American Journal of Plant Sciences
Vol.3 No.1(2012), Article ID:16640,19 pages DOI:10.4236/ajps.2012.31003
Molecular Adaptation of Peanut Metabolic Pathways to Wide Variations of Mineral Ion Composition and Concentration
![]()
CARC, Prairie View A & M University, Prairie View, USA.
Email: *cropyielddoublingbiotechnology@yahoo.com, goosuji@pvamu.edu
Received September 28th, 2011; revised October 25th, 2011; accepted November 29th, 2011
Keywords: Glutamate Dehydrogenase-Synthesized RNA; mRNA Silencing; Phosphate Translocator; Acetyl CoA Carboxylase; Cellulosic Biomass; Northern Analysis; HPLC; Fatty Acids
ABSTRACT
Plant evolution, nutritional genomics, and mineral nutrition have been well documented but no studies have focused on the molecular adaptation of crop metabolism to wide variations of mineral ion composition and concentration. Diversification of peanut species from primary centers of domestication in South America depended on metabolic adaptation to the mineral ion conditions of the newer habitats. Understanding the diversification molecular biology of peanut metabolic pathways will permit the synthesis of the best mineral ion combinations for doubling CO2 assimilation. Valencia and Virginia cultivars belong to different subspecies of the tetraploid Arachis hypogaea. They were planted in the absence and presence of up to 99 mM (equivalent to 166 moles per hectare) of different mineral ions. Molecular properties of the primary metabolic pathways were studied by Northern analyses using Valencia glutamate dehydrogenase (GDH)-synthesized RNAs as probes for Virginia mRNA and GDH-synthesized RNAs. Messenger RNAs are silenced by homologous RNAs synthesized by GDH. Peanut cellulose was analyzed by gravimetry; and fatty acids by HPLC. Complementary DNA probes made from Valencia GDH-synthesized RNAs hybridized perfectly to Virginia mRNAs and GDH-synthesized RNAs. Wide variations in mineral ion compositions and concentrations induced the GDHs of Valencia and Virginia to synthesize RNAs that differentially down-regulated the mRNAs encoding phosphate translocator, granule-bound starch synthase, phosphoglucomutase, glucosyltransferase, acetyl CoA carboxylase, nitrate reductase, and NADH-glutamate synthase so that the percent weights of oil (41.53 ± 8.75) and cellulose (30.29 ± 3.12) were similar in the control and mineral-treated peanuts. Therefore, RNA sequences that defined the molecular adaptation of mRNAs encoding the enzymes of primary metabolism were the same in the varietal types of A. hypogaea, in agreement with genetic data suggesting that tetraploid Arachis evolved relatively recently from the wild diploid ancestral species. Another molecular adaptation was to phosphate with or without K+ ion, and it prevented the silencing by GDH-synthesized RNAs of the mRNA encoding phosphate translocator resulting to doubling of cellulosic biomass yield (41323 kg/ha) compared with the N + P + K + S-treated positive control peanut (19428 kg/ha). Molecular adaptation of primary metabolic pathways at the mRNA level to 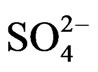 ion with or without
ion with or without 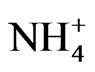 ion did not increase cellulosic biomass yields (27057 kg/ha) compared with negative control peanut because the mRNAs encoding granule-bound starch synthase, and NADH-glutamate synthase were not silenced by GDH-synthesized RNA in the N + S,
ion did not increase cellulosic biomass yields (27057 kg/ha) compared with negative control peanut because the mRNAs encoding granule-bound starch synthase, and NADH-glutamate synthase were not silenced by GDH-synthesized RNA in the N + S, 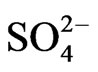 , and N + P + K + S-treated peanuts. These results could contribute towards further modeling at the mRNA level for improved mineral nutrient management of peanut production for fuel, fiber, feed, and food.
, and N + P + K + S-treated peanuts. These results could contribute towards further modeling at the mRNA level for improved mineral nutrient management of peanut production for fuel, fiber, feed, and food.
1. Introduction
Molecular adaptations of crop metabolism during crop diversification and domestication have been discussed under the physiological changes that led to improved germination and harvesting [1,2]. Such molecular adaptations were not redundant nor stress responses [3,4]. Crop domestication originated in about 24 regions [5] from where the crops dispersed mainly by human action. The wider the spread of a crop species, the more adaptable they are to different mineral ion conditions. Therefore, there is differential distribution of crop genera throughout the world, most landscapes being inhospitable for the growth of tuber and vegetable crop species, whilst cereal and leguminous crops [6] are more cultivated world-wide.
The widest dispersed seed legume is the Arachis peanut genus, with a primary center of domestication in southern Bolivia and northern Argentina [7] dated 8000 yr BP or earlier because it might have been domesticated by the predecessors of modern Arawak-speaking peoples [8]. There are six major geographical groups of cultivated peanuts in South America [7]. It is not certain how the geographical groups are related based on genetic maps [9]. However, the Virginia, Peruvian Runner, Valencia, and Spanish are the four basic varietal types of tetraploid Arachis hypogaea differentially distributed in the six geographic centers of diversification [7,10]. Tetraploid peanut is thought to have evolved relatively recently from diploid species [9,10]. Virginia and Runner peanuts are taxonomically related, whilst Valencia and Spanish are taxonomically related [8]. After Columbus, peanut quickly spread beyond South America to the Caribbean, Central America, Mexico, Philippines, Africa, Asia, and USA [8]; and is now cultivated in tropical, subtropical, and warm temperate zones [10] around the world on about 42 million acres between 40˚N and 40˚S latitudes. Plant mineral nutrition, domestication, and nutritional genomics have been well documented [3-5,11- 13], but no studies have focused on the molecular adaptation of crop metabolism to wide variations of mineral ion composition and concentration. Diversification of crop species from primary centers of domestication depends on metabolic adaptations to the mineral ion conditions of the newer habitats among other environmental factors. Understanding the molecular adaptations of peanut metabolism to mineral ions may illuminate the pattern of the diversification of its basic cultivars with respect to its diploid and allotetraploid genetics [8-10], and may permit the synthesis of best mineral ion combinations for modeling [14-16] the doubling of CO2 assimilation and fatty acid production.
Using the glutamate dehydrogenase (GDH) of crops as the target site of mineral ion action, it was demonstrated that higher ionic concentrations progressively disaggregated the subunit structure of the enzyme [17,18]; whereas different mineral ion compositions altered the electrical charges of the isoenzymes [19], with high total ion concentrations minimizing the accumulation of biomass. The link between GDH isomerization, mineral ions, and biomass lies in the metabolic pathway discrimination and integration functions of the RNAs synthesized by the enzyme [20]. GDH-synthesized RNA as a new gateway into biology has been applied to double cellulosic biomass and fatty acid yields [19-21] without application of kg quantities of fertilizers; and to explain some hitherto difficult biological phenomena in crop sciences including lack of crop yield responses to applied fertilizer [21], the inadequacy of genetic code-based probes/primers to screen physiological conditions [20] to mention a few. The GDH-synthesized RNAs silence mRNAs that are homologous to them, thereby controlling metabolism [20, 21]. But the molecular adaptation of metabolic pathways to mineral ion concentrations and compositions has not been described. GDH-synthesized RNA is ideal for investigating the molecular adaptation at the mRNA level of metabolic pathways to mineral ions because being metabolic but not genetic it is influenced only by environmental conditions. The logic of this research approach is that if the molecular metabolic adaptation of A. hypogaea was post species divergence (for example, after tetraploid formation); the GDH-synthesized RNAs of one varietal type may not be homologous to the mRNAs and GDH-synthesized RNAs of another varietal type. Conversely, if the molecular metabolic adaptation was prior to tetraploid formation, the GDH-synthesized RNAs of one varietal type may likely be homologous to the mRNAs and GDH-synthesized RNAs of another varietal type. Hereunder, the results indicate that mineral ions induced the GDHs of Valencia and Virginia peanuts to synthesize RNAs that are homologous to the mRNA encoding the regulatory enzymes of primary metabolism, thereby suggesting that molecular adaptation of metabolism to mineral ion conditions occurred prior to genetic divergence of the species. Peanuts are famous for their ability to produce large amounts of fatty acids, and are the most widespread oilseed food legume in the world [22]. GDH-synthesized RNAs could help to model and peer into what happened to peanut fatty acid biosynthesis at around 8000 yr BP or pre-history.
2. Materials and Methods
2.1. Treatment of Peanuts with Mineral Ion Solutions
Peanut (Arachis hypogaea L. Cv. Virginia) seeds were planted in boxes 243.84 × 243.84 × 30.48 cm (width × length × depth) [20], each filled with Metro-Mix 700 peat moss. About 100 - 110 seeds were planted per box. The applied mineral ion compositions mimicked and targeted the binomial subunit polypeptide compositions of the GDH isoenzymes [21]. The first box was left as the untreated (negative) control; the second box (N) was treated with 1 L of NH4Cl solution (25 mM), the third box (Pi) was treated with 1 L of Na2HPO4 solution (20 mM); the fourth box (S) was treated with 1 L of Na2SO4 solution (50 mM); the fifth box (K) was treated with 1 L KCl solution (4 mM); the sixth box (N + P + K + S) positive control was treated with 1 L of combined NH4Cl (25 mM), Na2HPO4 (20 mM), Na2SO4 (50 mM), and KCl (4 mM) solution; the seventh box (P + K) was treated with 1 L of combined Na2HPO4 (20 mM) and KCl (4 mM) solution; the eighth box (N + S) was treated with 1L of combined NH4Cl (25 mM) and Na2SO4 (50 mM) solution; the ninth box (P + N) was treated with 1 L of combined Na2HPO4 (20 mM) and NH4Cl (25 mM) solution; the tenth box (P + S) was treated with 1 L of combined Na2HPO4 (20 mM) and Na2SO4 (50 mM) solution as described before [20]. The boxes were watered every other day. Mineral ion solutions were applied sequentially, first at pre-flowering stage (2 weeks after seed germination), second at flowering, and thirdly at post-flowering. When the leaves turned yellow (peanut maturity), pods and shoots were harvested, allowed to dry on the greenhouse floor for about 2 weeks, and weighed. Pods were shelled by hand, and the kernels (seeds) weighed. Seeds were stored at –30˚C.
2.2. Analyses for Peanut Cellulose and Oil
Dry and milled (composited) seeds (100 g) per experimental treatment, sent to SGS North America Inc., St. Rose, Louisiana, USA were custom analyzed by HPLC for percent oil weight. Pulverized shoots (250 g) per experimental treatment, sent to Universal Testing, Quincy, IL, USA were custom analyzed for dry matter, and cellulose (neutral detergent fiber, and acid detergent fiber) using standard gravimetric method.
