World Journal of AIDS
Vol.2 No.2(2012), Article ID:19721,7 pages DOI:10.4236/wja.2012.22008
Genetic Diversity and Antiretroviral Drug Resistance among Drug-Naïve HIV-1 Infected Pregnant Women Attending Antenatal Clinics in Abidjan, Côte d’Ivoire*
![]()
1Faculty of Pharmaceutical and Biological Sciences, University of Cocody, Abidjan, Côte d’Ivoire; 2National Public Health Laboratory, Abidjan, Côte d’Ivoire; 3Faculty of Medical Sciences, University of Cocody, Abidjan, Côte d’Ivoire; 4Pasteur Institute, Abidjan, Côte d’Ivoire.
Email: gylouk@africaonline.co.ci, {#nangatchocht, ndalaurent, kouacle, lathrosergefr, cablanmianarsher, akouamc}@yahoo.fr
Received March 28th, 2012; revised April 20th, 2012; accepted May 18th, 2012
Keywords: HIV-1; drug naïve pregnant women; ARV resistance
ABSTRACT
To clarify the distribution of HIV-1 subtypes and drug resistance-related mutations, we collected and analysed serum from pregnant women who are ARV drug-naive in Abidjan. The prevalence of HIV-1 subtypes and mutations associated with antiretroviral drug resistance among drug-naïve HIV-1 infected pregnant women was investigated from plasma of 90 young pregnant primigravida. The HIV-1 pol and env genes were amplified by using primers recognizing conserved viral sequences and sequenced by employing BigDye chemistry. Positions 1 - 99 of the PR and 1 - 350 of the RT genes were analyzed for mutations based on the international AIDS society USA panel. In 39 strains which both genes were sequenced including CFR02_AG 30 (76.9%), subtype A 3 (7.7%), CFR06_cpx 2 (5.1%), CFR09_cpx 1 (2.6%), and discordant sequences suggesting the presence of a few number of recombinant involving CRF02-AG and subtype A 3 (7.7%). None of the major drug resistance mutations was detected. The frequent minor mutations associated drug resistance observed were M36I (52%/96.3%), L10I/R/V (19%/35.2%) and L63P (7%/12.9%). The M36I mutation was widespread in all subtypes. Our result demonstrated first a significant level of viral heterogeneity and then only the presence of minor resistance associated mutations. Our study emphasizes the need of HIV sentinel survey in Côte d’Ivoire and shows that pregnant women who are candidates for receiving antiretroviral drug therapies do not contain naturally occurring or preexisting drug resistance mutations. So such drug therapies are likely to be highly effective in this setting.
1. Introduction
To date, the remarkable viral diversity of HIV-1 results in the classification of the virus into types, groups, subtypes, sub-subtypes and over circulating recombinant forms (CRFs) [1]. Although the epidemic in the western world is primarily due to subtype B, most of the other subtypes and CFRs are found in Africa [2]. Subtypes A, C and CFR02 are predominating, but similarly to the global distribution of HIV-1 variants, their geographic distribution on the african continent is heterogenous and differs from country to country and even within certain countries [3]. Highly active antiretroviral therapy (HAART) has greatly reduced HIV/AIDS-related morbidity and mortality. However, the use of antiretroviral (ARV) has created a major public health concern about the possible emergence of drug-resistant mutants that would lead to treatment failure. Now that access to HIV treatment is expanded, surveillance of drug-resistant strains over time should be implemented with preference on population groups that represent individuals who were recently infected with HIV as recommended by WHO or for whom data on previous ARV treatment can be collected [4].
Amino acid mutations in protease (PR) and reverse transcriptase (RT) associated with resistance to antiretroviral drugs are identified when sequences are compared to the consensus pol sequence of wild-type HIVHXB2 (www.iasusa.org). Limited studies have been conducted in Sub-Saharan Africa (SSA) to identify mutations associated with antiretroviral drug resistance. It is imperative to investigate and describe mutations associated with resistance to antiretroviral drugs in the PR and RT genes of HIV-1 subtypes and CRF among treatment-naïve populations in SSA. Baseline information collected from these studies will be of importance in the development of algorithms for the interpretation of mutations conferring antiretroviral drug resistance in different geographical regions.
In West Africa, Côte d’Ivoire has the highest HIV-1 prevalence rate (3.4%) and 60% of HIV-infected patients are women, most of them of childbearing age [5]. In this country, HAART has been available since 1998 through the UNAIDS/Côte d’Ivoire program of access to HIV/ AIDS treatment drugs. Moreover, in December 2009, 72,000 patients under ARV treatment were indexed in the country whereas 368,000 others are waiting for ARV treatment (http://www.ipsinternational.org, update 2010). Thus, an increasing number of pregnancies is occurring in women initiating ART. The prevalence of HIV drug resistance mutation (HIVDRM) is scored as 5% - 6% in untreated recently infected patients [6].
