Advances in Enzyme Research
Vol.1 No.3(2013), Article ID:36964,9 pages DOI:10.4236/aer.2013.13006
Kinetic studies on recombinant stem bromelain
![]()
1Bioprocess and Molecular Engineering Research Unit, Department of Biotechnology Engineering, International Islamic University Malaysia, Kuala Lumpur, Malaysia; *Corresponding Author: hamzah@iium.edu.my
2Department of Biochemistry, Bayero University, Kano, Nigeria
3International Institute for Halal Research & Training (INHART) International Islamic University Malaysia, Kuala Lumpur, Malaysia
Copyright © 2013 Muntari Bala et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received 22 May 2013; revised 30 June 2013; accepted 8 July 2013
Keywords: Protease; Recombinant Bromelain; Substrates; Inhibitors; Activators
ABSTRACT
Stem bromelain is a plant thiol protease with several industrial and therapeutic applications. This current work presents kinetic studies of recombinant bromelain (recBM) expressed in Escherichia coli BL21-AI on four synthetic substrates, N-α-carbobenzoxy-L-alanyl-p-nitrophenyl ester (ZANPE), N-α-carbobenzoxy-L-arginyl-L-arginine-p-nitroanilide (ZAANA), N-α-carbobenzoxy-L-phenylalanyl-L-valyl-L-arginine-p-nitroanilide (ZPVANA) and L-pyroglutamyl-L-phenylalanyl-L-leucine-p-nitroanilide (PFLNA). Hydrolytic activities of recBM at various pH and temperature conditions were compared to that of commercial bromelain (cBM). Both enzymes demonstrated high activities at 45˚C and pH 5 - 8 for recBM and pH 6 - 8 for cBM. recBM showed marginally lower Km and slightly higher kcat/Km for ZAANA, ZANPE and ZPVANA in comparison to cBM. trans-Epoxysuccinyl-L-leucylamido {4- guanidino} butane (E-64) severely affected recBM and cBM hydrolysis of the synthetic substrates by competitive inhibition with Ki values of 3.6 - 5.1 µM and 5.5 - 6.9 µM for recBM and cBM, respectively. The evaluated properties of recBM including temperature and pH optima, substrate specificity and sensitivity to inhibitors or activators, satisfy the requisites required for food industries.
1. INTRODUCTION
Proteases are very significant industrial enzymes because they represent about 60% of all commercial enzymes worldwide. They are widely used in food, pharmaceutical and detergent industries [1]. Plant proteases have been gaining unique attention in the field of biotechnology and medicine due to their exploitable properties. The most recognized plant proteases with greater commercial values are papain from Carica papaya, ficin from Ficus spp. and bromelain from Ananas comosus [2].
Bromelain is a crude, aqueous extract from the stems and fruits of pineapples (Ananas comosus) derived from Bromeliaceae family. It contains a mixture of different proteases as well as phosphatase, glucosidases, peroxidases, cellulases and glycoproteins [3]. Stem bromelain (EC 3. 4. 22. 32) is the major protease present in extracts of pineapple stem while fruit bromelain (EC 3. 4. 22. 33) is the major enzyme present in pineapple fruit juice [3]. Some other minor thiol endopeptidases—ananain and comosain—are also present in the pineapple stem bromelain. All commercially available bromelain are derived from the stem. Stem bromelain is activated by cysteine while hydrogen sulphide and sodium cyanide are less effective [4]. The enzyme is inhibited by heavy metals such as mercury and silver as well as trans-epoxysuccinyl-L-leucylamido {4-guanidino} butane, commonly known as E-64.
Bromelain has numerous therapeutic, industrial and other applications. It has been widely used in food industry for baking processes, meat tenderization, clarification of beer, as food supplement and in prevention of browning of apple juice [5]. Additionally, it is used as active ingredient to provide mild peeling effects in cosmetic industries [6]. Furthermore, it has also been used in leather industries for skin pre-tanning, softening and bating [7]. In textile industries, bromelain is used for improving the dyeing qualities of protein fibers, decomposing or partially solubilizing protein fiber from silk and wool [8]. Similarly, bromelain is used as hydrolyzing agent for the release of antimicrobial peptides of leatherjacket’s insoluble proteins [9].
Stem bromelain is a highly accepted phytotherapeutic agent. It has anti-tumor and anti-inflammatory effects [4]. In addition, it exerts several inhibitory effects on platelet aggregation, bronchitis, angina pectoris, surgical traumas, sinusitis, thrombophlebitis and pyelonephritis. Moreover, it enhances absorption of drugs, especially antibiotics [10]. The advances in molecular genetics and genetic engineering in the last decades have made it possible to clone and express virtually any gene into a suitable microbial host, so that new enzymes from other microorganisms and also from higher organisms can be produced in convenient microbial hosts like bacteria, yeasts and fungi [11].
Considering the numerous applications of stem bromelain, it is imperative to produce the enzyme in recombinant form so as to achieve improved purification, biochemical characterization and formulation. In that regard, we have reported the cloning and expression of recombinant stem bromelain (recBM) [12], recovery from soluble and insoluble enzyme forms [13], lab-scale optimization of some of its culture cultivation conditions [14] and the enzyme thermal and storage stability [15]. The purpose of the current research is to further characterize the enzyme by focusing on the kinetic studies of recBM using four synthetic substrates; N-α-carbobenzoxy-L-phenylalanyl-L-valyl-L-arginine-p-nitroanilide (ZPVANA), N-α-carbobenzoxy-L-alanyl-p-nitrophenyl ester (ZANPE), N-α-carbobenzoxy-L-arginyl-L-arginine-p-nitroanilide
(ZAANA) and L-pyroglutamyl-L-phenylalanyl-L-leucine-p-nitroanilide (PFLNA). The results are then compared to that of commercial bromelain (cBM) as control.
2. MATERIALS AND METHODS
2.1. Reagents
Synthetic amide and peptide substrates, N-α-carbobenzoxy-L-alanyl-p-nitrophenyl ester (ZANPE), N-α-carbobenzoxy-L-arginyl-L-arginine-p-nitroanilide (ZAANA), N-α-carbobenzoxy-L-phenylalanyl-L-valyl-L-arginine-pnitroanilide (ZPVANA) and L-pyroglutamyl-L-phenylalanyl-L-leucine-p-nitroanilide (PFLNA) were purchased from Bachem, Germany. Luria Bertani (LB) was a product of Merck, Germany. A single lot (Ref. No. 61391) of stem bromelain (EC3. 4. 22. 32) was purchased from MP Biomedicals, LLC (France). Coomassie brilliant blue R-250, acrylamide, bisacrylamide and low-range molecular weight markers were obtained from Bio-Rad (Hercules, CA). All other reagents were purchased from Sigma (St. Louis, MO).
