture Conditions
Cupriavidus pauculus KPS 201 (MTCC 6280), used throughout the present study was maintained on slopes of Tris gluconate minimal medium [2] supplemented with 1.0 mM Ni(II) as chloride salt and grown at 28ºC for 24 h.
2.2. Growth and Nickel Uptake
Ni uptake by KPS 201 cells was conducted in Tris-minimal medium (50 ml per 250 ml Erlenmeyer flask) supplemented with Ni. The medium was inoculated with 18 h old inoculum, grown either in absence (non-induced cells) or in presence of 0.25 mM Ni (induced cells) and agitated on a rotary shaker (120 rpm) at 28℃. Growth was monitored by recording optical density changes at 540 nm (UV-Vis-spectrophotometer, Jenway 6505) and measuring total protein content per ml of the culture. For nickel estimation, growing cells were harvested by centrifugation in a Hermle Z36HK cold centrifuge (10,000 x g at 4℃) and the cell pellet was digested with 6 M HNO3 at 80℃ for 2 h. Protein and nickel content of biomass were estimated following the methods as described below.
2.3. Cell Fractionation
Ni-loaded cells were harvested, washed and fractionated following the modified procedure of Bernhard et al. [8]. Periplasmic fraction was obtained by osmotic shock method using sucrose and lysozyme. Sphaeroplasts obtained were lysed by sonication (Hielscher ultrasonic processor, UP 100H) and centrifuged at 10,000 x g (10 min at 4℃) to remove cell debris. The supernatant was further centrifuged at 90,000 x g for 45 min at 4℃ (Beckman Coulter Ultracentrifuge LE-80K) to separate membrane (pellet) and cytoplasmic (soup) fractions.
Protein from each fraction was precipitated with 10% (w/v) cold trichloroacetic acid, washed and dissolved in 10 mM Tris-Cl (pH 8.0).
2.4. Protein Estimation and Gel Electrophoresis
Total protein of biomass was extracted with 1% (w/v) SDS and precipitated with 25% cold trichloroacetic acid. Protein content was estimated by folin-phenol reagent using bovine serum albumin as standard [9]. Proteins in the isolated cell fractions were resolved in 10% (w/v) SDS-PAGE [10] and stained with silver nitrate [11].
2.5. Estimation of Nickel
Nickel content (mM Ni/g protein) of the digested biomass and cellular fractions was quantified using Atomic Absorption Spectrometer (Varian SpectrAA20 Plus) operating at 232 nm, 0.2 mm slit width and air-acetylene flame.
3. RESULTS
3.1. Nickel Resistance in Cupriavidus pauculus KPS 201
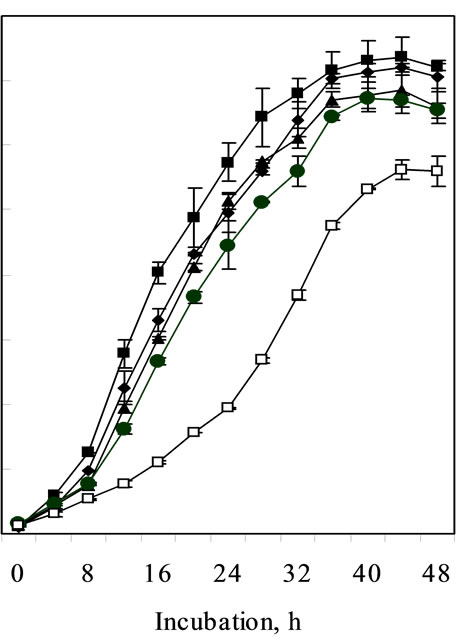
Growth kinetics of C. pauculus KPS 201 in Ni-supplemented (1.0-10.0 mM) Tris-minimal medium exhibited a typical metal-induced response for Ni resistance. Non-induced cells of KPS 201 when grown in presence of Ni showed a characteristic prolonged lag which increases with increasing metal concentration showing a maximum (22 h) in 10 mM Ni (Figure 1(a)). Under identical conditions, cells induced with 0.25 mM Ni grew exponentially without any distinguishable lag (Figure 1(b)).