The aim of the present study was to clarify the distribution of HIV-1 subtypes and the baseline prevalence of drug resistance-related mutations in antiretroviral drug-naive HIV-1 pregnant women living in three districts of Abidjan before the extensive use of antiretroviral drug is supplemented in the country.
2. Study Population and Methods
2.1. Study Subjects and Samples
Young pregnant primigravida women (17 - 25 years) were recruited in the antenatal clinic (ANC) sentinel surveillance conducted by Retrovirus Côte d’Ivoire (RETROCI) as recommended by the WHO resistance network [4]. Women came from three districts of Abidjan: Abobonord, Abobo-sud and Koumassi. Women were assumed to have acquired infection within the last few years and data on ART exposure were also collected.
All women underwent preand post-test counselling and signed informed consent form prior to participation in the study. Inclusion criteria were: 1) HIV-1 seropositive pregnant woman, 2) Pregnant women attending antenatal clinic for a first visit, 3) Not receiving highly active antiretroviral therapy (HAART), 4) Samples and demographic information were collected between September 2004 and January 2005. HIV-1 serology was performed on the basic of the Retrovirus Côte d’Ivoire (RETROCI) HIV testing algorithm using the three rapid assay confirmatory strategies for the detection of antibodies to HIV (www.who.int/hiv:pub). First, HIV antibody testing was performed by using two mixed (HIV-1 and HIV-2) rapid assays Enzynost Intergral (Dade Behring Marburg GmbH, Marburg, Germany), Murex 1.2.0 (Murex Biotech Ltd, Dartford, Kent, UK). Secondly Determine (Abbott Laboratories, IL, USA) was used to discriminate HIV-1 and HIV-2. Discordant blood samples were tested by the additional testing by using ELISA (Vironostika ELISA test; BioMérieux, France).
The study received ethical approval from the ethics committees of the Ivoirian National Program.
2.2. RNA Extraction, RT PCR, Secondary PCR and Sequencing
RNA was extracted from plasma samples using Amplicor HIV-1 monitor test (Roche, version 1.5) according to the manufacturer’s instructions. The genetic subtypes were identified in env and pol as previously described [3,7]. Briefly, RT PCR for the env and pol regions was performed by using Expand High Fidelity PCR system (Roche, France) with primer pair ED12/ED5 (env) and IN3/G25REV (pol). Secondary PCR was performed by using primer pair ES7/ES8 (env) and AV150/PolM4 (pol) under conditions recommended by the manufacturer. Ultrapure Water was used as a negative control in each run. The amplified fragment of approximately 670 base pairs (env) and 1770 base pairs (pol) were purified with GeneClean Turbo kit (Q-Biogenic, Illkirch, France) and were directly sequenced using Big Dye Terminator v3.1 (Applied Biosystems, France) according to the manufacturer’s instructions. Electrophoresis and data collections were performed on an Applied Biosystem 3100 genetic analyzer. The newly determined sequences were aligned with known representatives of the different subtypes and CFRs described in West and Central Africa by using CLUSTAL W and minor manual adjustments were made where necessary to accommodate reading frames. Regions that could not be aligned unambiguously, due to length or sequence variability, were omitted from the analysis. Phylogenetic analysis using the neighbourjoining method and reliability of the branching orders using boostrap approach were implemented with CLUSTAL W [8].
2.3. Drug Resistance Analysis
PT and RT resistance mutations were reported as listed by the international AIDS society USA panel, update Fall 2010 (www.iasusa.org) and the genotypic interpretation was performed with the ANRS AC11 algorithm, update 2011 (www.hivfrenchresistance.org).
2.4. Statistical Methods
We used Friedman test to analyse the significantly difference between groups regarding the frequency of detected mutations and strains. p < 0.05 was considered as statistically significant.
3. Results
3.1. Baseline Characteristic of 90 HIV-1 Infected Women
A total nine hundred and thirty six young pregnant primigravida women were recruited. The mean age was 19 years and extremes 17 - 25 years. Ninety women were infected by HIV-1, 42 (Abobo-nord), 26 (Koumassi) and 22 (Abobo-sud). The CD4 cells counts ranged 237 to 609 cells/mm3 (Table 1).
3.2. HIV-1 Detection and Subtyping
The genes pol and env were successfully detected from 54 and 39 patients respectively. The 39 samples for which both genes were sequenced included CFR02_AG (76.9%), subtype A (7.7%), CFR06_cpx (5.1%), CFR09_ cpx (2.5%), and discordant sequences (7.7%) involving CRF02-AG for pol gene and subtype A for env gene (Table 1). The pol gene fragment sequenced alone in 15 cases were subtypes A (n = 13), CFR02_AG (n = 1) and unclassified strain (n = 1). We isolated in Abobo-nord, the subtype A (14.3%), CFR02_AG (71.4%) and CFR- 06_cpx (14.3%). In Abobo-sud, CFR02_AG (76.4%) and subtype A (11.8%) were detected. In Koumassi, CFR02_ AG (80.2%), CFR06_cpx (6.6%) and CFR09_cpx (6.6%) were identified. The recombinant CFR02_AG predominated in all districts (71.4% - 80.2%) as shown in Table 2.