2.2. Microbial Strain
The Escherichia coli BL21-AI strain (Invitrogen, USA) harboring stem bromelain gene used in this study was as described in our earlier study [12]. Specifically, the gene encoding stem bromelain was initially amplified from pineapple’s stem cloned into pENTR/TEV/D-TOPO before being sub-cloned into the expression vector pDEST17 (Invitrogen, USA). The expression vector containing stem bromelain gene was then transformed into E. coli BL21-AI competent cells.
2.3. Enzyme Expression
Recombinant bromelain expression was conducted as described in our earlier studies [12-14]. E. coli BL21-AI cells harboring recombinant bromelain gene were grown in shake flasks overnight in LB media containing 100 μg/ml ampicillin. The overnight culture was then diluted 50 folds in a fresh LB media and was grown in a 2 liter bioreactor at 27˚C, air flow rate of 1.0 vvm with 250 rpm agitation until a cell density of OD600 nm = 0.6 was reached. This was followed by addition of L-arabinose (0.15% w/v, final concentration) and the induction was allowed to continue for 8 hours. Cells were harvested from the spent media by centrifugation (6000 × g) at 4˚C for 20 min and stored at −20˚C for further use. All the experiments were performed in triplicates.
2.4. Enzyme Purification
The harvested cells were subjected to sonication (sonicator, 150 v/t model, Biologics, Inc. USA) on ice using 6 - 10 sec burst, with 10 sec interval at high amplitude. This was followed by centrifugation (8000 × g) at 4˚C, for 30 min and the supernatant was collected and purified by AKTA purifier FPLC system (GE Healthcare BioSciences, USA) attached with a glass column (0.7 × 10 cm, Bio-Rad, USA) that is packed with 4 ml Ni-NTA His•bind resin (Novagen, Germany). Purification of recBM was conducted in accordance to the manufacturer’s instructions. The supernatant (10 ml) was loaded onto a pre-equilibrated Ni-NTA His•bind column in accordance to the manufacturer’s protocol. The FPLC system was set at a flow rate of 1 ml/min throughout the operation. Washing step was achieved by using a wash buffer (50 mM NaH2PO4 pH 8, 300 mM NaCl, 20 mM imidazole) while purified recBM was recovered using an elution buffer (50 mM NaH2PO4 pH 8, 300 mM NaCl and 500 mM imidazole).
2.4.1. SDS-PAGE and Western Blot
After each step of enzyme recovery and purification, the protein fractions were tested by SDS-PAGE in 12.5% polyacrylamide gels as described earlier [12]. Visualization was conducted by Coomassie staining [16].
The protein bands on the gel were transferred to polyvinylidene fluoride (PVDF) membrane (Bio-Rad Laboratory, Inc., USA) using Mini-Trans-Blot system (BioRad Laboratory, Inc., USA). The membrane was incubated at 4˚C (for one hour) with 5% dried milk powder in TBST buffer (0.1 M Tris-HCl, pH 7.4, 0.9% NaCl, and 0.1% Tween 20) so as to block non-specific binding. This was then followed by incubation with primary antibody (Anti-His Tag, clone HIS. H8, Millipore) in a ratio of 1:1000. The membrane was subsequently incubated with secondary antibody (Goat Anti-rabbit IgG (H + L)- HRP) conjugate, Bio-Rad Laboratory, Inc., USA) at the same ratio as described above. The membrane was later incubated with BCIP (5-bromo-4-chloro-3-indolyl phosphate) color development solution for 4 hours. Finally, the PVDF membrane was air died and the images of the Western blot were visualized and analyzed using AlphaImager software (Alpha Innotech, USA).
2.5. Determination of Protein Concentration
The protein concentration of the purified recombinant bromelain was estimated by Bradford’s method [17].
2.6. Bromelain Activity towards Synthetic Substrates at Various pH and Temperature
Bromelain hydrolytic activity was assessed against all the synthetic substrates listed earlier so as to determine the maximum activity of the enzyme under optimum pH conditions. The enzyme activity was further assessed on the substrate that yielded the highest bromelain hydrolytic activity for determination of optimum temperature condition. The results obtained were then used for indepth kinetic studies. This is further elaborated in the following sections.
2.7. Enzymatic Activity Measurement
2.7.1. Determination of Bromelain Activity at Different pH Values
The bromelain activity against the four synthetic substrates was determined over a pH range of 2 - 12 in the following 0.1 M buffer systems: glycine/HCl (pH 2 - 4.), sodium phosphate (pH 5 - 8), Tris-HCl (pH 8 - 10) and sodium bicarbonate (pH 10 - 12). Prior to the addition of the substrate, 0.3 ml of bromelain preparations were preincubated with 1.4 ml of each of the buffer solution (containing 4 mM EDTA and 8 mM DTT) at 45˚C for 5 min. This was followed by the addition of 0.3 ml substrate solution into the mixture and incubated for further 5 min. For ZANPE, changes in the absorbance were measured at 405 nm for the p-nitrophenol produced [18]. On the on the hand, for the remaining three substrates (ZAANA, ZPVANA and PFLNA) changes in the absorbance were measured at 410 nm for the p-nitroaniline generated [19,20].
2.7.2. Determination of Bromelain Activity at Different Temperature
The optimum temperature of recBM and cBM was determined by using ZPVANA as the substrate under its determined optimum pH. The assay was carried out in 0.1 M sodium phosphate buffer (pH 8) as described above for this substrate (2.7.1) at various temperature range of 4˚C to 75˚C.
2.8. Determination of Kinetic Parameters
Kinetic parameters (kcat, Km and Vmax) were determined on ZANPE, ZAANA, ZPVANA and PFLNA substrates. The concentrations of substrates used ranged from 0.1 to 0.6 mM in the reaction mixture. From the product concentrations as a function of time curves, instantaneous rates were calculated at several different substrate concentrations under described conditions. Kinetic parameters of hydrolysis by bromelain were calculated using linear regression analysis by means of Lineweaver-Burk plots.