2.3. Purification and Assay of GDH
GDH charge isomers were extracted from peanut seeds from the control or mineral-treated boxes, and purified by electrophoresis as described before [23]. RNA synthetic activity of GDH isoenzymes was assayed in combined deamination and amination substrate solutions of 0.1 M Tris-HCl buffer (pH 8.0) containing the four NTPs (0.6 mM each), CaCl2 (3.5 mM), L-glu (3.23 µM), NAD+ (0.375 µM), NH4Cl (0.875 mM), α-ketoglutarate (10.0 mM), NADH (0.225 mM), 5 Units RNase inhibitor, 1 Unit DNase 1, and 5 µg of actinomycin D as described before [20]. Reaction was started by adding 0.2 mL of whole gel-eluted GDH charge isomers containing 3 - 9 µg protein per mL. Final volume of the reaction was brought to 0.4 mL with 0.1M Tris-HCl buffer pH 8.0. Reactions were incubated at 16˚C overnight and stopped by phenol-chloroform (pH 5.5) extraction of the enzyme. RNA was precipitated with ethanol, and dissolved in minimum volume of molecular biology quality water. RNA yield and quality were determined by agarose gel electrophoresis, and photometry. Assays were carried out in duplicate to verify the reproducibility of the results.
2.4. Total RNA
Total RNA was extracted from peanut seeds using the acid-equilibrated (pH 4.7) phenol: chloroform 5:1 method [24].
2.5. cDNA Synthesis, Cloning, Characterization, and Probe Selection
cDNAs were synthesized with 2 µg of each product RNA synthesized by Valencia Cultivar GDH charge isomers using random hexamer primer [19,23,25,26]. Restriction fragment PCR amplification; adapter ligation; sequencing gel fractionation; and purification of cDNA fragments [21] were conducted according to the methods of Display Systems Biotech, Vista, CA, USA. Selected cDNA fragments were subcloned into pCR4-TOPO vector and transformed into TOP10 One Shot Chemically Competent Escherichia coli (Invitrogen, Carlsbad, CA), followed by overnight growth on selective plates. Up to ten positive transformant colonies were picked per plate and cultured overnight in LB medium containing 50 µg/mL of kanamycin. Plasmid DNA was purified with a plasmid kit (Novagen, Madison, WI). The insert cDNA was sequenced with T3 and T7 primers by Genemed Synthesis, Inc. (South San Francisco, CA, USA), and Functional Biosciences, Inc. (Madison, WI, USA). To identify the GDH-synthesized RNAs that were homologous to mRNAs encoding the primary metabolic enzymes, the cDNA sequences were used as queries to search the NCBI nucleotide-nucleotide (excluding ESTs) BLAST (blastn), and non-redundant protein translation (blastx) databases. Complementary DNAs that displayed the highest alignment scores with mRNAs encoding the enzymes of photosynthesis, glycolysis, cellulose, fatty acid, nitrogen assimilation, and nucleotide biosyntheses [20] were selected as the probes.
2.6. Northern Blot Analysis
Total RNA (15 µg), and RNAs (10 µg) synthesized by Virginia Cultivar GDH charge isomers from the control and mineral ion-treated peanuts were loaded, briefly electrophoresed on 2% agarose gels, stained with ethidium bromide, and photographed to verify RNA quality. RNA was electro-transferred from the electrophoresed gel onto Brightstar-Plus nylon membrane (Applied Biosystems, Foster City, CA, USA) as described before [20].
The cDNAs that were used as Northern probes were those homologous to mRNAs encoding phosphate translocator, granule-bound starch synthase (GBSS), phosphoglucomutase (PGM), glucosyltransferase, acetyl CoA carboxylase (ACCase), NADH-glutamate synthase (GOGAT), nitrate reductase (NR), glycinamide ribonucleotide (GAR) synthetase/GAR transformylase. For the labeling of the cDNA probes, cDNA inserts were amplified by PCR from the corresponding plasmids (15 ng) using M13 forward and M13 reverse primers (2 µM each), [32P]-dATP (6000 Ci/mmol, 20 mCi/mL), dCTP/dGTP/ TTP mix 50 mM, (2 µL), and Taq polymerase (1U), in a final volume of 50 µL. Amplification was according to Display Systems Biotech (Vista, CA, USA) “touchdown” PCR procedure (denature: 94˚C, 1 min. For the first 10 cycles: 94˚C, 30 sec; anneal: 60˚C, 30 sec for the first cycle, then reduced the temperature 0.5˚C each cycle until an annealing temperature of 55˚C was reached after 10 cycles; extension: 72˚C, 1 min. Continued another 25 cycles with 94˚C, 30 sec; 55˚C, 30 sec; 72˚C, 1 min; final extension 72˚C, 5 min). Nylon membranes with immobilized RNA were prehybridized with ULTRAhyb buffer and hybridized with 32P-labeled cDNA inserts as probes overnight at 68˚C as described before [21]. Solutions of labeled cDNA were first heated in boiling water bath for 10 min before adding to the prehybridized membrane. After hybridization, the membranes were washed (30 min, 68˚C) with NorthernMax (Applied Biosystems, Foster City, CA, USA) low stringency wash solution followed by NorthernMax high stringency wash solution (30 min, 68˚C). The membrane was autoradiographed by exposure to X-ray film within intensifying screens at –80˚C. Northern band intensities were digitalized using UN-SCAN-IT gel digitalizing software (Silk Scientific, Inc., Orem, Utah, USA).
3. Results
3.1. Mineral Ion Composition and Concentration
Mineral ion compositions (Table 1) were formulated to target the subunit polypeptides of GDH. In addition to the N, Pi, K, and S added, there were trace amounts of Ca, B, Mg, Fe, Mn, and Si in the peat moss. Therefore, the peanut was exposed to a wide spectrum of mineral ion composition. The mineral ion treatments did not induce any deficiency symptoms because the older and younger leaves alike remained green until maturity. But there were visible differences in the rate of growth. Applied total mineral ion concentration was up to 99 mM (equivalent to 166.5 moles per hectare). Therefore, this was different from kg agronomic quantities of mineral fertilizers that are applied to field crops to target and saturate all macromolecules in the crop. The hexameric subunit structure of peanut GDH was stable up to 100 mM ion concentration [18]. The mineral ion compositions and concentrations made for internal repeats in the treatments thus limiting the random variability in applied treatments, and imposed firm control on the number of experimental repeats. The peat moss was common to all the experimental boxes, thereby minimizing variability in soil physical and chemical characteristics. The control box without applied mineral ions was the negative control; whilst the N + P + K + S (NPKS)-treated box was the positive control. The experimental conditions induced and synchronized the isomerization of GDH, synthesis of RNA by GDH, and silencing of mRNAs by the homologous GDH-synthesized RNA [21]. GDH isomerization [27] and synthesis of RNA as the target sites of mineral ion action are due to the binomial distribution of its three subunits in the hexameric isoenzymes, on the basis of the twin nonallelic GDH1 and GDH2 gene structure, with the gene (GDH1) encoding the more acidic subunits (α, and a) being heterozygous and codominant, whereas the other gene (GDH2) encoding the less acidic subunit (β) is homozygous [28]. Because the peanut seeds germinated under the untreated control soil condition followed later by mineral solution treatments, the outcomes of the treatments were overrides of the untreated control.
3.2. Bioscience of Metabolic Adaptation to Mineral Ions
The Northern blot results (Figures 1, 3-9) described the molecular adaptation of primary metabolic pathways
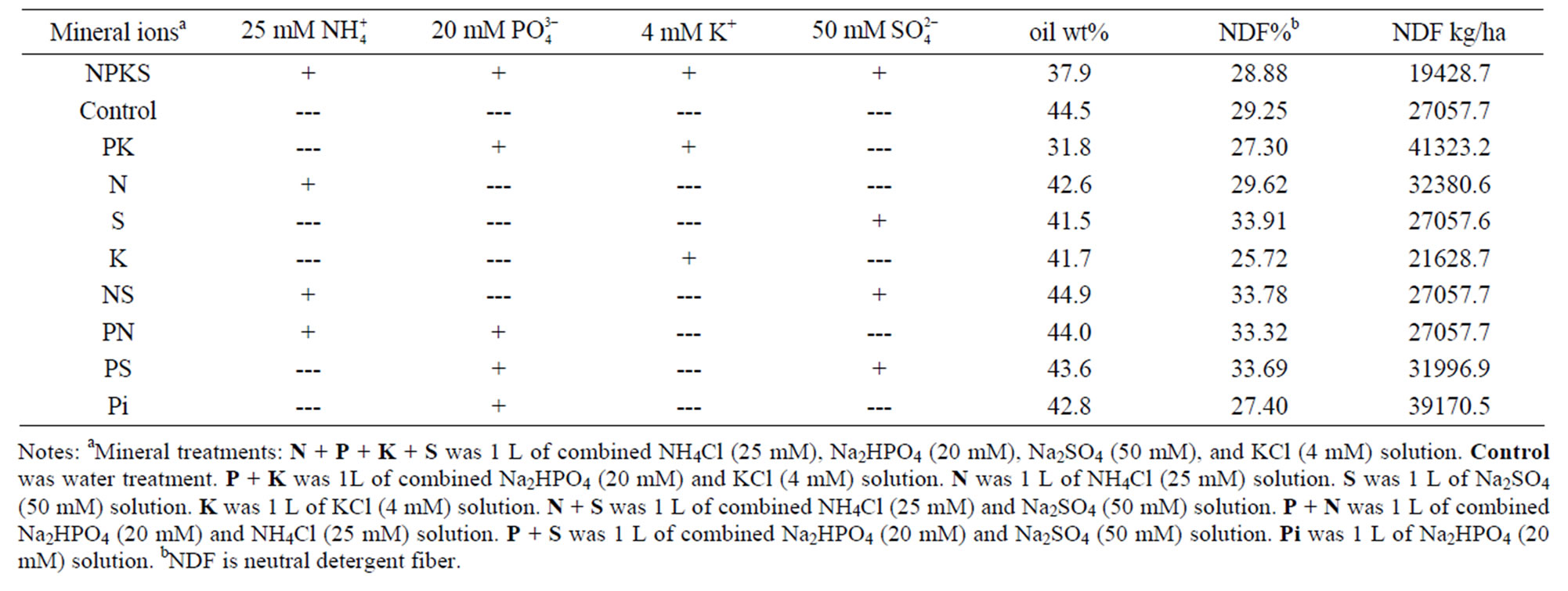
Table 1. Mineral salt compositions and concentrations, peanut oil and cellulose contents.