3.3. Protease Inhibitor (PI) Resistance-Associated Mutations
The amino acid mutations in protease (PR) and reverse
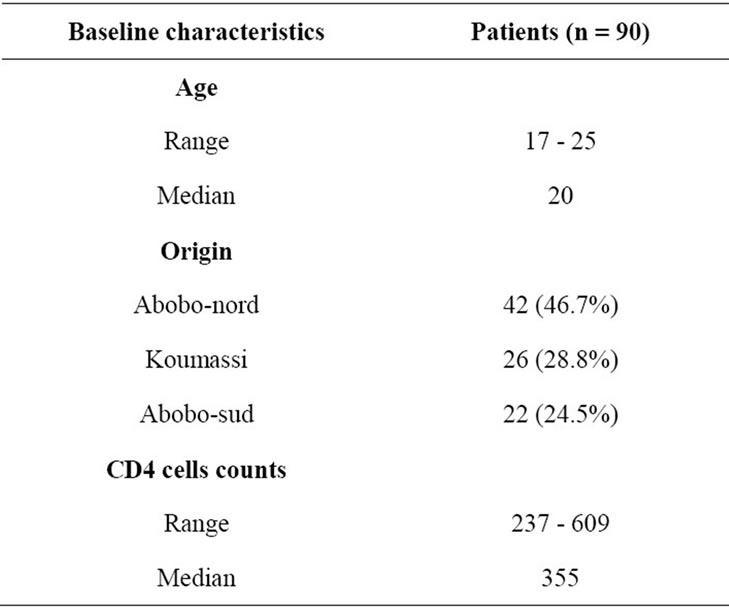
Table 1. Baseline characteristics of 90 ARV-naïve HIV-1 infected pregnant women.
transcriptase (RT) associated with resistance to antiretroviral can be classified into major or primary and secondary or accessory mutations None of the known primary mutations associated with PI resistance were detected among the PR sequences. Secondary mutations were observed. Frequent mutations detected were retrieved at position 36 (M36I, 52%/96.3%), position 10 (L10I/R/V, 19%/35.2%) and position 63 (L63P, 7%/12.9%) (Figure 1).
3.4. Reverse Transcriptase Inhibitor (RTI) Resistance-Associated Mutations
No mutation associated with non nucleoside reverse transcriptase inhibitor (NNRTI) was found. No primary mutation associated with nucleoside reverse transcriptase inhibitor (NRTIs) was detected while three secondary mutation associated with NRTIs resistance were observed S68G (1%/1.8%), V118I (1%/1.8%) and Y115F (1%/1.8%) as shown in Figure 1.
4. Discussion
The aim of the present study was to clarify the distribution of HIV-1 subtypes and the baseline prevalence of drug resistance-related mutation in antiretroviral drugnaive HIV-1. Among 90 HIV-1 strains collected in young pregnant primigravida women, we were unable to amplify 36 strains. The amplification failure could be attributed to the viral diversity at primer binding sites. Another reason that could explain this amplification failure was that the viral load could be low or undetectable. A study on viral load could help to confirm or reject such cause. But we disposed no materials to lead the study.
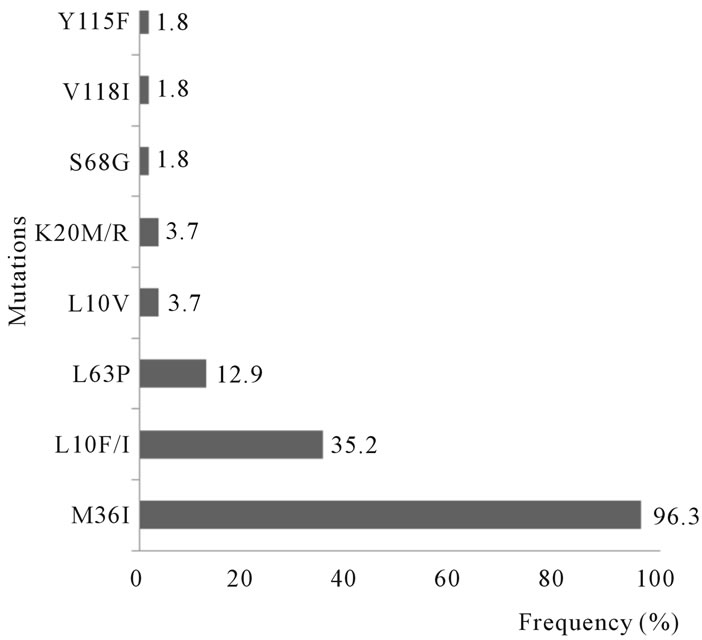
Figure 1. Frequency of drug mutations among 54 ARVnaïve HIV-1 infected pregnant women. The frequencies were calculated from 54 women in whom the gene pol was successfully detected.