2.9. Inhibition Kinetics of E-64 on Bromelain
Ki determinations were carried out by lowering the enzyme and inhibitor concentrations to obtain a nonlinearity of dose-response curves. Bromelain solution was pre-incubated in an activation buffer with increasing concentrations of E-64 inhibitor (1 to 5 µM) for 10 min at 45˚C. Residual activity of the enzyme was measured on the four different synthetic substrates as described above (section 2.7.1). Control reactions in the absence of inhibitor were also carried out. Inhibitory constants (Ki) were determined as the intersection on the x-axis of the secondary plots of the slopes (Km/Vmax) obtained from the Lineweaver-Burk lines against E-64 concentrations. All experiments were conducted in triplicate.
3. RESULTS AND DISCUSSION
3.1. SDS-PAGE and Western Blot
Following cell lysis and removal of cell debris, a single purification step involving Ni-NTA His•bind column chromatography afforded highly purified recBM (Figure 1). A single homogeneous protein band of approximately 43 kDa was observed on SDS-polyacrylamide gel and Western blot analysis also indicated a single labeled protein of identical molecular size. The observed size of 43 kDa for recBM is greater than ~25 kDa of mature stem bromelain due to the presence of signal peptide, propep-
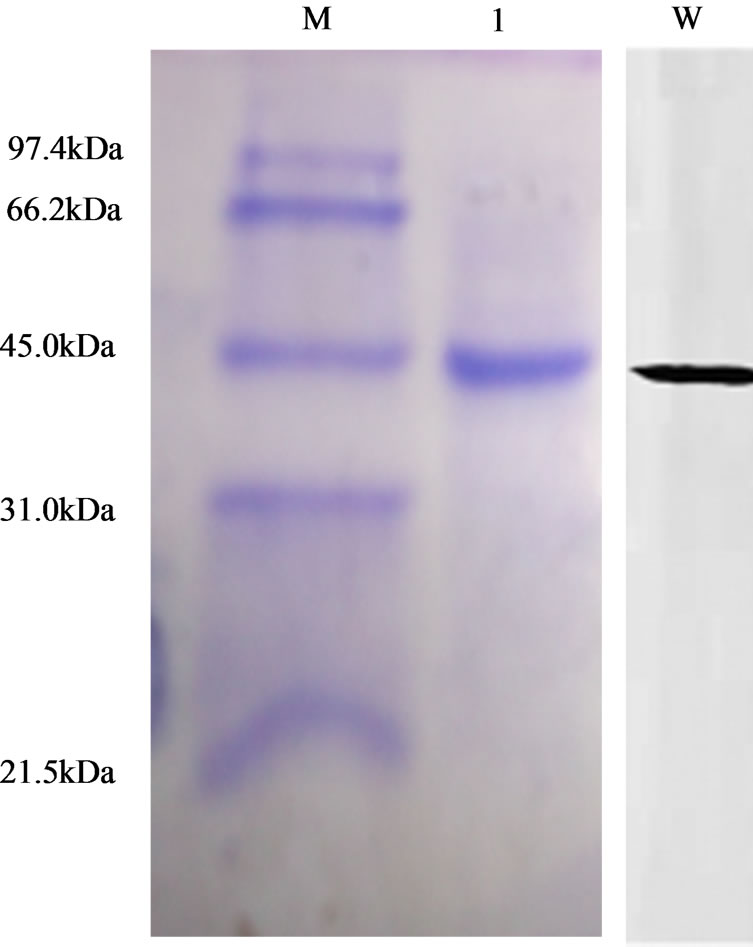
Figure 1. SDS-PAGE and Western blot analysis of the recombinant bromelain (recBM) expressed in E. coli BL21-AI. SDSPAGE and Western blot showing purified soluble recBM. Lane M: protein molecular weight marker; lane 1: purified soluble recBM, and lane W: Western blot.
tide and additional amino acids generated from His6x tag that was introduced during the cloning of the bromelain gene [12].
3.2. Activity of Bromelain at Various pH and Temperature
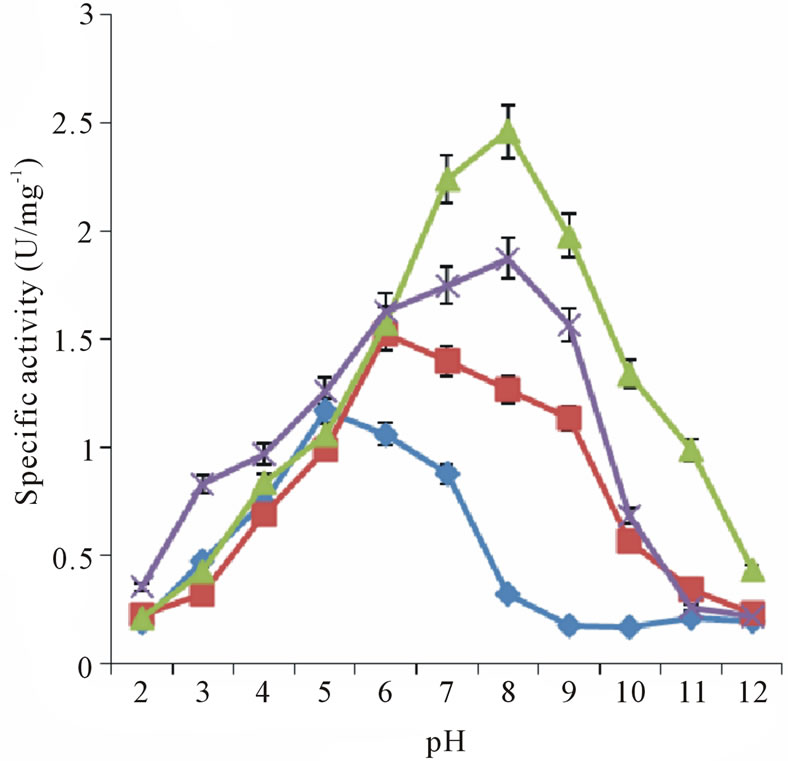
Stem bromelain is known to have broad substrate specificity and hydrolyzes a large number of natural and synthetic substrates [4]. Thus, bromelain activity can be assayed using synthetic substrates. The activities of recBM were compared with those of cBM with a variety of synthetic substrates at different pH values (2 - 12). The results are shown in Table 1, Figures 2(a) and 2(b). It can be observed that the pH optimum ranges for recBM and cBM were found to be 5 - 8 and 6 - 8, respectively. Most importantly, both enzymes have maximum activities at optimum temperature of 45˚C (Figure 2(c)). This implied that the recBM has slight broader pH
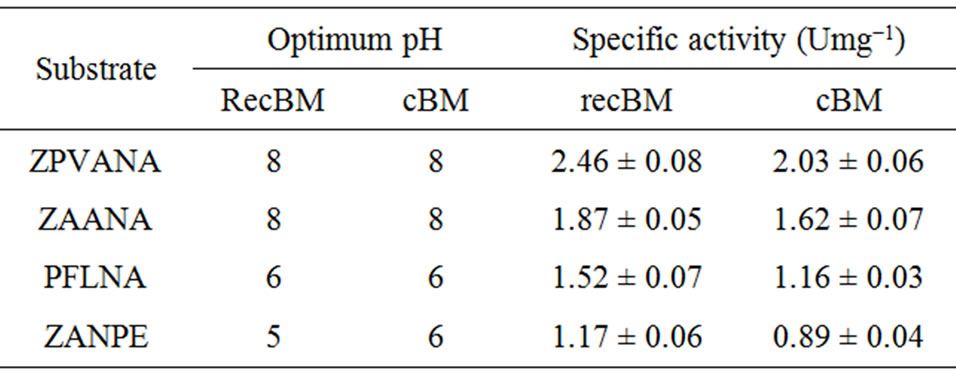
Table 1. Specific activities of bromelains towards four synthetic substrates measured at different pH values.