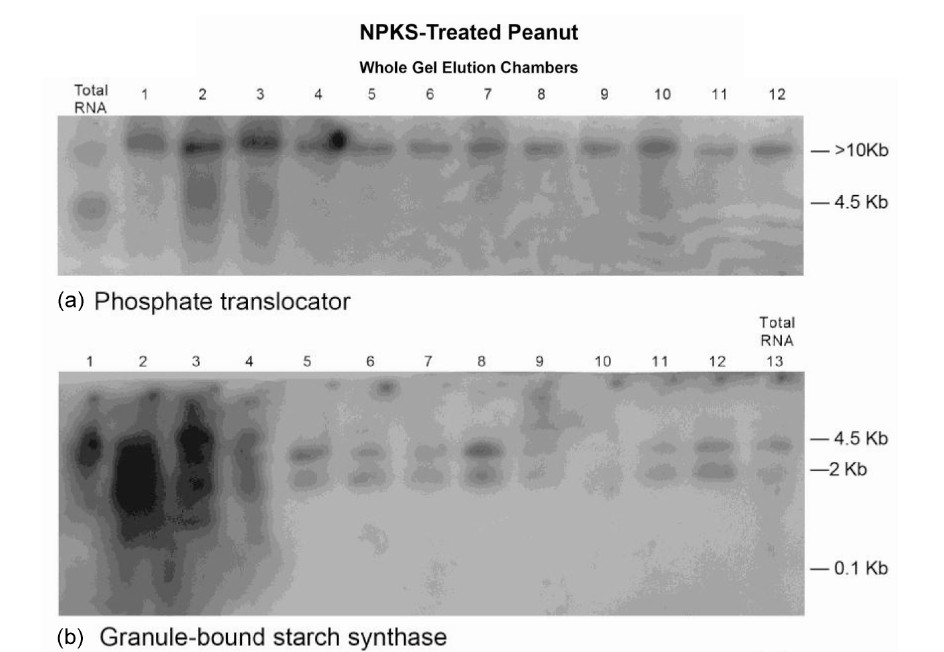
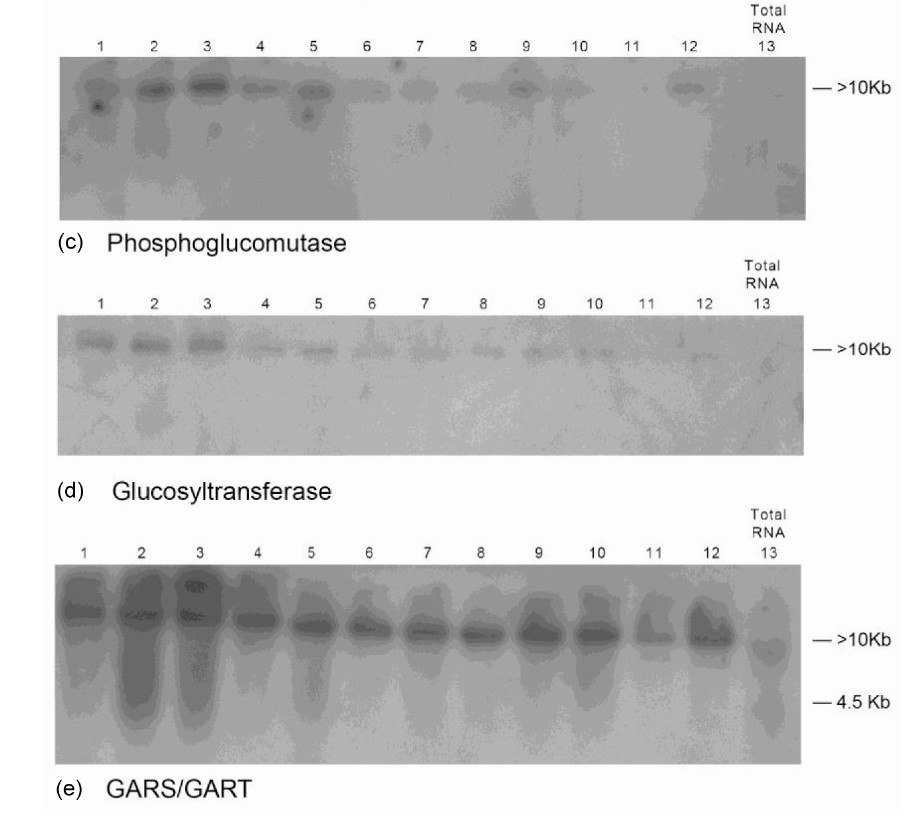
Figure 1. Molecular adaptation of peanut metabolism to N + P + K + S mineral salts combination: RNAs (lanes 1 - 12) synthesized by the charge isomers of whole-gel purified GDH of the N + P + K + S-treated peanut; and total RNA (lane 13) isolated from N + P + K + S-treated peanut were electrophoresed through 2% agarose gel. The electrophoresed gels were transblotted onto nylon membranes followed by membrane screening with 32P-labeled cDNAs of the GDH-synthesized RNAs that were homologous to the mRNAs encoding the indicated enzymes. The membranes were washed with high stringency solutions and autoradiographed.
to mineral ion composition and concentration. The Northern bands demonstrated the population distribution of short sequences of Virginia GDH-synthesized RNAs that were homologous to the Valencia GDH-synthesized RNAs; the Virginia mRNAs that were homologous to the short sequences of Valencia GDH-synthesized RNAs; the Virginia GDH-synthesized RNAs that were present in total RNA; the threshold quantities of probe sequences per Virginia GDH-synthesized RNA; and confirmed the absence of total RNA from Virginia GDH-synthesized RNA preparations. The blots showed that the GDH-synthesized RNAs fell into different molecular size groups: 0.1, 2.0, 4.5, and >10 kb bands [20]. Low molecular weight oligos (25 - 50 nucleotides long) synthesized by GDH have also been detected [29]. GDH-synthesized RNAs are metabolic but not genomic therefore they are not translatable [23]. The high molecular weight GDHsynthesized RNAs are suggested to undergo degradation to the low molecular weight oligos [21], which are the effective siRNA molecules.
Previous results [21,25,26] identified phosphate translocator, GBSS, PGM, glucosyltransferase, ACCase, GARS/ GART, NR, and NADH-GOGAT as the enzyme steps that control vegetable oil contents and CO2 assimilation in peanut. The mRNAs encoding the primary metabolic enzymes were the target sites of the action of the GDH-synthesized RNAs. The target mRNAs either were fully silenced (down-regulated), not silenced (not downregulated), or partially silenced (partial down-regulated) by the homologous GDH-synthesized RNAs [20]. Phosphate translocator is a chloroplast membrane protein antiport system using Pi and phosphorylated C3, C5, or C6 compounds as counter substrates [30] in CO2 assimilation pathways. Starch synthases catalyze the formation of α-(1-4)-linked linear glucosyl chains [31] in CO2 assimilation pathways. PGM partitions carbon between the pathways of starch synthesis and glycolysis [32]. Glucose is converted to hexose phosphates for metabolism, and uridine 5’-diphosphate-glucose destined for cellulose biosynthesis [33]. It has been suggested [34] that cellulose biosynthesis involves chain initiation, elongation and termination, with the participation of glucosyltransferase in the chain initiation reaction [35]. Pi translocator, GBSS, PGM, and glucosyltransferase control CO2 assimilation and saccharide metabolism in peanut [20]. ACCase catalyzes the ATP-dependent carboxylation of acetyl-CoA to form malonyl-CoA, the precursor for fatty acid synthesis [36]. ACCase controls the production of fats in peanut [21]. Purine biosynthesis, nitrate reduction, and GS-GOGAT cycle cooperate to assimilate nitrogen in peanut [20]. The purine metabolic pathway is employed in tropical legumes to assimilate and detoxify ammonia [37]. GAR synthetase catalyzes the second step, and GAR transformylase catalyzes the third step in purine biosynthetic pathway [38]; and they regulate purine metabolism in peanut [21]. Nitrate reductase catalyzes the reduction of nitrate to nitrite which is then reduced by nitrite reductase to 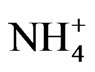 ion [39], followed by incorporation into amino acids by GS-GOGAT cycle [40]. NADH-GOGAT is an important enzyme in the salvage of
ion [39], followed by incorporation into amino acids by GS-GOGAT cycle [40]. NADH-GOGAT is an important enzyme in the salvage of 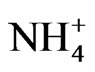 ion in peanut [20]. The above are the primary biochemical systems that are the target sites of mineral ion action for the molecular adaptation of metabolism. The molecular modifications changed according to the prevalent composition and concentration of mineral ions (Figures 1-9). The probes used for screening the Northern blots were the cDNAs (restriction fragments) of short sequences of GDH-synthesized RNAs (Table 2) that were homologous to the mRNAs encoding the enzymes of primary metabolism in peanut [20,21]. Complementary DNA probes made from Valencia GDH-synthesized RNAs hybridized perfectly under high stringent conditions, to Virginia mRNAs and GDH-synthesized RNAs.
ion in peanut [20]. The above are the primary biochemical systems that are the target sites of mineral ion action for the molecular adaptation of metabolism. The molecular modifications changed according to the prevalent composition and concentration of mineral ions (Figures 1-9). The probes used for screening the Northern blots were the cDNAs (restriction fragments) of short sequences of GDH-synthesized RNAs (Table 2) that were homologous to the mRNAs encoding the enzymes of primary metabolism in peanut [20,21]. Complementary DNA probes made from Valencia GDH-synthesized RNAs hybridized perfectly under high stringent conditions, to Virginia mRNAs and GDH-synthesized RNAs.