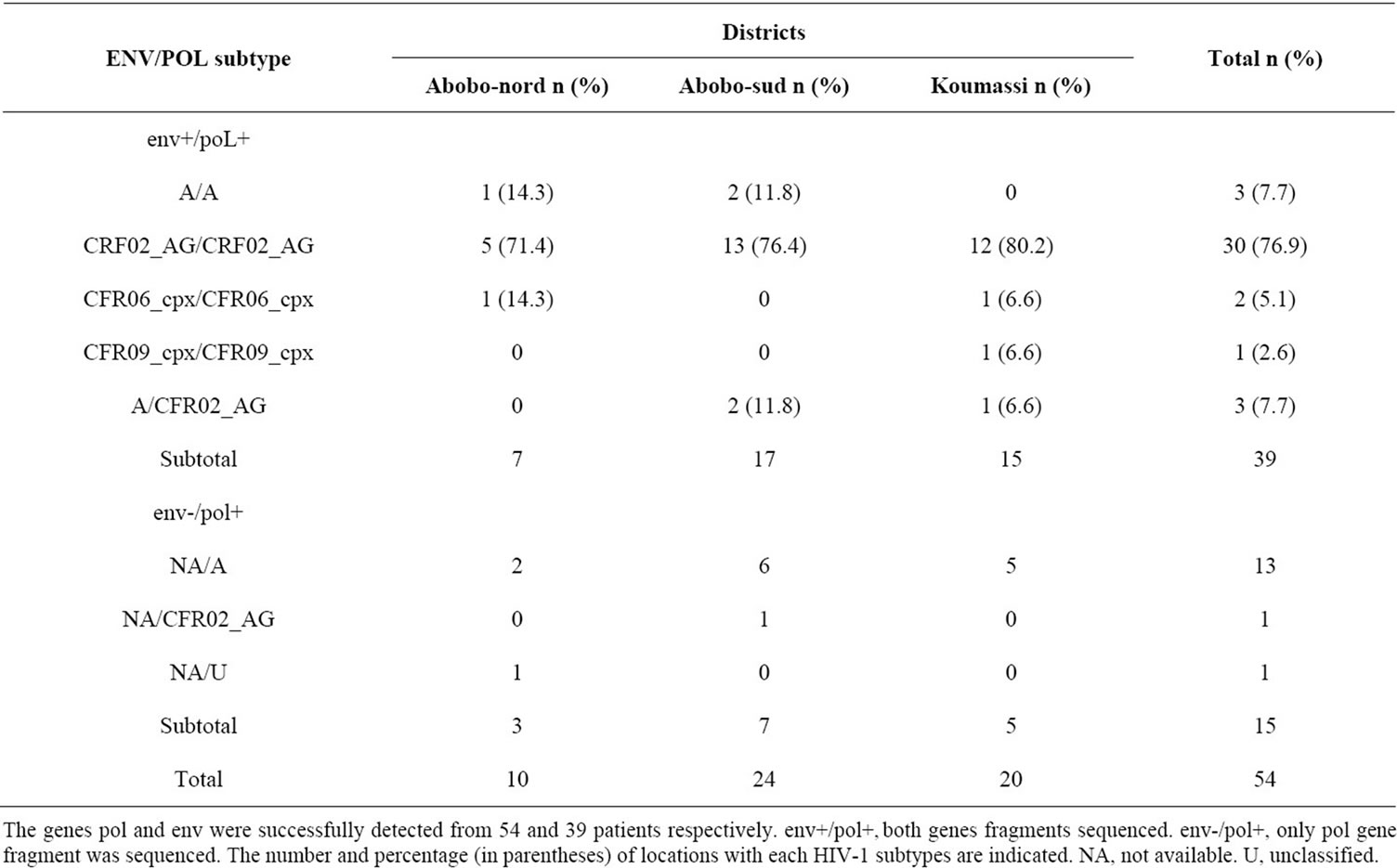
Table 2. Distribution of various HIV-1 subtypes among ARV-naïve HIV-1 infected pregnant women according to the districts.
The results obtained revealed that CFR02_AG was highly predominant in Abidjan where a few recombinant inter-subtypes also circulated. The results obtained are similar to the previous studies conducted in Abidjan and in others West Africa countries [8-11]. The results obtained suggested a complex and possible changing proportion of HIV subtypes contributing to the HIV epidemic in Abidjan. The proportion of HIV subtypes varied significantly between the three districts of Abidjan even though two of the districts (Abobo-nord and Abobo-sud) are adjacent to each other (p < 0.05). The fact that the HIV subtypes varied between different locations in Abidjan, suggested that HIV diversification may be ongoing.