 (a)
(a)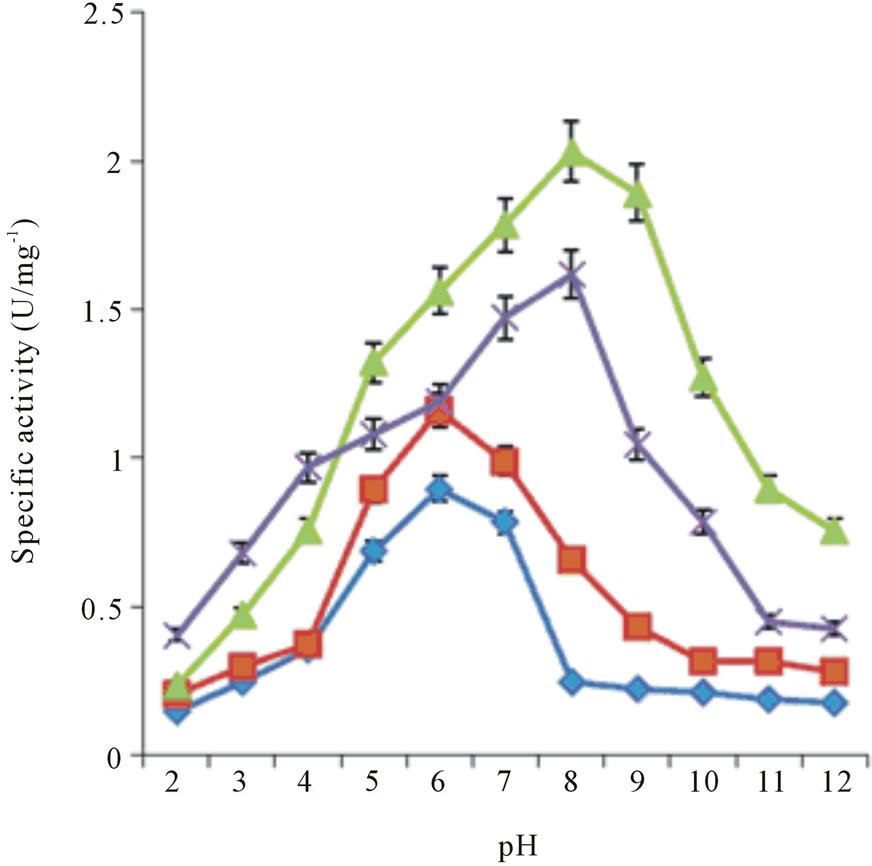 (b)
(b)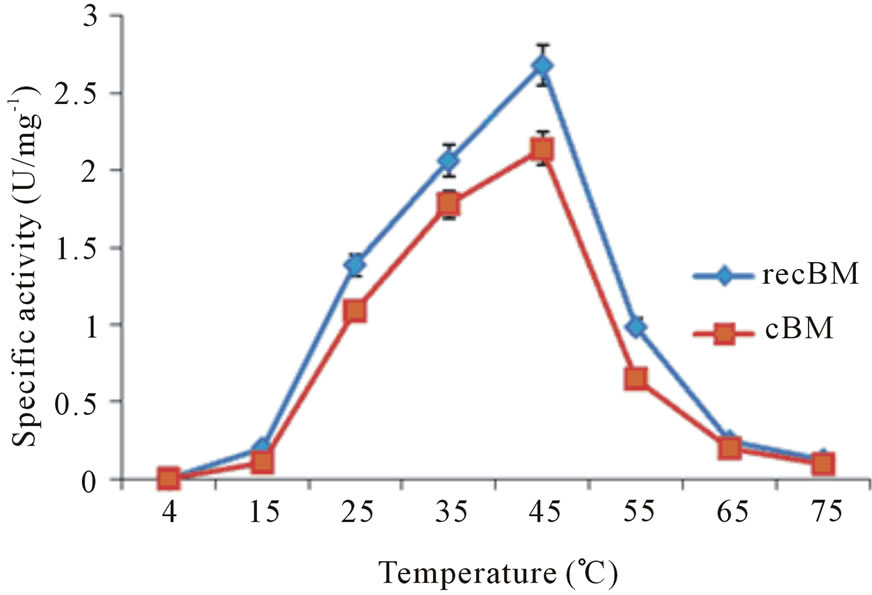 (C)
(C)
Figure 2. Specific activity of bromelains at different temperature and pH: (a) recBM and (b) cBM activities at different pH measured on four synthetic substrates: ZANPE (♦), ZPVANA (▲), ZAANA (×) and PFLNA (■). (c) recBM and cBM activities against temperature measured on ZPVANA.
range than cBM. The activities of recBM and cBM started to decline drastically beyond pH 8 and the enzymes were almost completely inactivated at pH 12. Moreover, bromelain activity-pH profile obtained is consistent with that of most thiol proteases belonging to the papain family. In pH range of 5 - 8, the enzymes have hydrogen bond between the thiol and imidazole functional group (Cys-His) that is critical for catalytic activity [21]. The order of susceptibility for the hydrolysis of synthetic substrates and the substrate specificity showed by recBM and cBM are apparent. recBM was found to have markedly higher hydrolytic activities than cBM against all the substrates tested. In addition, both enzymes had broad specificities, with ZPVANA being most susceptible and ZANPE being the least susceptible. The optimum pH of 8 observed for ZPVANA and ZAANA substrates is similar to that reported by Benucci, et al. [22] on stem bromelain. However, the authors obtained higher specific activities with ZAANA instead of ZPVANA. Besides, our study yielded higher bromelain specificity on these substrates (ZAANA and ZPVANA). The observed variations might be due to the differences in incubation temperature and the type of buffer used as their experiments were conducted at 25˚C in McIlvaine buffer.