The Northern band populations (Figure 1) showed that the GDH of N + P + K + S (NPKS)-treated Virginia peanut synthesized RNAs that were homologous to the mRNAs encoding phosphate translocator, GBSS, PGM, glucosyltransferase, GARS/GART, and NR. The Northern blot pattern (not shown) using the GDH-synthesized
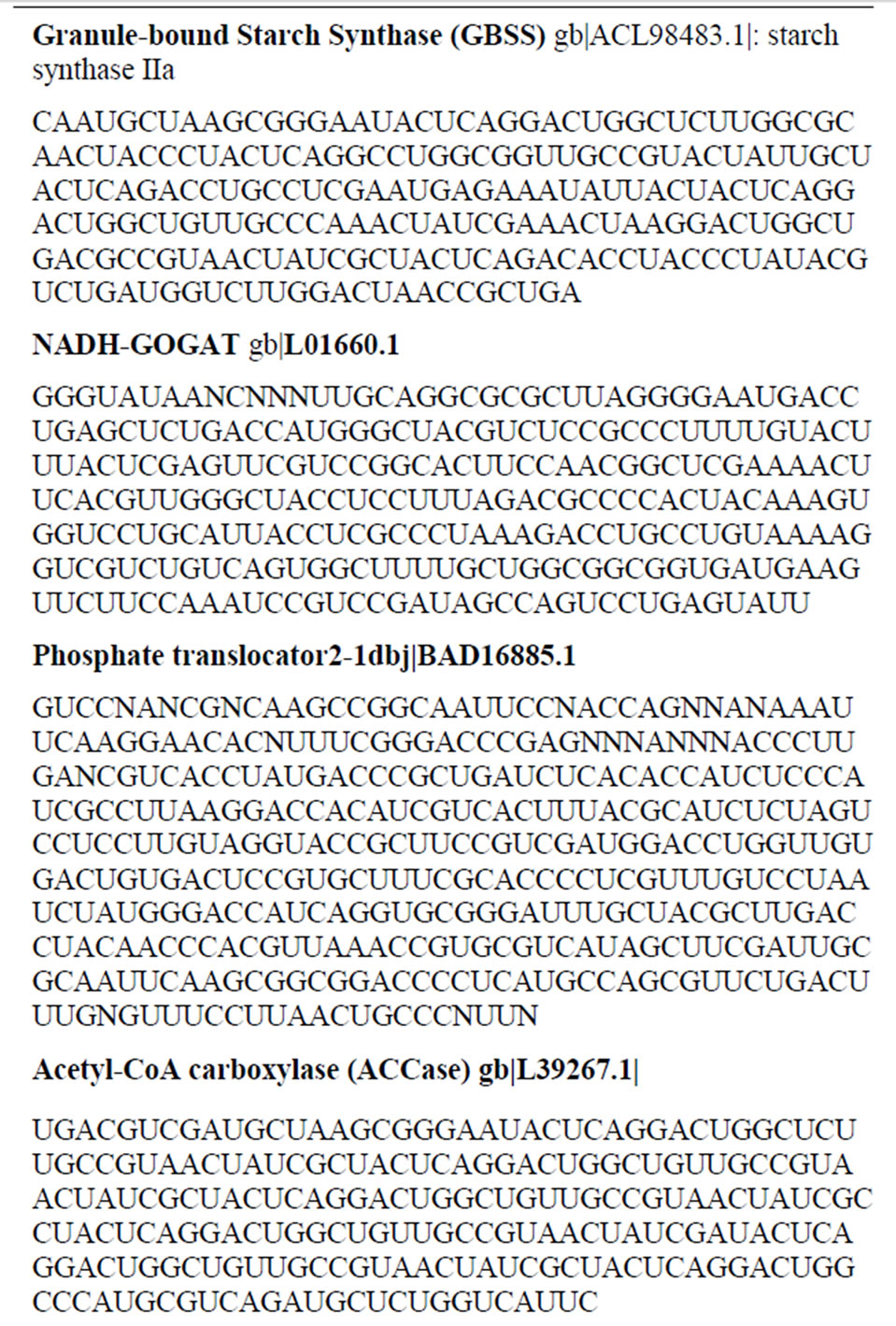
Table 2. Some peanut GDH-synthesized RNA sequence targets of molecular adaptation to mineral ion conditions. The RNA sequences are homologous to the mRNAs encoding some primary metabolic enzymes of peanut.
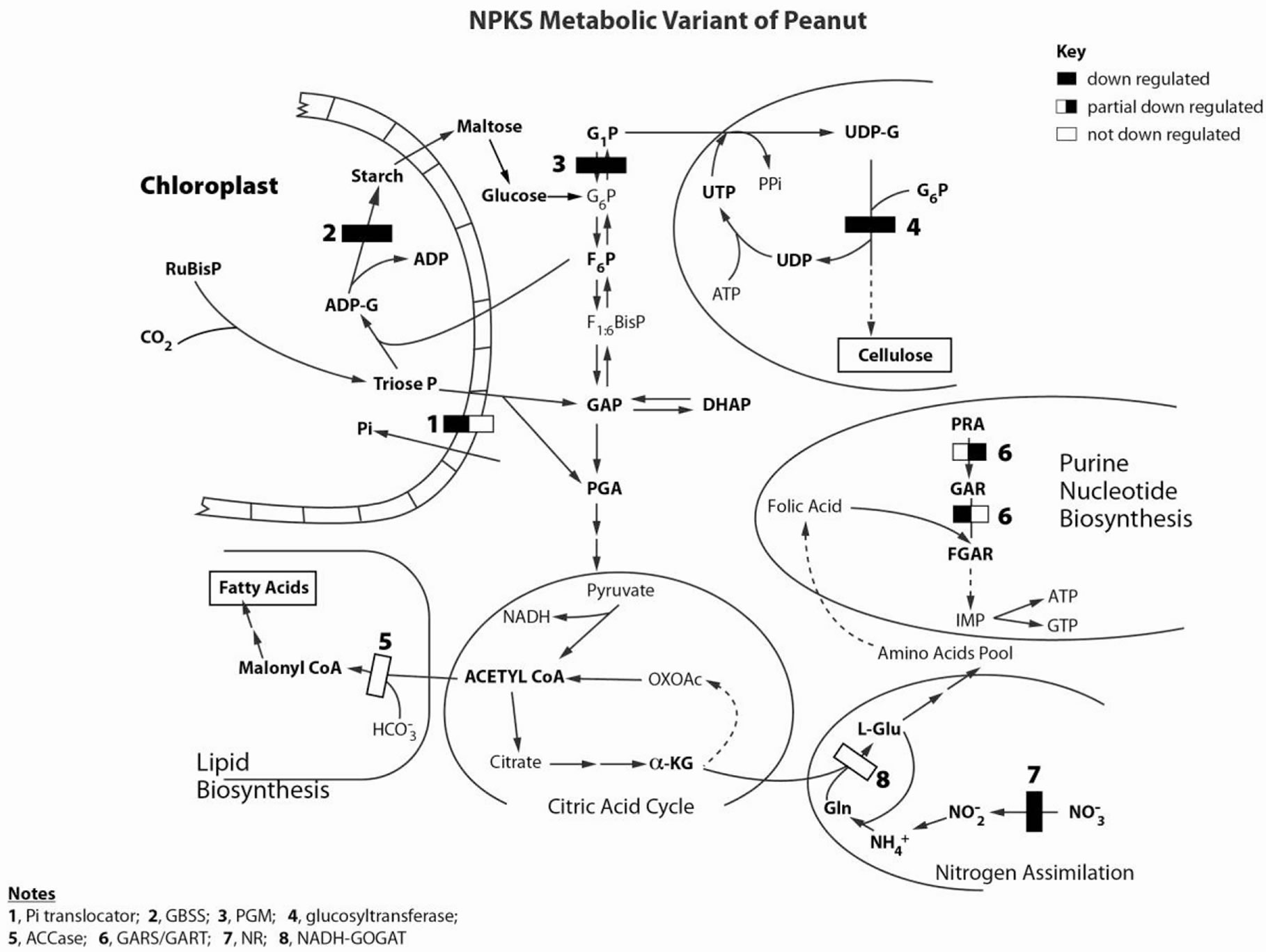
Figure 2. Molecular metabolic adaptation in N + P + K + S-treated peanut. Sketch of simplified metabolic pathways highlighting the enzymes whose encoding mRNAs were permuted by the RNAs synthesized by the GDH of the N + P + K + Streated experimental peanut to achieve production of cellulose and fatty acids. Only the chloroplastic GBSS is shown. Only the cytosolic PGM is shown. Only the plastidial ACCase is also shown. Because under the NPKS mineral ion concentrations, the mRNA {1} encoding Pi translocator was partially down-regulated, there was limited Pi translocator activity to import Pi into chloroplasts; mRNA {2} encoding GBSS was down-regulated, phosphorylated C3, C5, or C6 compounds [30] were not expended in starch synthesis thereby starving glycolysis and cellulose biosynthesis of intermediates to power fatty acid and cellulose production [20] resulting to very low yields per hectare of NDF cellulosic biomass and fatty acids (Table 1). Nitrogen assimilation is important in doubling peanut yield, therefore NPKS mineral ion composition ensured that the three mRNAs {6, 7, and 8} encoding GARS/GART, NR, and NADH-GOGAT respectively were not simultaneously down-regulated by the GDH-ynthesized RNAs [20,21]. This is an aspect of the model sequential permutation of metabolism at the mRNA level by GDH-synthesized RNA to normalize production of fatty acids and carbohydrates under NPKS mineral ion conditions. Some abbreviations: G1P, glucose-1-phosphate; G6P glucose-6-phosphate; α-KG, alpha-ketoglutarate.
RNA homologous to the mRNA encoding NR as probe was similar to that for glucosyltransferase (Figure 1(d)). Residual quantities of GDH-synthesized RNAs, as well as the mRNAs encoding Pi translocator, and GARS/ GART were detected in the Northern blots in the corresponding total RNA lanes (Figure 1). This suggested that in those cases, the GDH-synthesized RNAs only partially down-regulated the homologous mRNAs. But the homologous GDH-synthesized RNAs for the mRNAs encoding GBSS, PGM, NR, and glucosyltransferase did not detect residual mRNA in the total RNA lanes (Figure 1). Northern assays with the probes for GDH-synthesized RNAs homologous to mRNAs encoding ACCase, and NADH-GOGAT gave no bands meaning that the GDH of NPKS-treated peanut did not synthesize RNAs that were homologous to the two mRNAs. The milieu and permutation of down-regulated, partially down-regulated, and not down-regulated primary metabolic pathways of the NPKS-treated peanut (Figure 2) gave 37.9% oil weight, and 28.88% shoot NDF cellulose (Table 1).