The induced cells also showed high Ni resistance
 (a)
(a)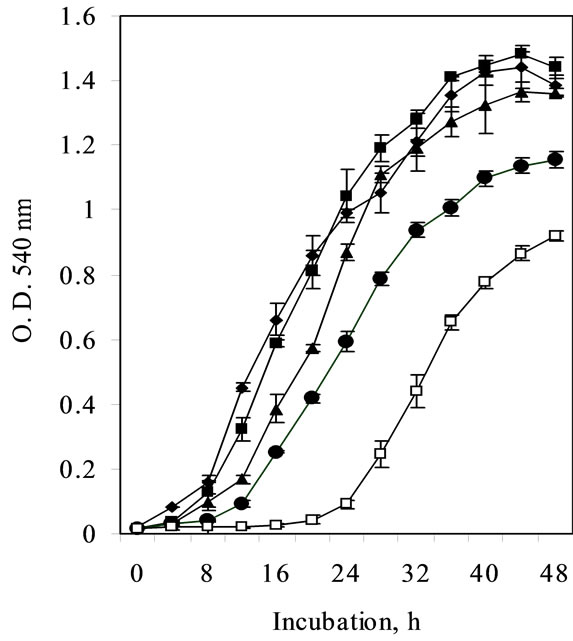 (b)
(b)
Figure 1. Growth of non-induced (a) and Ni-induced (b) cells of Cupriavidus pauculus KPS 201 in absence (-®-) and presence of 1.0 mM (–¾–), 2.0 mM (––), 5.0 mM (–·–) and 10.0 mM (–£–) Ni.
(MIC 21.3 mM) contrary to non-induced cells (15 mM Ni) (Figure 2).
3.2. Nickel Uptake during Growth
In view of enhanced growth performance in Ni and high degree of metal resistance, nickel uptake studies were conducted using Ni-induced (pre-grown in 0.25 mM Ni) KPS 201 cells. During growth in presence of non-toxic concentrations of Ni (1.0 – 10.0 mM), KPS 201 cells accumulated metal and attained equilibrium at late exponential to early stationary phase. The extent of Ni uptake in the biomass increased with increasing Ni concentration in the growth medium, attaining maxima (29.3 µM Ni/g protein) after 48 h of incubation in 5 mM Ni (Figure 3).
3.3. Cellular Compartmentalization of Nickel
Metal-loaded cells of KPS 201 (29.3 µM Ni/g protein) were disrupted and separated into periplasmic, membrane and cytosolic fractions [8] and their purity was confirmed by the assay of marker enzymes like alkaline phosphatase [12], gluconate dehydrogenase [13] and

Figure 2. Ni resistance in Cupriavidus pauculus KPS 201 cells grown in Tris-minimal medium. The medium was inoculated with 18 h grown culture prepared in the same medium either without Ni (non-induced) or supplemented with 0.25 µM Ni (induced). The flasks were incubated on a rotary shaker (120 rpm) at 30ºC for 24 h and growth was monitored by recording optical density changes at 540 nm.