3.3. Kinetic Parameters Estimations
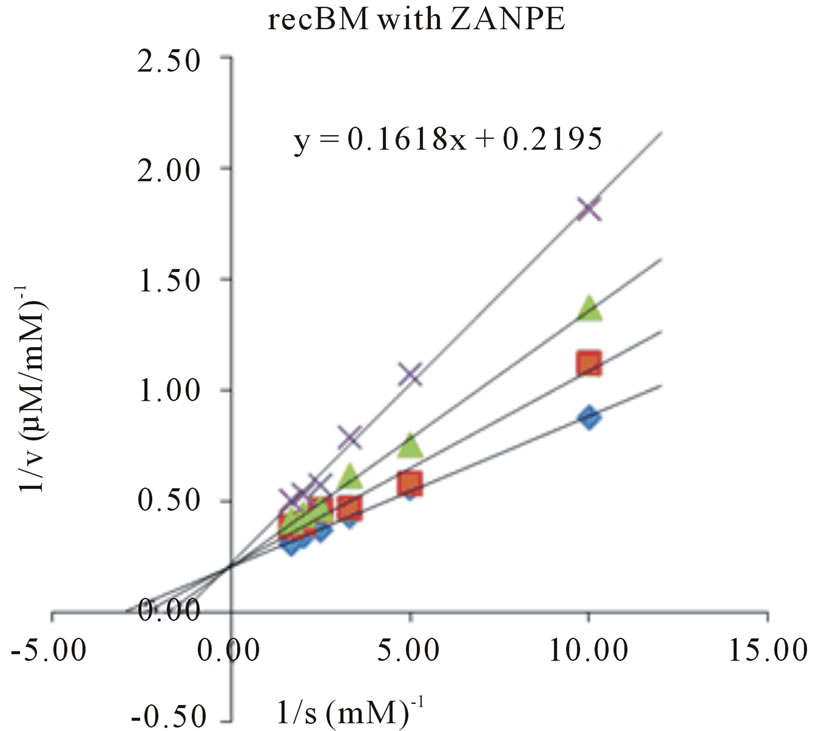
Determination of kinetic parameters of an enzyme is an essential tool in elucidating the entire catalytic process. The Michaelis-Menten constant (Km), maximum reaction velocity (Vmax), turnover number (kcat) and kcat/ Km for all the four synthetic substrates were determined by plotting the activity data obtained under pH and temperature optima, as a function of substrate concentration in Lineweaver-Burk plots (Figures 3 and 4). The Km
 (a)
(a) (b)
(b)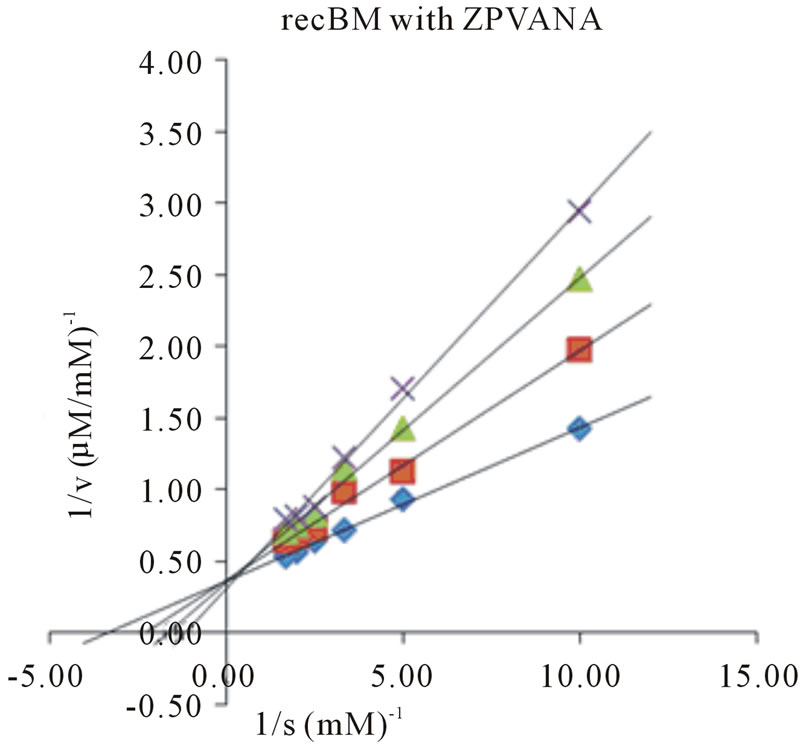 (c)
(c) (d)
(d)
Figure 3. Lineweaver-Burk plots of recBM in the absence and presence of E-64. Four different synthetic substrates were used: (a) ZANPE (b) ZAANA (c) ZPVANA and (d) PFLNA. Measurement was made at different E-64 concentrations (0 - 5 µM): control, 0 µM (♦), 1 µM (■), 3 µM (▲) and 5 µM (X).