The control Virginia peanut Northern blots [20] showed that the GDH-synthesized RNAs down-regulated the mRNAs encoding Pi translocator, PGM, NR, and GARS/ GART; but did not down-regulate the mRNAs encoding GBSS, glucosyltransferase, ACCase, and NADH-GOGAT. There were no partially down-regulated primary metabolic pathways in the control peanut. The milieu of the control peanut’s primary metabolic pathways gave 44.5%

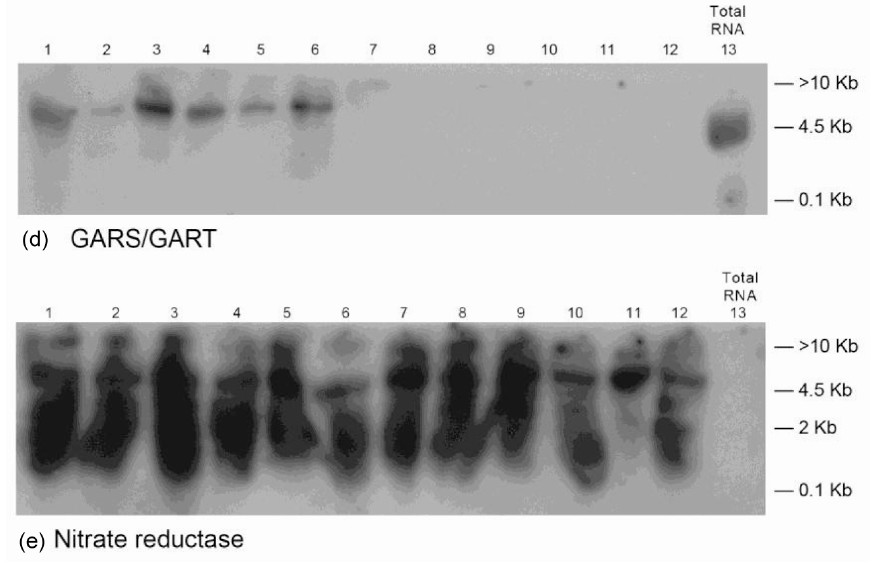
Figure 3. Molecular adaptation of peanut metabolism to P + K mineral salts combination: RNAs (lanes 2 - 13 in panel (a), lanes 1 - 12 in panels (b)-(e)) synthesized by the charge isomers of whole-gel purified GDH of the P + K-treated peanut; and total RNA (lane 1 in panel (a), lane 13 in panels (b)-(e)) from P + K-treated peanut were electrophoresed through 2% agarose gel. The electrophoresed gels were trans-blotted onto nylon membranes followed by membrane screening with 32P-labeled cDNAs of the GDH-synthesized RNAs that were homologous to the mRNAs encoding the indicated enzymes. The membranes were washed with high stringency solutions and autoradiographed.
oil weight, and 29.25% NDF cellulose. The GDH-synthesized RNAs adaptively differentiated the primary metabolic pathways at the mRNA level so as to allow the same fundamental biochemical processes to be sustained in the negative control, and NPKS-treated peanuts despite the absent mineral ions and the wide differences in mineral ion composition and concentration. This may be part of the molecular metabolic adaptation that enabled
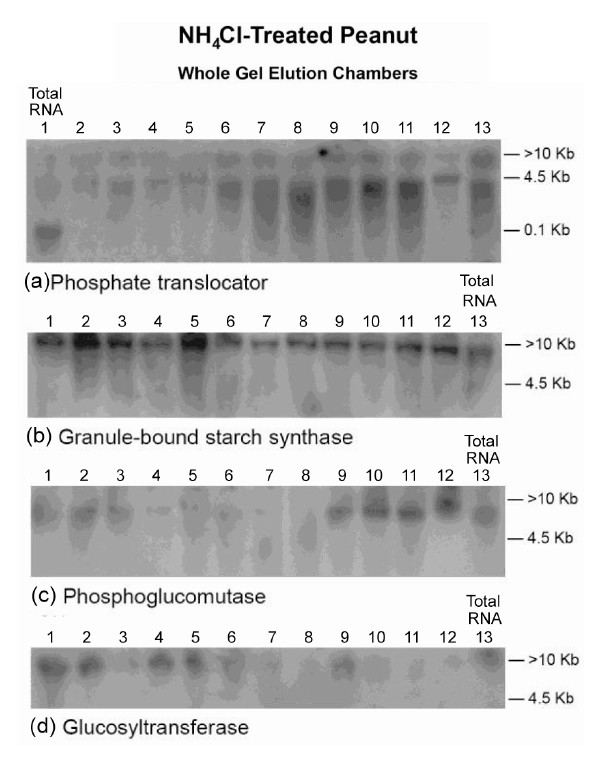
Figure 4. Molecular adaptation of peanut metabolism to NH4Cl: RNAs (lanes 2 - 13 in panel (a), and lanes 1 - 12 in panels (b)-(d)) synthesized by the charge isomers of wholegel purified GDH of NH4Cl-treated peanut; and total RNA (lane 1 in panel (a), and lane 13 in panels (b)-(d)) from NH4Cltreated peanut were electrophoresed through 2% agarose gel. The electrophoresed gels were trans-blotted onto nylon membranes followed by membrane screening with 32P-labeled cDNAs of the GDH-synthesized RNAs that were homologous to the mRNAs encoding the indicated enzymes. The membranes were washed with high stringency solutions and autoradiographed.
peanut cultivation to spread world-wide in warm temperate regions, tropics, and sub-tropics [22].This confirms the chemistry and operation of the metabolic pathways. The molecular metabolic adaptations in the NPKS-treated peanut metabolic variant as schematically presented (Figure 2), is a multidimensional reaction model for fatty acid and cellulosic biomass accumulation in peanut. The modeling of primary metabolism at the mRNA level presented in Figure 2 is however different from computational approximation of global metabolism that is based on enzyme kinetic fluxes assembled from different studies [15,16].
The Northern band populations (Figure 3) showed that the GDH of P + K (PK)-treated Virginia peanut synthesized RNAs that were homologous to the mRNAs encoding GBSS, PGM, glucosyltransferase, GARS/GART, and NR. The Northern result for the NR-encoded probe was remarkable in that all the GDH-synthesized RNA had sequences that were homologous to the probe. Multiple repeats of a sequence are typical of GDH-synthe-
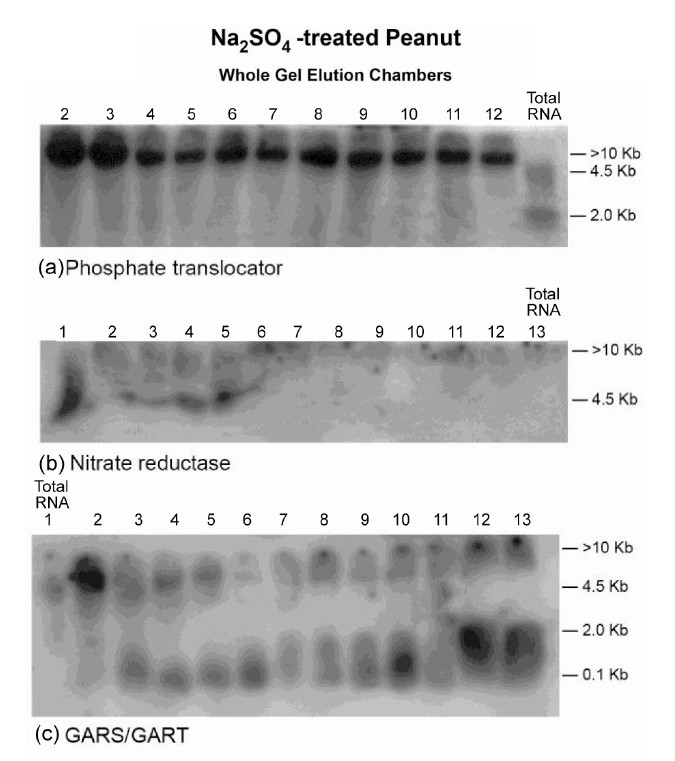
Figure 5. Molecular adaptation of peanut metabolism to Na2SO4: RNAs (lanes 1 - 12 in panels ((a) and (b)), lanes 2 - 13 in panel (c)) synthesized by the charge isomers of wholegel purified GDH; and total RNA (lane 13 in panels ((a) and (b)), lane 1 in panel (c)) from Na2SO4-treated peanut were electrophoresed through 2% agarose gel. The electrophoresed gels were trans-blotted onto nylon membranes followed by membrane screening with 32P-labeled cDNAs of the GDHsynthesized RNAs that were homologous to the mRNAs encoding the indicated enzymes. The membranes were washed with high stringency solutions and autoradiographed.
sized RNAs [26]. Residual band of mRNA (partial downregulation) was present in the total RNA lane of the GARS/GART Northern result. Northern assays with the probes for GDH-synthesized RNAs homologous to the mRNAs encoding Pi translocator, ACCase, and NADHGOGAT gave no bands meaning that the GDH of the PK-treated peanut did not synthesize RNA that was homologous to the three mRNAs. The milieu and permutation of down-regulated, partially down-regulated, and not down-regulated primary metabolic pathways of the PKtreated peanut (Figure 3) gave 31.8% oil weight, and 27.3% shoot NDF cellulose (Table 1). Therefore, the GDH-synthesized RNAs adaptively discriminated and integrated the primary metabolic pathways at the mRNA level so as to allow the same fundamental biochemical processes to be sustained in the PK-treated peanuts as in the negative control despite the wide differences in mineral ion composition and concentration.
The Northern results (Figure 4) showed that the GDH of NH4Cl-treated Virginia peanut synthesized RNAs that
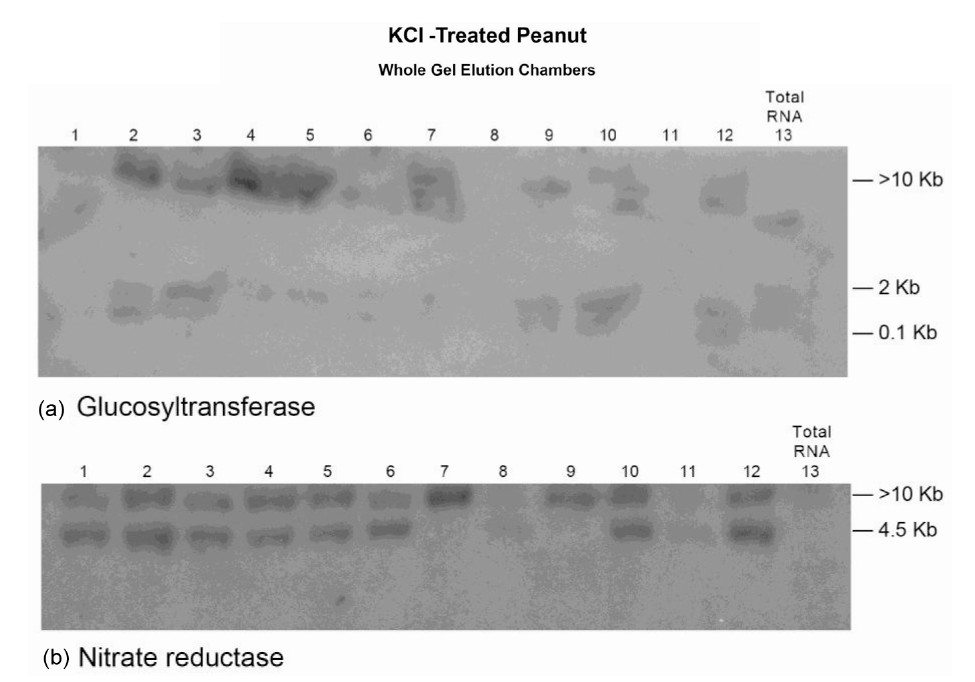
Figure 6. Molecular adaptation of peanut metabolism to KCl: RNAs (lanes 1 - 12) synthesized by the charge isomers of whole-gel purified GDH of KCl-treated peanut; and total RNA (lane 13) from KCl-treated peanut were electrophoresed through 2% agarose gel. The electrophoresed gels were trans-blotted onto nylon membranes followed by membrane screening with 32P-labeled cDNAs of the GDH-synthesized RNAs that were homologous to the mRNAs encoding the indicated enzymes. The membranes were washed with high stringency solutions and autoradiographed.
were homologous to the mRNAs encoding Pi translocator, GBSS, PGM, and glucosyltransferase. Residual quantities of GDH-synthesized RNAs, as well as the mRNAs encoding Pi translocator, and GBSS (partial down-regulation) were detected in the Northern blots in the corresponding total RNA lanes. Northern assays with the probes for GDH-synthesized RNAs homologous to mRNAs encoding ACCase, GARS/GART, NR, and NADH-GOGAT gave no bands. The unique cooperation and permutation of down-regulated, partially down-regulated, and not downregulated primary metabolic pathways of the NH4Cltreated peanut (Figure 4) gave 42.6% oil weight, and 29.62% shoot NDF cellulose (Table 1). Therefore, the GDH-synthesized RNAs adaptively discriminated and integrated the primary metabolic pathways at the mRNA level to allow the same fundamental biochemical processes to be sustained in the NH4Cl-treated peanuts as in the controls despite the absent mineral ions and the wide difference in mineral ion composition.