An unclassified virus was observed in one patient from Abobo-nord. The appearance of new variants is the result of a co-infection and it occurs in the areas where several variants circulate. This new variants have a significant effect on the epidemic of the HIV infection in particular on the tests of serological diagnostic and viral load. Further, this may represent a significant obstacle to a vaccine development. Actually an efficient vaccine against HIV, should take into account the different circulating subtypes and recombinant forms isolated in Côte d’Ivoire. It appears necessary to characterize as soon as possible the variants into circulation in Abidjan.
HIV-1 drug resistance arises from mutations in the genes that encode the molecular targets for the drugs, i.e., the RT and protease pol gene products. This viral polymorphism is due to the high rate of HIV-1 replication and the low fidelity of RT [12]. The emergence of amino acid substitutions associated with resistance to RT and PIs has been extensively characterized [13], and these substitutions can be classified into major and accessory (modifying) mutations. Major mutations lead to a several fold decrease in sensitivity to one or more antiretroviral drugs [14]. Accessory mutations may not result in a significant decrease in sensitivity but are associated with an increase in viral fitness (replication capacity) [13]. Thus, the appearance of a major mutation in a genome already containing accessory mutations could influence the speed with which highly resistant viruses are selected during therapy. The results obtained suggest that any major mutations conferring resistance to protease inhibitors (PIs) were not seen. There was no evidence of major mutations in young pregnant ART naïve women with recent infection. The absence of major mutations in the studied population was consistent with other studies that were conducted in settings that were currently scaling up ART in Africa [7,10,11,14,15]. These studies have reported prevalence levels of <5%, the WHO detection threshold limit in antiretroviral naïve populations. However, a higher frequency of HIVDRM has recently been reported from a large study in Rwanda, South Africa, Uganda and Zambia where slightly more than 5% drug resistance was identified in samples collected from 2006 through 2009 [16].
In contrast to major mutations, 96.3% of the strains had two or more minor mutations associated with PIs resistance; only 3.7% of strains (mainly subtype CRF02_ AG strains) had no minor mutations. M36I is the predominant minor mutation and was observed in all subtypes, CFR02_AG (n = 8), A (n = 3), CFR09_cpx (n = 3), CFR06_cpx (n = 2) and unclassified strain (n = 1). Amino acid substitutions associated with PI resistance were reported as natural non-B HIV-1 variants in treatment-naive patients [17,18]. The prevalence of substitutions obtained was similar to those previously described [19]. In a previous study conducted in Abidjan, the authors have detected key drug resistance mutation in recently HIV-1 infected adults [9]. This difference with our results may be explained by the fact that previous studies concerned the general population. The proportion of resistant strains in the general population did not reflect recently transmitted resistance, as it is not known whether the HIV-positive individuals are completely ARV naive.
The mutations leading to resistance to nucleoside reverse transcriptase inhibitor (NRTIs) and non nucleoside reverse transcriptase inhibitor (NNRTIs) were well defined and differ within the two classes of RT inhibitors [13]. No NNRTI mutation was detected. No major mutation conferring resistance to NRTIs was seen, but three accessory mutation, S68G (1.8%), V118I (1.8%) and Y115F (1.8%) were found. Selection for S68G mutation were observed after virologic failure on tenofovir, abacavir, didanosine or stavudine containing regiments and in some cases linked with K65R selection. The S68G mutation followed the initial selection for K65R and M184V [20]. The V118I mutation was associated with resistance to zidovudine when they appeared together with E44D but the significance of V118I when it occurred alone in isolation is unknown [21,22]. Several observations suggested that S68G and V118I may represent a preexisting polymorphism [23-25]. The S68G mutation was present in around 3% of untreated patients and the prevalence of V118I in naive drug patients varied from 2.5% to 8.3% [26]. The Y115F mutation was associated to resistance to abacavir and did not contribute singly to resistance to abacavir but in combination with other mutations (K65R, L74V, M184V) enhanced NRTIs resistance [27].
There were several limitations encountered in this study. The women were recruited only in three districts of Abidjan. The prevalence of drug resistance mutations in the study population was very low, and thus there was no ability to estimate the frequency of transmitted drug resistance. According to the information collected, most of the individuals were diagnosed at an advanced stage of infection (1 - 4 years after infection). The exact lapse of time between the moments of infection and sample collection is unknown. Therefore, the results obtained may have been influenced by this period. Theoretically, newly infected patients are more likely to harbour resistant HIV strain variants than chronically treatment-naïve patients. This might be explained by the fact that wild type strains may become predominant in the absence of selective drug pressure, when individuals are infected with both wild type and mutant viruses. We didn’t dispose information concerning the viral load. However, the focus on young, largely primigravida pregnant women was expected to identify recent infections. The younger age, significantly higher CD4 cell numbers and low or undectable viral load related to recent infection may provide some evidence for the results obtained.