 (a)
(a) (b)
(b)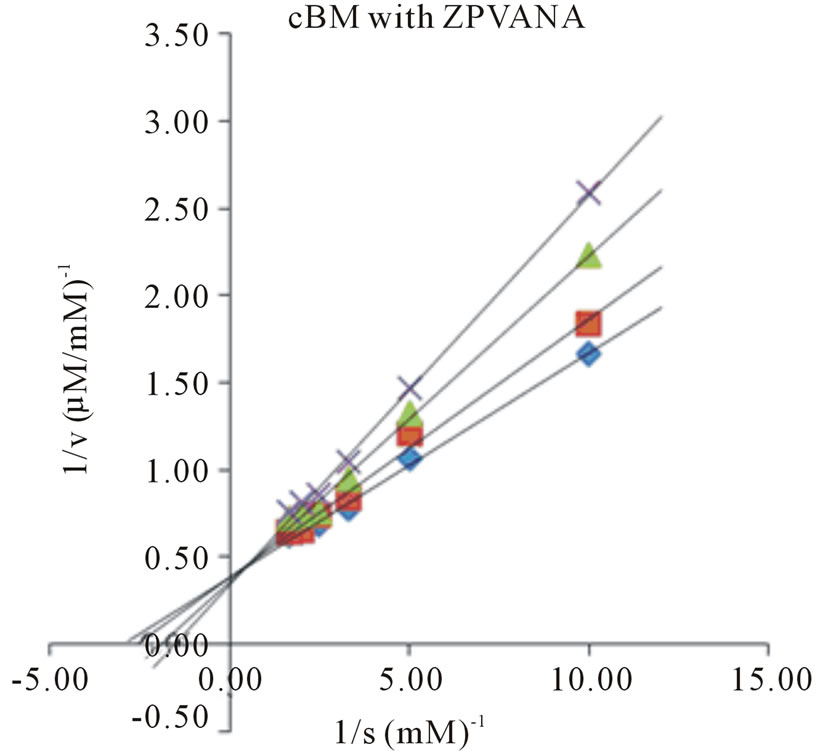 (c)
(c)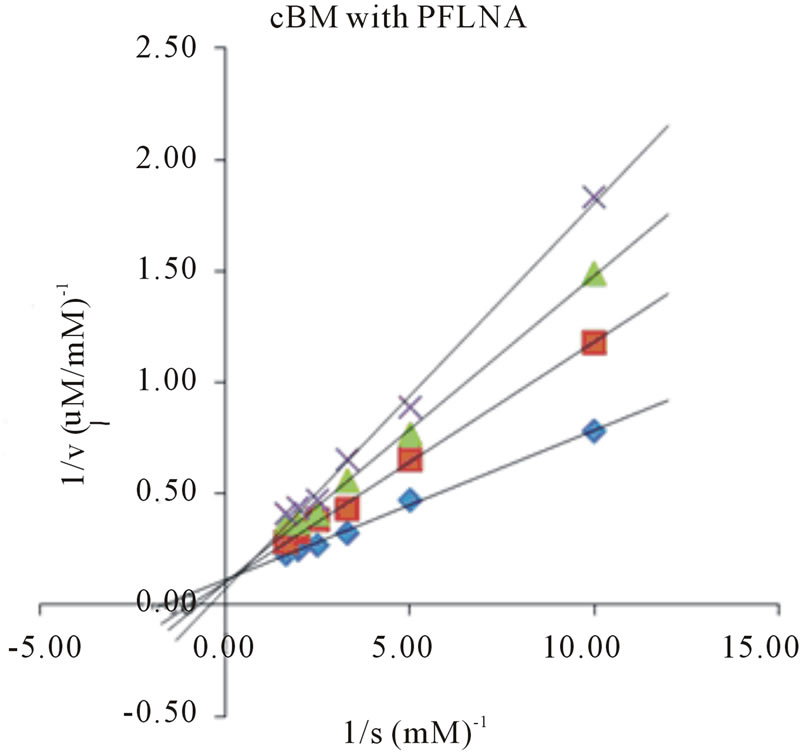 (d)
(d)
Figure 4. Lineweaver-Burk plots of cBM in the absence and presence of E-64. Four different synthetic substrates were used: (a) ZANPE (b) ZAANA (c) ZPVANA and (d) PFLNA. Measurement was made at different E-64 concentrations (0 - 5 µM): control, 0 µM (♦), 1 µM (■), 3 µM (▲) and 5 µM (X).
values varied significantly for both bromelains with recBM having the lowest Km values (0.29 - 0.8 mM) in all the substrates studied (with the exemption of PFLNA) as compared to cBM (0.34 - 0.68 mM) (Table 2). The lowest Km values for both cBM and recBM are recorded on ZPVANA (0.34 & 0.29 mM), while higher values were seen on PFLNA (0.60 & 0.80 mM) and ZANPE (0.68 & 0.34 mM), respectively. The kcat/Km ratio sometimes called the specificity constant is the best way to compare the catalytic efficiency of enzyme [23]. PFLNA was found to have the highest kcat/Km (49.57 & 52.53 mM−1s−1) as such this substrate is highly recommended for the determination of the bromelain activity at optimal experimental conditions. PFLNA has been a classic substrate for thiol endopeptidases [20].
Marginal increases in kcat/Km values (28.41 - 52.53 mM−1s−1) in all the 4 substrates were found for recBM as compared to kcat/Km values (17.86 - 49.57 mM−1s−1) for cBM. This suggests that recBM exhibited similar to slightly better catalytic efficiency than cBM towards the studied substrates. Hence, there are only minor but noticeable differences in the specificity of recBM and cBM to the substrates. The data obtained in this study is comparable to the kinetic data reported for cysteine proteases including stem bromelain [20,24] granulosain [21], ascepalin [25], hieronymain II [26] and penduliforain I [27]. Based on kinetic data obtained in this study, it can be inferred that the kinetic behavior of recBM (containing His6x tag, signal and propeptide) is not much different to that of mature cBM.