The Northern band populations (Figure 5) showed that the GDH of Na2SO4-treated Virginia peanut synthesized RNAs that were homologous to the mRNAs encoding Pi translocator, NR, and GARS/GART. Residual quantities of GDH-synthesized RNAs, as well as the mRNAs encoding Pi translocator (partial down-regulation), GARS/ GART and NR were detected in the Northern blots in the respective total RNA lanes. The probe for the mRNA encoding GARS/GART hybridized to the GDH-synthesized RNA bands to give a population of Northern bands (Figure 5(c)) that was similar to the pattern of GDHsynthesized RNA bands [20] thus demonstrating multiple repeat of the sequence. Northern assays with the probes for GDH-synthesized RNAs homologous to the mRNAs encoding GBSS, PGM, glucosyltransferase, ACCase, and NADH-GOGAT gave no bands. The cooperation and permutation of down-regulated, partially down-regulated, and not down-regulated primary metabolic pathways of the Na2SO4-treated peanut normalized the fundamental biochemical processes by yielding 41.5% oil weight, and 33.91% shoot NDF cellulose similar to the control peanuts (Table 1).
The Northern band populations (Figure 6) showed that the GDH of KCl-treated Virginia peanut synthesized RNAs that were homologous to the mRNAs encoding glucosyltransferase, NR, and Pi translocator. The Northern band pattern (not shown) using the GDH-synthesized RNA probe that was homologous to the mRNA encoding Pi translocator was similar to that for glucosyltransferase (Figure 6(a)). Residual quantities of GDH-synthesized RNA and of the mRNAs encoding Pi translocator and glucosyltransferase were detected in the Northern blots in the corresponding total RNA lanes. Northern assays with the probes for GDH-synthesized RNAs homologous to the mRNAs encoding GBSS, PGM, ACCase, GARS/ GART, and NADH-GOGAT gave no bands. The milieu and permutation of down-regulated, partially down-regu-
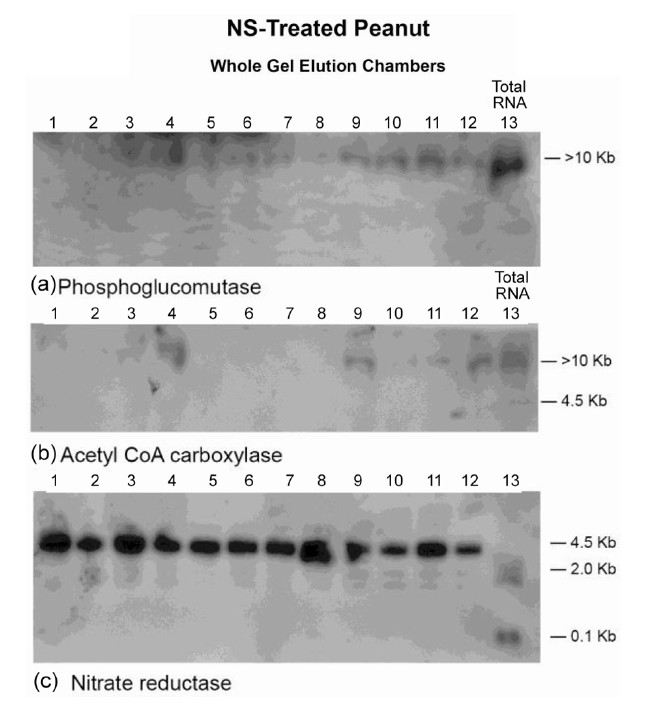
Figure 7. Molecular adaptation of peanut metabolism to N + S mineral salts combination: RNAs (lanes 1 - 12) synthesized by the charge isomers of whole-gel purified GDH of the N + S-treated peanut; and total RNA (lane 13) isolated from the N + S-treated peanut were electrophoresed through 2% agarose gel. The electrophoresed gels were trans-blotted onto nylon membranes followed by membrane screening with 32P-labeled cDNAs of the GDH-synthesized RNAs that were homologous to the mRNAs encoding the indicated enzymes. The membranes were washed with high stringency solutions and autoradiographed.
lated, and not down-regulated primary metabolic pathways of the KCl-treated peanut normalized the fundamental biochemical processes by yielding 41.7% oil weight, and 25.72% shoot NDF cellulose similar to the control peanuts (Table 1).
The Northern band populations (Figure 7) showed that the GDH of N + S (NS)-treated Virginia peanut synthesized RNAs that were homologous to the mRNAs encoding PGM, ACCase, NR, and glucosyltransferase (not shown). The Northern band pattern using the GDH-synthesized RNA probe that was homologous to the mRNA encoding glucosyltransferase was similar to that for PGM (Figure 7(a)). Residual quantities of GDH-synthesized RNAs, as well as mRNA encoding PGM, ACCase, and NR (partial down-regulation) were detected in the Northern blots in the respective total RNA lanes. Northern assays with the probes for GDH-synthesized RNAs homologous to the mRNAs encoding Pi translocator, GBSS, GARS/GART, and NADH-GOGAT gave no bands. The consortium and permutation of down-regulated, partially down-regulated, and not down-regulated primary metabolic pathways of the NS-treated peanut normalized the fundamental biochemical processes by yielding 44.9% oil weight, and 33.7% shoot NDF cellulose similar to the control peanuts (Table 1) despite the dissimilarity in mineral ion composition and concentration.
The Northern band populations (Figure 8) showed that the GDH of P + N (PN)-treated Virginia peanut synthesized RNAs that were homologous to the mRNAs encoding Pi translocator, GBSS, PGM, glucosyltransferase, and ACCase. Residual quantities of GDH-synthesized RNA bands, as well as the mRNAs encoding Pi translocator, GBSS, ACCase, and PGM (partial down-regulation) were detected in the Northern blots in the corresponding total RNA lanes. Northern hybridization with the probes for GDH-synthesized RNAs homologous to the mRNAs encoding GARS/GART, NR, and NADHGOGAT gave no bands. The milieu and permutation of down-regulated, partial down-regulated, and not downregulated primary metabolic pathways of the PN-treated peanut normalized the fundamental biochemical processes by yielding 44.0% oil weight, and 33.32% shoot NDF cellulose similar to the control peanuts (Table 1) despite the dissimilarity in mineral ion composition and concentration.
The Northern band populations (Figure 9) showed that the GDH of P + S (PS)-treated Virginia peanut synthesized RNAs that were homologous to the mRNAs encoding Pi translocator, PGM, GBSS, glucosyltransferase, NR, NADH-GOGAT, and GARS/GART. The Northern band patterns (not shown) using the probes that were homologous to the mRNAs encoding NADH-GOGAT, and NR were similar to that for glucosyltransferase (Figure 9(d)). Northern hybridization with the probes for GDH-synthesized RNAs homologous to the mRNA encoding ACCase gave no bands. Residual quantities of GDH-synthesized RNA, as well as of mRNAs encoding Pi translocator, PGM, glucosyltransferase, GBSS, and NADH-GOGAT were detected in the Northern blots in the respective total RNA lanes. The conglomerate and permutation of down-regulated, partial down-regulated, and not down-regulated primary metabolic pathways of the PS-treated peanut normalized the fundamental biochemical processes by yielding 43.6% oil weight, and 33.69% shoot NDF cellulose similar to the controls and the other metabolic variants of peanut (Table 1) despite the dissimilarity in mineral ion composition and concentration.