5. Conclusion
The results obtained show a high viral heterogeneity and the lack of primary mutations associated with antiretroviral resistance. But the minor mutations were detected among the drug-naive pregnant women in Abidjan. The data obtained suggested that routine resistance testing before the initiation of therapy in this initial stage of the treatment program may not be necessary in this setting. Moreover, our results suggested that the HIV first line treatment was likely to be highly effective in drug patients women and the possibility to reduce perinatal transmission risk. As transmission of ARV resistance mutation is an important public health and has clinical implications, it is necessary to conduct extensive surveys covering pregnant women in the all districts in Abidjan, with a sufficient number of samples, to fully assess the molecular epidemiology of HIV in Abidjan and to guide antiretroviral treatment and vaccine strategies.
6. Authors’ Contributions
Loukou Y.G., Akoua-Koffi M.C. conceived the study Zinzendorf N.Y., Djé C. and Kouadio H. coordinated data collection and performed laboratory, data analyses and interpretation of results, Zinzendorf N.Y., Djé coordinated laboratory analysis, Kouadio H., Lathro J., Cablan A., Dje K. participated in laboratory analysis, Loukou Y.G., Zinzendorf N.Y. participated in study design, interpretation of results. All authors contributed to the writing, read and approved the final manuscript.
7. Acknowledgements
We thank the ANC teams of the three districts Abobonord, Abobo-sud and Koumassi. We are gratefull to women who participated in this research. This work was supported by RETROCI.
REFERENCES
- D. L. Robertson, J. P. Anderson, J. A. Bradac, J. K. Carr, B. Foley, R. K. Funkhouser, F. Gao, B. H. Hahn, M. L. Kalish, C. Kuiken, G. H. Learn, T. Leitner, F. McCutchan, S. Osmanov, M. Peeters, D. Pieniazek, M. Salminen, P. M. Sharp, S. Wolinsky and B. Korber, “HIV-1 Nomenclature Proposal,” Science, Vol. 288, No. 5463, 2000, pp. 55-56. doi:10.1126/science.288.5463.55d
- M. Peeters, C. Toure-Kane and J. N. Nkengasong, “Genetic Diversity of HIV in Africa: Impact on Diagnosis, Treatment, Vaccine Development and Trials,” AIDS, Vol. 17, No. 18, 2003, pp. 2547-2560. doi:10.1097/00002030-200312050-00002
- N. Vidal, C. Mulanga, S. E. Bazepeo, J .K. Mwamba, J. W. Tshimpaka, M. Kashi, N. Mama, C. Laurent, F. Lepira, E. Delaporte and M. Peeters, “Distribution of HIV-1 Variants in the Democratic Republic of Congo Suggests Increase of Subtype C in Kinshasa between 1997 and 2002,” Journal of Acquired Immune Deficiency Syndromes, Vol. 40, No. 4, 2005, pp. 456-462. doi:10.1097/01.qai.0000159670.18326.94
- S. Bertagnolio and D. Sutherland, “WHO Approach to Track HIV Drug Resistance Emergence and Transmission in Countries Scaling up HIV Treatment,” AIDS, Vol. 19, No. 12, 2005, pp. 1329-1330. doi:10.1097/01.aids.0000180107.19006.02
- S. Toure, B. Kouadio, C. Seyler, M. Traore, N. DakouryDogbo, J. Duvignac, N. Diakite, S. Karcher, C. Grundmann, R. Marlink, F. Dabis, X. Anglaret and Aconda Study Group, “Rapid Scaling-Up of Antiretroviral Therapy in 10,000 Adults in Cote d’ivoire: 2-Year Outcomes and Determinants,” AIDS, Vol. 22, No. 7, 2008, pp. 873-882. doi:10.1097/QAD.0b013e3282f768f8
- A. Ayouba, T. T. Lien, J. Nouhin, L. Vergne, A. F. Aghokeng, N. Ngo-Giang-Huong, H. Diop, C. T. Kane, D. Valéa, F. Rouet, D. Joulia-Ekaza, T. D. Toni, E. Nerrienet, E. M. Ngole, E. Delaporte, D. Costagliola, M. Peeters and M. L. Chaix, “Low Prevalence of HIV Type 1 Drug Resistance Mutations in Untreated, Recently Infected Patients from Burkina Faso, Côte d’Ivoire, Senegal, Thailand, and Vietnam: The ANRS 12134 Study,” AIDS Research Human Retroviruses, Vol. 25, No. 11, 2009, pp. 1193-1196. doi:10.1089/aid.2009.0142
- L. Vergne, S. Diagbouga, C. Kouanfack, A. Aghokeng, C. Butel, C. Laurent, N. Noumssi, M. Tardy, A. Sawadogo, J. Drabo, H. Hien, L. Zekeng, E. Delaporte and M. Peeters, “HIV-1 Drug-Resistance Mutations among Newly Diagnosed Patients before Scaling-Up Programmes in Burkina Faso and Cameroon,” Antiviral Therapy, Vol. 11, No. 5, 2006, pp. 575-579.