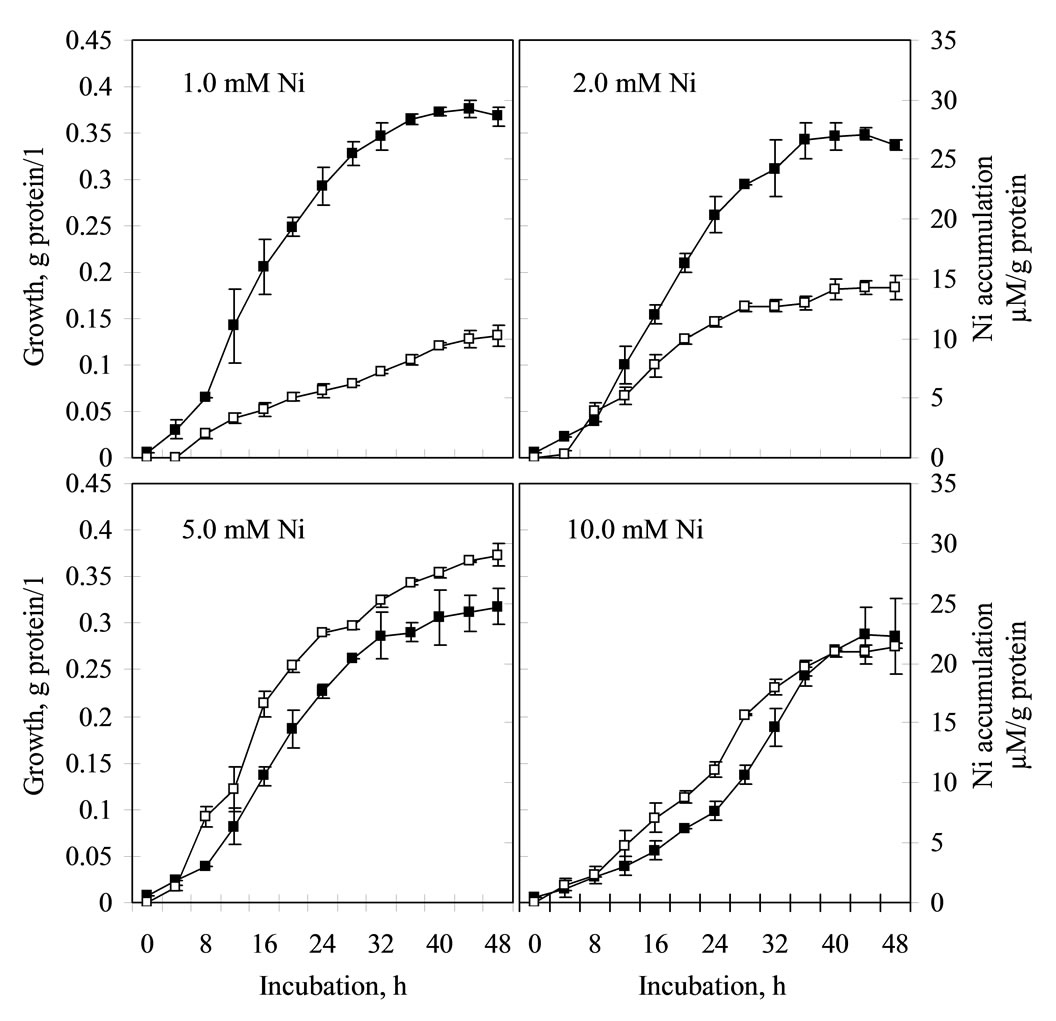
Figure 3. Growth (–¾–) associated Ni accumulation (–£–) in Cupriavidus pauculus KPS 201. Cells previously induced in 0.25 mM Ni were grown in Tris-minimal medium supplemented with 1–10 mM Ni. Total protein (g/l) of biomass was estimated along with amount of accumulated Ni (µM/g protein) following folin phenol assay and Atomic Absorption Spectrometry respecttively.
PHA depolymerase [14] respectively. A major part (90.62%) of the accumulated metal was retained in the periplasm (26.58 µM Ni/g protein), while only 0.51% Ni (0.15 µM Ni/g protein) was bound to the cell membrane. Cytoplasmic fraction contained 8.83% of the total Ni accumulated by intact cells (Table 1).