The Northern band populations [20] showed that the GDH of Pi-treated peanut synthesized RNAs that were homologous to the mRNAs encoding PGM, glucosyltransferase, ACCase, GARS/GART, and NADH-GOGAT. Residual quantities of the mRNAs encoding PGM, ACCase, and glucosyltransferase were detected in the corresponding Northern blots. Northern assays with the probes for GDH-synthesized RNAs homologous to the
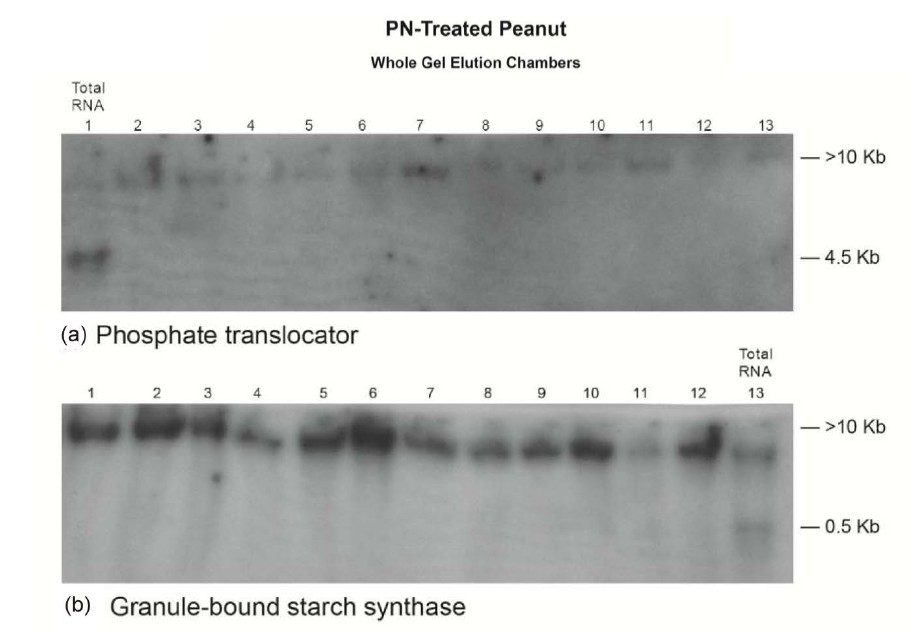
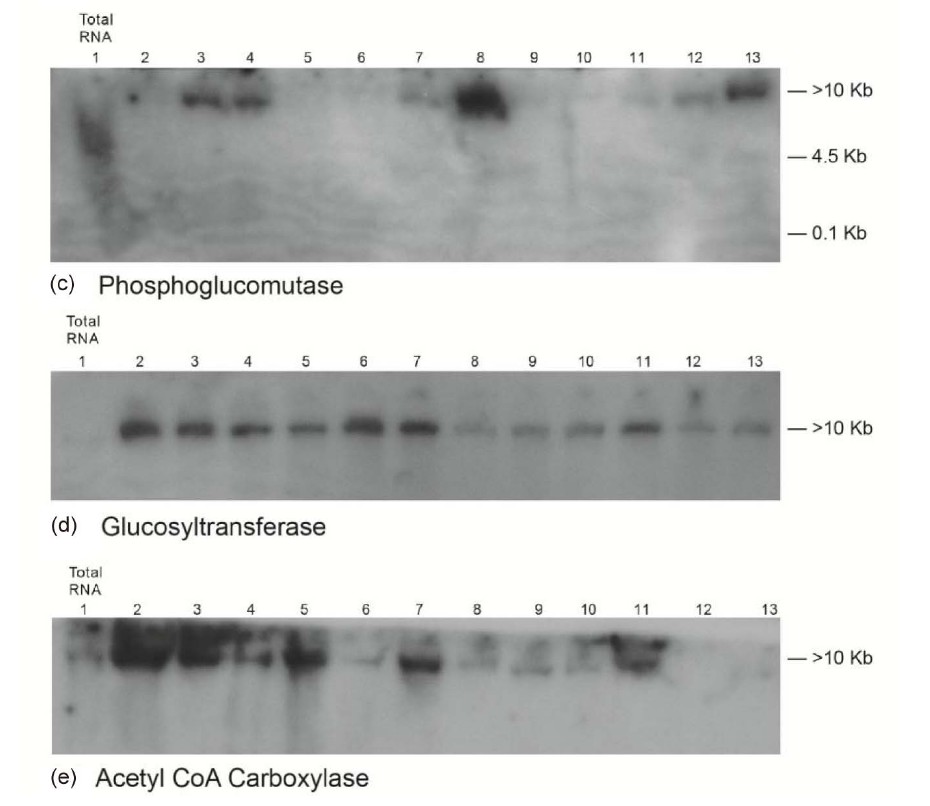
Figure 8. Molecular adaptation of peanut metabolism to P + N mineral salts combination: RNAs (lanes 2 - 13 in panels ((a), and (c)-(e)) and lanes 1 - 12 in panel (b)) synthesized by the charge isomers of whole-gel purified GDH of P + N-treated peanut; and total RNA (lane 1 in panels (a), and (c)-(e)) and lane 13 in panel (b)) isolated from P + N-treated peanut were electrophoresed through 2% agarose gel. The electrophoresed gels were trans-blotted onto nylon membranes followed by membrane screening with 32P-labeled cDNAs of the GDH-synthesized RNAs that were homologous to the mRNAs encoding the indicated enzymes. The membranes were washed with high stringency solutions and autoradiographed.
mRNAs encoding Pi translocator, GBSS, and NR gave no bands. The consortium and permutation of down-regulated, partial down-regulated, and not down-regulated primary metabolic pathways of the Pi-treated peanut normalized the fundamental biochemical processes by yielding 42.8% oil weight, and 27.4% shoot cellulose similar to the control peanut and the other metabolic variants of peanut (Table 1) despite the dissimilarity in
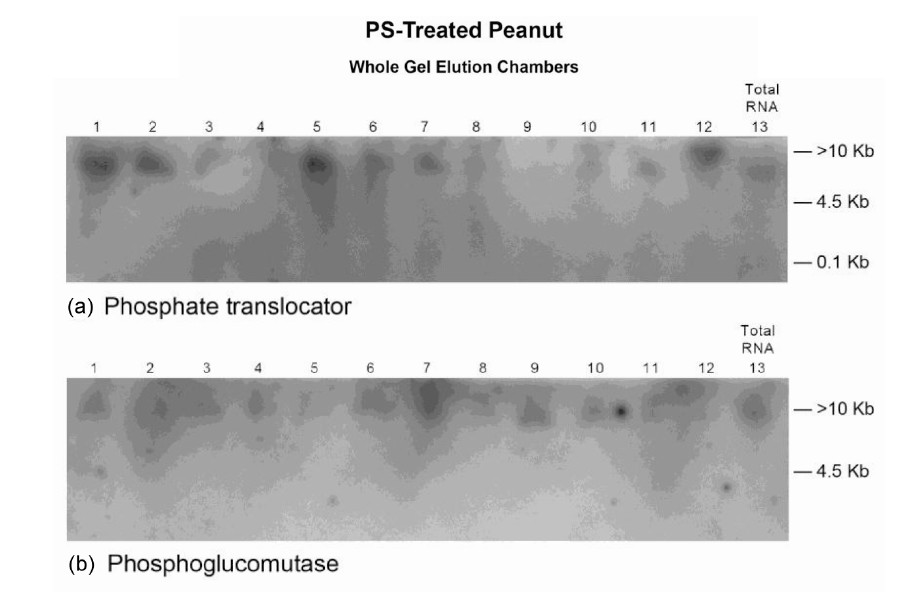
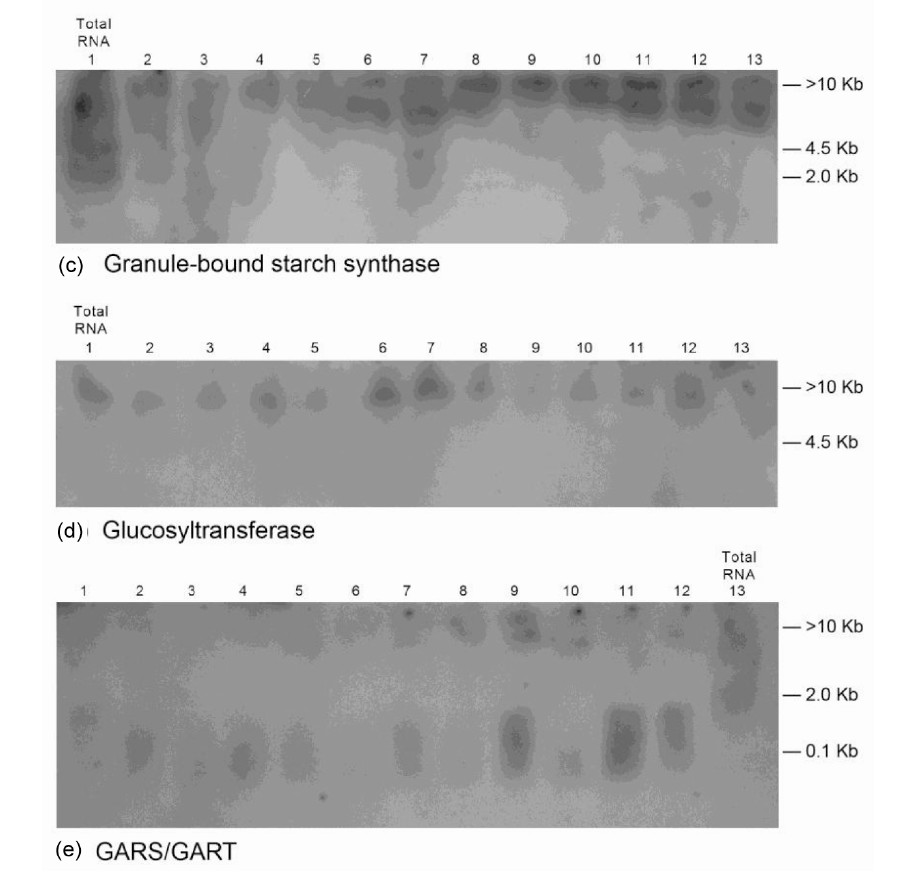
Figure 9. Molecular adaptation of peanut metabolism to P + S mineral salts combination: RNAs (lanes 1 - 12 in panels ((a), (b), and (e)), lanes 2 - 13 in panels ((c) and (d))) synthesized by the charge isomers of whole-gel purified GDH of P + S-treated peanut; and total RNA (lane 13 in panels ((a), (b), and (e), lane 1 in panels (c) and (d)) from P + S-treated peanut were electrophoresed through 2% agarose gel. The electrophoresed gels were trans-blotted onto nylon membranes followed by membrane screening with 32P-labeled cDNAs of the GDH-synthesized RNAs that were homologous to the mRNAs encoding the indicated enzymes. The membranes were washed with high stringency solutions and autoradiographed.
mineral ion composition and concentration.
From the fore-going, wide variations in mineral ion compositions and concentrations induced the GDHs of Valencia and Virginia peanuts to synthesize RNAs that differentially down-regulated the mRNAs encoding Pi translocator, GBSS, PGM, glucosyltransferase, ACCase, GARS/GART, NR, and NADH-GOGAT so that the percent weights of oil (41.53 ± 8.75) and cellulosic biomass (30.29 ± 3.12) were similar in the control and mineral treated peanuts. Therefore, RNA sequences (Table 2) [20] that defined the molecular adaptation of mRNAs encoding the enzymes of primary metabolism were the same in Valencia and Virginia cultivars; meaning that the molecular adaptation of peanut metabolism to mineral ion concentration and combination probably occurred prior to species divergence. This agrees with the suggestions [41] that the allotetraploid was fairly nearly related to the wild diploid ancestors, and [9] that the genetic divergence to tetraploidy was relatively recent. GDH-synthesized RNAs have helped to peer into what happened to peanut fatty acid biosynthesis pre-history.