3.4. Determination of Inhibitory Constant (Ki)
In this study, the E-64 concentration was held at different constant values (less than that for complete inacti-
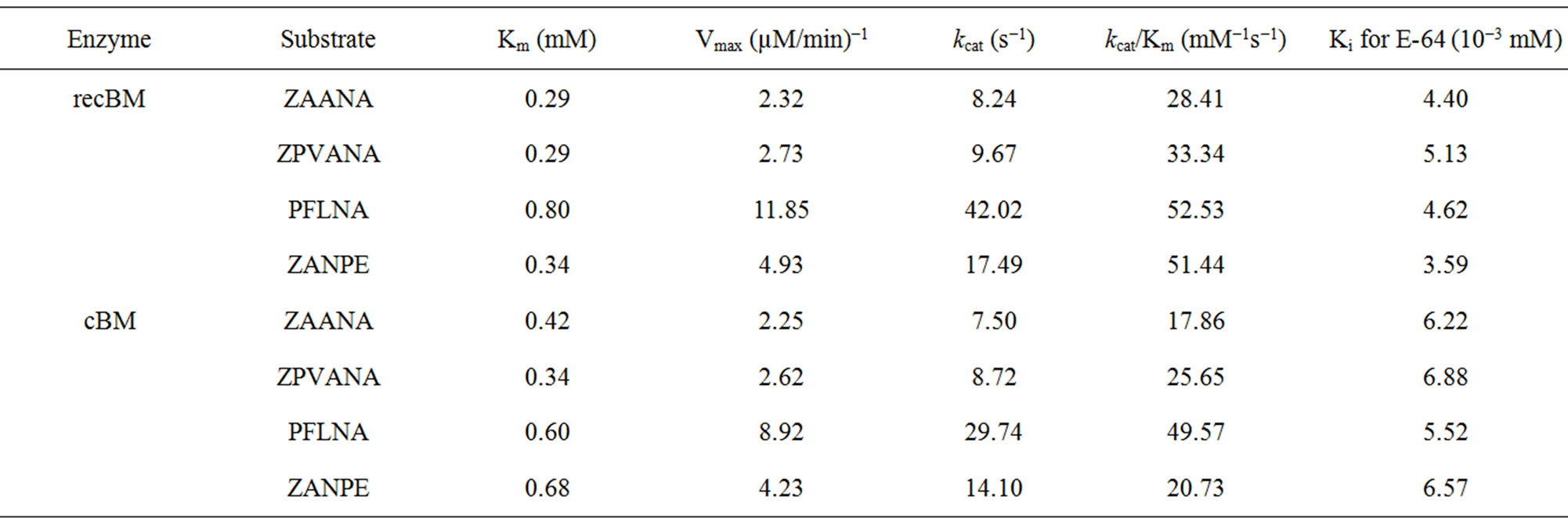
Table 2. Kinetic parameters obtained for bromelains towards synthetic substrates and inhibitor.
vation), while the substrate concentrations were varied, permitting measurement of the effect of increasing substrate concentration on the initial reaction rate. Lineweaver-Burk plots yielded coinciding intercepts on the reciprocal initial reaction rate axis (Figures 3 and 4). The secondary replot of the slope (Vmax/Km) against E-64 concentrations was linearly fitted suggesting that bromelain has a single inhibition site for E-64. The results indicated that the value of Vmax remained the same and the value of Km increased with increasing E-64 inhibitor concentrations suggesting competitive inhibition to both recBM and cBM (Table 2). This observation is not unique to bromelain only because E-64 has been reported to competitively inhibit trypsin-like proteinases [28], α-gingivain, a thiol protease from Porphyromonas gingi [29] and clostripain, a thiol protease from Clostridium histolyticum [30].
RecBM recorded lower Ki values (3.59 − 4.40 × 10−3 mM) against all the studied substrates as compared to cBM (5.52 − 6.68 × 10−3 mM) which signifies that recBM has similar or slightly better affinity to E-64 than does cBM, and thus, more sensitive to the inhibitor. In addition, ZPVANA has the highest E-64 Ki values for both enzymes, thus, less competitively inhibited by E-64 as compared to other tested substrates. This suggests the existence of differences in the nature of E-64 inhibitory effects on bromelains which to some extent depends on the type of substrate used.
4. Conclusion
In this study, recombinant bromelain (recBM) had been expressed and successfully purified to apparent homogeneity through a single chromatography purification step. The recBM enzyme showed slightly higher activeties than commercial bromelain (cBM) against all the substrates tested at various pH and temperatures. Both Both enzymes were found to have maximum activities at optimum temperature of 45˚C. The studied inhibitor induced competitive inhibition on bromelains and recBM was more sensitive to the E-64 inhibition. PFLNA was found to have the highest kcat/Km as such this substrate is highly recommended for the determination of the bromelain activity. Furthermore, recBM has more affinities than cBM towards the studied substrates. The recBM containing His6x tag, signal and propeptide used in this study is not much different to the mature cBM.
5. ACKNOWLEDGEMENTS
The authors are most grateful to the Department of Biotechnology Engineering, International Islamic University Malaysia, for providing all the required laboratory facilities to successfully conduct the research work. In addition, we appreciate the financial assistance from the Ministry of Higher Education, Malaysia (FRGS grant no.11-04-0162) granted to Azura Amid and Hamzah Mohd. Salleh.
REFERENCES
- Lucia, F. and Tomas, G.V. (2010) Native and biotechnologically engineered plant proteases with industrial applications. Food and Bioprocess Technology, 4, 1066- 1088.
- Dubey, V.K. and Jagannadham, M.V. (2003) Procerain, a stable cysteine protease from the latex of Calotropis prcera. Phytochemisty, 62, 1057-1071. Hdoi:10.1016/S0031-9422(02)00676-3
- Kelly, G.S. (1996) Bromelain: A literature review and discussion of its therapeutic applications. Alternative Medicine Review, 1, 243-257.
- Maurer, H.R. (2001) Bromelain: Biochemistry, pharmacology and medical uses. Cellular and Molecular Life Sciences, 58, 1234-1245. Hdoi:10.1007/PL00000936H
- Tochi, B.N., Wang, Z., Xu, S.Y. and Zhang, W. (2009) Effect of stem bromelain on the browning of apple juice, American Journal of Food Technology, 10, 1-8.
- Aehle, W. (2007) Enzymes in Industry: Production and Applications. Wiley-VCH/Verlag GmbH and Co., Weinheim, 6.
- Walsh, G. (2002) Protein Biochemistry and Biotechnology. John Wiley and Sons, New York, 312.
- Koh, J., Kang, S.M., Kim, S.J., Cha, M.K. and Kwon, Y.J. (2006) Effect of pineapple protease on the characteristics of protein fibers. Fibers and Polymers, 7, 180-185. Hdoi:10.1007/BF02908264
- Ketnawa, S., Rawdkuen, S. and Chaiwut, P. (2010) Two phase partitioning and collagen hydrolysis of bromelain from pineapple peel Nang Lae cultivar. Biochemical Engineering Journal, 52, 205-211. Hdoi:10.1016/j.bej.2010.08.012
- Yuan, G., Wahlqvist, M.L., He, G., Yang, M. and Li, D. (2006) Natural products and anti-inflammatory activity. Asia Pacific Journal of Clinical Nutrition, 15,143-152.