3.4. Protein Profile
Protein profiles of periplasmic, membrane and cytoplasmic fractions of Ni-induced KPS 201 cells were compared with those of the non-induced control following SDS-PAGE. Periplasmic protein fractions (Figure 4) of Ni-induced cells grown in presence of 1 to 10 mM Ni (Lane 3-6) showed two distinct bands of approximately
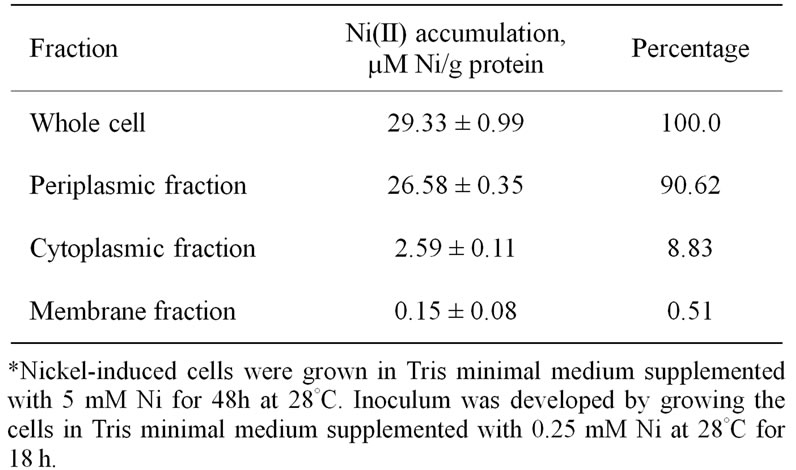
Table 1. Cellular compartmentalization of Ni in metal-loaded Cupriavidus pauculus KPS 201 grown in nickel supplemented medium *

Figure 4. Electrophoretic analysis of periplasmic proteins of Ni-induced Cupriavidus pauculus KPS 201 cells grown in presence of 1–10 mM Ni. Lane 1: Marker proteins (kDa), Lane 2: Non-induced control, Lane 3: 1.0 mM Ni, Lane 4: 2.0 mM Ni, Lane 5: 5.0 mM Ni, Lane 6: 10.0 mM Ni.
74 and 66 kDa, but similar protein bands were missing in cells not exposed to nickel (Lane 2). Cytosolic and membrane protein profiles of Ni-grown and control cells, however, have failed to show any visible variation in their banding patterns (data not shown).
4. DISCUSSION
Nickel resistance in bacteria from nickel-polluted ecosystem is reported to be inducible [4,15]. Similar induction of Ni resistance system in Burkholderia 32W-2, native to New Caledonian ultramafics was evident at 1.0 mM Ni [3]. On the contrary, induction of Ni resistance in Cupriavidus pauculus KPS 201 (Figure 1) was achieved at a much lower concentration of Ni (0.25 mM). Under such induced state the MIC in Tris minimal medium (21.3 mM Ni, Figure 2) was much lower compared to that of non-induced cells grown in enriched medium [7]. Such a variation could be attributed to complexation of Ni with organic constituents of the enriched medium [16].
Although uptake of Ni (29.33 µM Ni/g protein) by induced KPS 201 cells during growth was dependent on initial metal concentration up to 5 mM Ni, it declined at higher concentration (Figure 3). Such a decline could be attributed to induction of KPS 201 cells with low Ni concentration as well as toxicity imparted at high metal concentration leading to inhibition of RNA and protein synthesis [17]. It was further evident that in C. pauculus preferential accumulation (90.62%) of Ni in the periplasm (Table 1) was coupled with the expression of two periplasmic proteins of molecular weights 74 and 66 kDa under Ni induced condition (Figure 4). Similar low molecular weight periplasmic proteins involved in Ni resistance have also been demonstrated in E. coli K-12 (56 kDa) [5]; P. putida (18 kDa) [6] and C. necator H16 (33.1 kDa) [18]. Further research is needed to establish the Ni-sequestering property of these periplasmic proteins in C. pauculus KPS 201.
5. ACKNOWLEDGEMENTS
Financial support to A. Pal from Department of Science and Technology, New Delhi, India (DST Fast Track Young Scientist Scheme; SR/FT/L-125/2005 dated 12.07.06) is duly acknowledged.