- J. D. Thompson, D. G. Higgins and T. J. Gibson, “CLUSTAL W: Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice,” Nucleic Acids Research, Vol. 22, No. 22, 1994, pp. 4673-4680. doi:10.1093/nar/22.22.4673
- T. Toni, B. Masquelier, A. Minga, X. Anglaret, C. Danel, A. Coulibaly, H. Chenal, F. Dabis, R. Salamon, H. J. Fleury and Primo-CI ANRS 1220 Study Group, “HIV-1 Antiretroviral Drug Resistance in Recently Infected Patients in Abidjan, Côte d’Ivoire: A 4-Year Survey, 2002- 2006,” AIDS Research and Human Retroviruses, Vol. 23, No. 9, 2007, pp. 1155-1160. doi:10.1089/aid.2007.0072
- D. A. Yaotsè, V. Nicole, N. F. Roch, P. D. Mireille, D. Eric and P. Martine, “Genetic Characterization of HIV-1 Strains in Togo Reveals a High Genetic Complexity and Genotypic Drug-Resistance Mutations in ARV Naive Patients,” Infection, Genetic and Evolution, Vol. 9, No. 4, 2009, pp. 646-652. doi:10.1016/j.meegid.2009.04.002
- J. L. Sankalé, S. Langevin, G. Odaibo, S. T. Meloni, A. I. Ojesina, D. Olaleye and P. Kanki, “The Complexity of Circulating HIV Type 1 Strains in Oyo State, Nigeria,” AIDS Research Human Retroviruses, Vol. 23, No. 8, 2007, pp. 1020-1025. doi:10.1089/aid.2006.0304
- B. D. Preston, B. J. Poiesz and L. A. Loeb, “Fidelity of HIV-1 Reverse Transcriptase,” Science, Vol. 242, No. 4882, 1988, pp. 1168-1171. doi:10.1126/science.2460924
- M. S. Hirsch, H. F. Günthard, J. M. Schapiro, F. BrunVézinet, B. Clotet, S. M. Hammer, V. A. Johnson, D. R. Kuritzkes, J. W. Mellors, D. Pillay, P. G. Yeni, D. M. Jacobsen and D. D. Richman, “Antiretroviral Drug Resistance Testing in Adults Infected with Human Immunodeficiency Virus Type 1: 2008 Recommendations of an International AIDS Society-USA Panel,” Clinical Infections Disease, Vol. 47, No. 2, 2008, pp. 266-285. doi:10.1086/589297
- M. Tshabalala, J. Manasa, L. S. Zijenah, S. Rusakaniko, G. Kadzirange, M. Mucheche, S. Kassaye, E. Johnston and D. Katzenstein, “Surveillance of Transmitted Antiretroviral Drug Resistance among HIV-1 Infected Women Attending Antenatal Clinics in Chitungwiza, Zimbabwe,” PLoS One, Vol. 6, No. 6. 2011, p. e21241.
- H. Diop-Ndiaye, C. Toure-Kane, N. Leye, N. F. NgomGueye, C. Montavon, M. Peeters and S. Mboup, “Antiretroviral Drug Resistance Mutations in AntiretroviralNaive Patients from Senegal,” AIDS Research and Human Retroviruses, Vol. 26, No. 10, 2010, pp. 1133-1138. doi:10.1089/aid.2009.0295
- R. L. Hamers, M. Siwale, C. L. Wallis, M. Labib, R. Van Hasselt, W. S. Stevens, R. Schuurman, A. M. Wensing, M. Van Vugt, T. F. Rinke De Wit and PharmAccess African Studies to Evaluate Resistance, “HIV-1 Drug Resistance Mutations Are Present in Six Percent of Persons Initiating Antiretroviral Therapy in Lusaka, Zambia,” Journal of Acquired Immune Deficiency Syndromes, Vol. 55, No. 1, 2010, pp. 95-101. doi:10.1097/QAI.0b013e3181e544e0
- D. Pieniazek, M. Rayfield, D. J. Hu, J. Nkengasong, S. Z. Wiktor, R. Downing, B. Biryahwaho, T. Mastro, A. Tanuri, V. Soriano, R. Lal and T. Dondero, “Protease sequences from HIV-1 Group M Subtypes A-H Reveal Distinct Amino Acid Mutation Patterns Associated with Protease Resistance in Protease Inhibitor-Naive Individuals Worldwide. HIV Variant Working Group,” AIDS, Vol. 14, No. 1, 2000, pp. 1489-1495. doi:10.1097/00002030-200007280-00004
- L. Vergne, M. Peeters, E. Mpoudi-Ngole A. Bourgeois, F. Liegeois, C. Toure-Kane, S. Mboup, C. Mulanga-Kabeya, E. Saman, J. Jourdan, J. Reynes and E. Delaporte, “Genetic Diversity of Protease and Reverse Transcriptase Sequences in Non-Subtype-B Human Immunodeficiency Virus Type 1 Strains: Evidence of Many Minor Drug Resistance Mutations in Treatment-Naive Patients,” Journal of Clinical Microbiology, Vol. 38, No. 11, 2000, pp. 3919-3925.