- Illanes A. (2008) Enzyme Biocatalysis. Springer Science and Business Media V.B., Brazil, 69. Hdoi:10.1007/978-1-4020-8361-7
- Amid, A., Ismail, N., Yusof, F. and Salleh, H.M. (2011) Expression, purification and characterization of a recombinant stem bromelain from Ananas comosus. Process Biochemistry, 46, 2232-2239. Hdoi:10.1016/j.procbio.2011.08.018
- Bala, M., Amid, A., Mel, M., Jami, S. and Salleh, H.M. (2011) Recovery of recombinant bromelain from Escherichia coli BL21-AI. African Journal of Biotechnology, 10, 8829-18832. Hdoi:10.5897/AJB11.2761
- Muntari, B., Salleh, H.M., Amid, A., Mel, M. and Jami, M.S. (2012) Recombinant bromelain production in Escherichia coli: Process optimization in shake flask culture by Response Surface Methodology. AMB Express, 2, 1-9. Hdoi:10.1186/2191-0855-2-12
- Ismail, N. and Amid, A. (2012). Differential scanning calorimetry as tool in observing thermal and storage stability of recombinant bromelain. Food Research International, 19, 727-731.
- Laemmli, U. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 237, 680-685. Hdoi:10.1038/227680a0
- Bradford, M.M. (1976) A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry, 72, 248-254. Hdoi:10.1016/0003-2697(76)90527-3
- Silverstein, R.M. (1974) The assay of the bromelains using N-CBZ-L-lysine-p-nitrophenylester and N-CBZ-Lglycine-p-nitrophenyl ester as substrates. Analytical Biochemistry, 62, 478-484. Hdoi:10.1016/0003-2697(74)90180-8
- Filippova, I., Lysogorskaya, E.N., Oksenoit, E.S., Rudenskaya, G.N., Stepanov, V.M. (1984) L-Pyroglutamyl-lphenylalanyl-l-leucine-p-nitroanilide—A chromogenic substrate for thiol proteinase assay. Analytical Biochemistry, 143, 293-297. Hdoi:10.1016/0003-2697(84)90665-1
- Rowan, A.D. and Buttle, D.J. (1994) Pineapple cysteine endopeptidases. Methods in Enzymology, 244, 555-568. Hdoi:10.1016/0076-6879(94)44040-9
- Valle, D.M., Bruno, M., Laura, L.M.I., Caffini, N.O., Cantera, A.M.B. (2008) Granulosain I, a Cysteine Protease isolated from ripe fruits of solanum granuloso-leprosum (Solanaceae). The Protein Journal, 27, 267-275. Hdoi:10.1007/s10930-008-9133-4
- Benucci, I., Liburdi, K., Garzillo, A.M.V. and Esti, M. (2011) Bromelain from pineapple stem in alcoholic-acidic buffers for wine application. Food Chemistry, 124, 1349- 1353. Hdoi:10.1016/j.foodchem.2010.07.087
- Morcelle, S.R., Trejo, A.S., Canals, F., Avilés, F.X., Priolo, N.S. (2004) Funastrain c II: A cysteine endopeptidase purified from the latex of Funastrum clause. The Protein Journal, 23, 205-215. Hdoi:10.1023/B:JOPC.0000026416.90134.7b
- Napper, A.D., Bennett, P., Borowski, M., Holdridge, M.B., Leonard, M.J.C., Rogers, E.E., Duan, Y., Laursen, R.A., Reinhold, B.S. and Hames, S.L. (1994) Purification and characterization of multiple forms of the pineapplestem-derived cysteine proteinases ananain and comosain. Biochemical Journal, 301, 727-735.
- Liggieri, M., Arribe, C., Trejo, S.A., Canals, F., Avile, F.X. and Priolo, N.S. (2004) Purification and biochemical characterization of Asclepain c I from the latex of Asclepias curassavica L. constanza. The Protein Journal, 23, 403-411. Hdoi:10.1023/B:JOPC.0000039554.18157.69
- M Bruno, M.A., Trejo, S.A., Aviles, X.F., Caffini, N.O. and Lopez, L.M. (2006). Isolation and characterization of Hieronymain II, another peptidase isolated from fruits of Bromelia hieronymi Mez (Bromeliaceae). The Protein Journal, 25, 224-223. Hdoi:10.1007/s10930-006-9005-8
- Perez, A., Carvajal, C., Trejo, S., Torres, M.J., Martin, M.I., Lorenzo, J.C., Natalucci, C.L. and Hernandez, M. (2010) Penduliflorain I: A cysteine protease isolated from Hohenbergia penduliflora (A.Rich.) Mez (Bromeliaceae). The Protein Journal, 29, 225-233. Hdoi:10.1007/s10930-010-9243-7
- Novillo, C., Casta, P. and Ortego, F. (1997) Inhibition of digestive trypsin-like proteases from larvae of several lepidopteran species by the diagnostic cysteine protease inhibitor E-64. Insect Biochemistry and Molecular Biology, 27, 247-254. Hdoi:10.1016/S0965-1748(96)00092-6
- Sreedharan, S.K., Patel, H., Smith, S., Gharbia, S.E., Shah, H.N. and Brocklehurst, K. (1993) Additional evidence for the cysteine proteinase nature of gingivain the extracellular proteinase of Porphyromonas gingivalis. Biochemical Society Transactions, 21, 218S-218S.
- Sreedharan, S.K., Verma, C., Caves, L.S.D., Brocklehurst, S.M., Gharbias, S.E., Shah, H.N. and Brocklehurst, K. (1996) Demonstration that 1-trans-epoxysuccinyl-L-leucylamido-(4-guanidino) butane (E-64) is one of the most effective low Mr inhibitors of trypsin-catalysed hydrolysis, Characterization by kinetic analysis and by energy minimization and molecular dynamics simulation of the E-64-13-trypsin complex. Biochemical Journal, 316, 777- 786.
ABBREVIATIONS
BAPNA, N-α-benzoyl-DL-arginine-p-nitroanilide: BCIP, 5-bromo-4-chloro-3-Indolyl phosphate: BZ-Phe-Val-Argp-NA, N-α-CBZ-L-phenylalanyl-L-valyl-L-arginine-p-nitroanilide: cBM, commercial bromelain: CBZ, carbobenzoxy: E-64, trans-epoxysuccinyl-L-leucyl-amido (4-guanidino) butane: N-α-CBZ-Ala-pNE, N-α-CBZ-alanyl-p-nitrophenyl ester: PFLNA, L-pyroglutamyl-L-phenylalanyl-L-leucine-p-nitroanilide: PMSF, Phenylmethanesulfonyl fluoride: PVDF, polyvinylidene difluoride: recBM, recombinant bromelain: SDS-PAGE, sodium dodecylsulfate-polyacrylamide gel electrophoresis: Z-Arg-Arg-p-NA, N-α-CBZ-L-arginyl-L-arginine-p-nitroanilide.