REFERENCES
- Brooks, R.R. (1987) Serpentine and its vegetation, a multidisciplinary approach. Croom Helm, London.
- Schlegel, H.G., Cosson, J.P. and Baker, A.J.M. (1991) Nickel hyperaccumulating plants provide a niche for nickel resistant bacteria. Botanica Acta, 104, 18-25.
- Stoppel, R.D. and Schlegel, H.G. (1995) Nickel resistant bacteria from anthropogenically nickel polluted and naturally nickel percolated ecosystems. Applied and Environmental Microbiology, 61(6), 2276-2285.
- Nies, D.H. and Silver, S. (1989) Plasmid determined inducible efflux is responsible for resistance to cadmium, zinc and cobalt in Alcaligenes eutrophus. Journal of Bacteriology, 171(2), 896-900.
- De Pina, K., Navarro, C., Mcwalter, L., Boxer, D.H., Price, N.C., Kelly, S.M., Mandrand-Berthelot, M. and Wu, L. (1995) Purification and characterization of nickel-binding protein NikA of Escherichia coli K 12. European Journal of Biochemistry, 227(3), 857-865.
- Tripathi, V.N. and Srivastava, S. (2006) Extracytoplasmic storage as the nickel resistance mechanism in a natural isolate of Pseudomonas putida S4. Canadian Journal of Microbiology, 52(4), 287-292.
- Pal, A., Wauters, G. and Paul, A.K. (2007) Nickel tolerance and accumulation by bacteria from rhizosphere of nickel hyperaccumulators from serpentine soil of Andaman, India. Plant and Soil; 293(1-2), 37-48.
- Bernhard, M., Friedrich, B.R. and Siddiqui, R.A. (2000) Ralstonia eutropha TF93 is blocked in Tat-mediated protein export. Journal of Bacteriology, 182(3), 581-588.
- Lowry, O.H., Rosebrough, N.J. and Farr, A.L. (1951) Protein measurement with the folin-phenol reagent. Journal of Biological Chemistry, 193(1), 265-275.
- Laemmli, U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227(5259), 680-685.
- Shevchenko, A., Wilm, M., Vorm, O. and Mann, M. (1996) Mass spectrometric sequencing of proteins from silver stained polyacrylamide gels. Analitycal Chemistry, 68(5), 850-858.
- McAlister, T.J., Costertan, J.W., Thompson, L., Thompson, J. and Ingram, J.N. (1972) Distribution of alkaline phosphatatse within the periplasmic space of Gram-negative bacteria. Journal of Bacteriology, 111(3), 827-832.
- Matsushita, K., Shinagawa, E., Adichi, O. and Ameyama, M. (1979) Membrane-bound D-gluconate dehydrogenase from Pseudomonas aeruginosa: purification and structure of cytochrome binding form. Journal of Biochemistry, 85(5), 1173-1181.
- Kobayashi, T., Nishikoriand, K. and Saito, T. (2004) Properties of an intracellular poly (3-hydroxybutyrate) depolymerase (PhaZ1) from Rhodobacter sphaeroides. Current Microbiology, 49(3),199-202.
- Rubikas, J., Matulis, D., Leipus, A. and Urbaitiene, D. (1997) Nickel resistance in Escherichia coli V38 is dependent on the concentration used for induction. FEMS Microbiology Letters, 155(2), 193-198.
- Angle, J.S. and Chaney, R.L. (1989) Cadmium resistance screening in nitrilotriacetate-buffered minimal media. Applied and Environmental Microbiology, 55(8), 2101- 2104.
- Mulrooney, S.B. and Hausinger, R.P. (2003) Nickel uptake and utilization by microorganisms. FEMS Microbiology Reviews, 27(2-3), 239-261.
- Wolfram, L., Friedrich, B. and Eitinger, T. (1995) The Alcaligenes eutrophus protein HoxN mediates nickel transport in Escherichia coli. Journal of Bacteriology, 177(7), 1840-1843.