- J. D. Church, S. E. Hudelson, L. A. Guay, S. Chen, D. R. Hoover, N. Parkin, S. A. Fiscus, F. Mmiro, P. Musoke, N. Kumwenda, J. B. Jackson, T. E. Taha and S. H. Eshleman, “HIV Type 1 Variant with Nevirapine Resistance Mutations Are Rarely Detected in Antiretroviral Drug-Naive African Women with Subtypes A, C and D,” AIDS Research and Human Retroviruses, Vol. 23, No. 6, 2007, pp. 764-768. doi:10.1089/aid.2006.0272
- B. T. Røge, T. L. Katzenstein, N. Obel, H. Nielsen, O. Kirk, C. Pedersen, L. Mathiesen, J. Lundgren and J. Gerstoft, “K65R with and without S68: A New Resistance Profile in Vivo Detected in Most Patients Failing Abacavir, Didanosine and Stavudine,” Antiviral Therapy, Vol. 8, No. 2, 2003, pp. 173-182.
- L. Romano, G. Venturi, S. Bloor, R. Harrigan, B. A. Larder, J. C. Major and M. Zazzi, “Broad Nucleoside-Analogue Resistance Implications for Human Immunodeficiency Virus Type 1 Reverse-Transcriptase Mutations at Codons 44 and 118,” Journal of Infections Diseases, Vol. 185, No. 7, 2002, pp. 898-904. doi:10.1086/339706
- M. Girouard, K. Diallo, B. Marchand, S. McCormick and M. Götte, “Mutations E44D and V118I in the Reverse Transcriptase of HIV-1 Play Distinct Mechanistic Roles in Dual Resistance to AZT and 3TC,” Journal of Biological Chemistry, Vol. 278, No. 36, 2003, pp. 34403- 34410. doi:10.1074/jbc.M303528200
- E. Domingo and J. J. Holland, “Mutation Rates and Rapid Evolution of RNA Viruses,” In: S. S. Morse, Ed., The Evolutionary Biology of Viruses, Raven Press, New York, 1994, pp. 161-184.
- W. M. Nadembega, S. Giannella, J. Simpore, F. CeccheriniSilberstein, V. Pietra, A. Bertoli, S. Pignatelli, M. C. Bellocchi, J. B. Nikiema, G. Cappelli, A. Bere, V. Colizzi, C. P. Perno and S. Musumeci, “Characterization of DrugResistance Mutations in HIV-1 Isolates from Non-HAART and HAART Treated Patients in Burkina Faso,” Journal of Medical Virology, Vol. 78, No. 11, 2006, pp. 1385- 1391. doi:10.1002/jmv.20709
- R. M. Viani, K. Hsia, P. Hubbard, J. Ruiz-Calderon, R. Lozada, J. Alvelais, M. Gallardo and S. A. Spector, “Prevalence of Primary HIV-1 Drug Resistance in Pregnant Women and in Newly Diagnosed Adults at Tijuana General Hospital, Baja California, Mexico,” International Journal of STD & AIDS, Vol. 18, No. 4, 2007, pp. 235- 238. doi:10.1258/095646207780658962
- J. G. García-Lerma, P. J. Gerrish, A. C. Wright, S. H. Qari and H. Walid, “Evidence of a Role for the Q151L Mutation and the Viral Background in Development of Multiple Dideoxynucleoside-Resistant Human Immunodeficiency Virus Type 1,” Journal of Virology, Vol. 74, No. 20, 2000, pp. 9339-9346. doi:10.1128/JVI.74.20.9339-9346.2000
- V. Miller, M. Ait-Khaled, C. Stone, P. Griffin, D. Mesogiti, A. Cutrell, R. Harrigan, S. Staszewski, C. Katlama, G. Pearce and M. Tisdale, “HIV-1 Reverse Transcriptase (RT) Genotype and Susceptibility to RT Inhibitors during Abacavir Monotherapy and Combination Therapy,” AIDS, Vol. 14, No. 2, 2000, pp. 163-171. doi:10.1097/00002030-200001280-00012
NOTES
*Sequence data: The PR and RT sequences are available in GenBank.
Competing interests: The authors declare that they have no competing interests.
#Corresponding author.