The percent yields of peanut oil and cellulose (Table 1) differed by less than ~12% compared with the controls, thus rendering the differences economically insignificant when the costs of the mineral salts are factored into the operational costs. Therefore, the mineral ion compositions and concentrations did not increase CO2 assimilation per unit weight of peanut. This was the molecular adaptation in part that normalized peanut yield despite variations in mineral ion concentration and composition. GDH-synthesized RNAs have helped to model peanut metabolic reactions for fatty acid and cellulosic biomass production. Na2SO4 with or without NH4+ ion combination produced the same dry matter cellulose yields as the negative control peanut (27057 Kg) per unit area (ha) of land (Table 1). Therefore, there was neither synergism nor antagonism in the molecular adaptation of metabolic pathways to mineral ion conditions. The molecular adaptation that normalized dry matter yield per unit land area was due in part to the ability of Na2SO4, and N + S to prevent GDH charge isomers from synthesizing RNA sequences that are homologous to the mRNAs encoding GBSS, and NADH-GOGAT in the Na2SO4 (Figure 5), and N + S-treated (Figure 7) peanuts (Table 2). These normalizations were likely part of the molecular adaptations of primary metabolism at the mRNA level that enabled peanut cultivation to spread throughout the world [22] irrespective of adversities in soil mineral ion compositions and concentrations. Phosphate treatment with or without K+ ion combination doubled the NDF dry matter yields (41323 kg) compared with the N + P + K + S-treated positive control peanut (19428 kg) per unit area (ha) of land (Table 1). The molecular adaptation for doubling cellulosic biomass yield per unit land area could be due in part to the ability of Pi, and P+K mineral ions to prevent GDH charge isomers from synthesizing RNA sequence that is homologous to the mRNA encoding Pi translocator (Table 2) in the Pi [20], and P + Ktreated (Figure 3) peanuts. Compared with the N + P + K + S-treated peanut that had the lowest vegetable oil yield per hectare (Figure 2), the additional adaptation for doubling vegetable oil per hectare could be due in part to the ability of the P + K mineral solution to prevent GDH isomers from synthesizing RNA sequence that is homologous to the mRNA encoding Pi translocator [20]. This highlighted the great importance of Pi translocator to mobilize triose phosphates to glycolysis. Peanuts are famous for their ability to produce vegetable oil. The adaptation to produce oil under wide variations of mineral ion composition and concentration could be due in part to the ability of control, NPKS, K+, 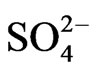 ,
, 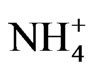 , PK, PS etc to prevent GDH charge isomers from synthesizing the RNA sequence that is homologous to the mRNA encoding ACCase (Table 2) in the control, and N PKS [20], K+ (Figure 6),
, PK, PS etc to prevent GDH charge isomers from synthesizing the RNA sequence that is homologous to the mRNA encoding ACCase (Table 2) in the control, and N PKS [20], K+ (Figure 6), 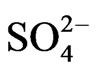 (Figure 5),
(Figure 5), 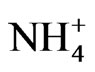 (Figure 4), PK (Figure 3), and PS-treated (Figure 9) peanuts.
(Figure 4), PK (Figure 3), and PS-treated (Figure 9) peanuts.
Digital (UN-SCAN-IT) quantitation of the Northern bands showed that GDH synthesized the required RNA sequences in sufficient quantities to meet the thresholds for silencing the target homologous mRNAs. The minimum 2:1 ratio between the GDH-synthesized RNA to the target mRNA was the requisite threshold for the silencing reaction [17]. The ratio was the baseline control platform applied for unbiased judgment of the Northern bands across experimental repeats and mineral ion treatments. The Northern analyses in which total RNAs were loaded side-by-side in the agarose gel wells to the GDH synthesized RNAs (Figures 1, 3-9) made for easier interpretation of the efficiency of transcript silencing by the GDH-synthesized RNAs. The definitive detection of the high molecular weight (>10 kb) RNAs synthesized by GDH in the total RNA lanes in the Northern hybridizations was an evidence that GDH synthesizes RNAs in vivo, although they are not translatable. The Northern results further highlighted the importance of the preponderant >10 kb GDH-synthesized RNA in the efficiency of mRNA silencing. The >10 kb GDH-synthesized RNA assured a maximum structural interaction with the homologous mRNAs, a crucial tertiary stabilization for the initiation of silencing [42].
Most of the hybridization reactions (Figures 1, 3-9) gave more than a product per RNA lane. This was caused by incomplete silencing of some mRNAs by homologous RNAs synthesized by GDH, presence of GDH-synthesized RNAs in total RNA preparations, and the isomeric nature of GDH-synthesized RNAs [21]. Therefore, highthrough-put screening procedures (microarray, quantitative PCR etc) were inadequate for analyzing the hybridization reaction products (Figures 1, 3-9).
As a new gateway into biology, procedures for analyzing transcript silencing by GDH-synthesized RNAs [20] are slightly different from classical siRNA approaches in that there are no transfection and reverse transcription steps, and the GDH-synthesized RNAs are endogenously elicited. Transcript silencing by GDH-synthesized RNA takes place as a biochemical reaction at the molecular level; and it is readily inducible. Experimentation on GHD-synthesized RNA embodies many layers of technical repeats, normalization, and standardization. GDH purification by sub-zero temperature whole gel electro-elution into the 14 chambers of Bio-Rad’s mini whole gel eluter gave 14 instead of one preparation of the enzyme. The binomial assembly of the subunit polypeptides in the hexameric isoenzymes did not permit the combination of the 14 fractions to make a single preparation. Therefore, in these protocols, there were 14 repeats of the purified GDH, 14 repeats of RNA synthesis reaction, and 14 repeats of RNA loading into agarose gel. The extensive repetitive steps in the GDH experimentation are vividly portrayed by the similarities in the Northern blot lanes (Figures 1, 3-9). More than 90% of the RNA lanes (Figures 1, 3-9) showed that RNA repetitive loadings onto gels were consistent, efficient and reproducible across mineral-treated peanuts. Such results evoke confidence in the research approach. The similarities of the GDH-synthesized RNA patterns on agarose gels and Northern blots notwithstanding, when the GDHsynthesized RNAs are converted to cDNAs followed by restriction enzyme digestion, the resulting fragments are homologous to hundreds of mRNAs [23]. Therefore, GDHsynthesized RNAs are different in the arrangement of the repeating and recurrent isomeric sequences [25,26]. Northern blots were performed in duplicate using each cDNA of GDH-synthesized probe, thereby making a total of 28-repeat RNA tracks per treated peanut. There were several lines of evidence for internal control reactions in the transcript silencing including the plus/plus and/or plus/minus sequence similarities among the GDHsynthesized RNAs [20], partial silencing of mRNAs encoding distinct enzymes, GDH-synthesized RNA present in total RNA, absence of total RNA from GDH-synthesized RNA etc. Messenger RNAs encoding several housekeeping enzymes were not useful references in the Northern assays because Northern results [29] in which GDHsynthesized RNAs were used as probes suggested the housekeeping mRNAs were also reprogrammed under the experimental regimen. Therefore, the ratio of 2:1 between the GDH-synthesized RNA and the target mRNA was adopted as the minimum normalization and standardization factor for silencing. Although the ideal reference internal mRNA is one that does not vary as a function of experimental treatment, it has been difficult to identify a single mRNA that meets the criterion [43]. Where the mRNAs were fully silenced and there were no residual mRNAs as in the Northern blots for PGM (Figure 1(c)), nitrate reductase (Figure 3(e)) etc, it was assumed that the 2:1 ratio was satisfied. Where the mRNAs were partially silenced as in the Northern blots for Pi translocator (Figure 1(a)), GARS/GART (Figure 3(d)), nitrate reductase (Figure 7(c)) etc, it was assumed that the residual mRNA constituted 50% of the total mRNA. The full amount of mRNA was determined when the GDH of the peanut did not synthesize the homologous RNA and the mRNA was the lone band in the Northern assay. The limits of the biological comprehinsiveness of the GDH-synthesized RNA experimentation were defined by the peanut without any mineral ion treatment representing the negative control [20]; and the peanut with the full mineral ion concentration and composition (NPKS) representing the positive control (Figures 1 and 2). The positive control is important to provide the line of verification for the molecular mechanism of GDH action, and for result verification, confirmation and validation.
Although all the experimental peanuts were green, the control [20], NPKS (Figure 1), sulfate (Figure 5), potassium (Figure 6), N + S (Figure 7), and P + N (Figure 8) treated peanuts exhibited low growth vigor. The said comparative reluctance to grow was clearly manifested by the low qualities of the corresponding Northern blots for their GDH-synthesized RNAs. Figures 1, 3-9 are the first time that Northern analyses have been conducted on RNA that is not genomic total RNA. Therefore, the quality of the GDH-synthesized RNA Northern blot results is somehow related to the vegetative growth vigor of the peanut, a relationship that genomic total RNA does not possess.
4. Discussion
A comparison of the molecular adaptations of the primary pathways in the NH4Cl (Figure 4), Na2SO4 (Figure 5), and N + S-treated (Figure 7) peanuts shows that the actions of mineral ions on GDH were specific; not redundant [3], and not stress responses [4]. The bioscience (Figures 1-9) of the GDH-synthesized RNAs was therefore a regular molecular basis for understanding the shifts in metabolic adaptation from the negative control to a mineral ion-treated peanut. It also explained in the same molecular terms the shifts from mineral ion compositions to concentrations, an evidence for the evolutionary importance of the adaptation. Successful Northern screening of the Virginia peanut’s total RNA and GDH-synthesized RNA with the Valencia’s GDH-synthesized RNA as probes (Figures 1, 3-9), further suggested that the two cultivars shared a common primary center of diversity prior to their morphological diversification [7-9]. The Valencia and Virginia varietal types belong to two different subspecies [7]. The peanut allotetraploid is believed to have derived from wild diploid ancestors [8]. That the Arachis metabolism was molecularly adapted to wide variations in mineral ion composition prior to domestication diversification agrees with the earlier suggestion [9,41] that the allotetraploid was fairly recent and nearly related to the wild diploid ancestors. The molecular adaptation of the Arachis primary metabolic pathways to produce oil under wide differences in mineral ion contents and concentrations could be judged as the major chemical characteristic and advantage that led to the early selection of the genus for domestication. This molecular adaptation was not studied much earlier probably because several metabolic adaptations may not have led to morphological and phenotypic changes [44], the adaptations having resulted from unconscious selective pressures that acted like natural selection under the imposed culture of the cultivated landscape. Therefore, molecular adaptations of crop metabolism to soil mineral concentrations and compositions may not readily be amenable to archeological evolutionary logic of investigation.
The GDH-synthesized RNAs that were homologous to the mRNAs encoding the enzymes of the primary biochemical processes shared extensive plus/plus and plus/ minus sequence similarities [20,21,25,26]. The sequence similarities enabled the GDH-synthesized RNAs to discriminate and integrate the metabolic processes through pathways permutation [20] of the homologous mRNA abundances. In the evolutionary normalization of the peanut primary biochemical systems to give similar yields of fatty acids and cellulose (Table 1) per unit weight of peanut in response to the wide range of mineral ion composition and concentration, the consortium of down-regulated, not down-regulated, and partially down-regulated pathways were permuted [20]. The similar weights of cellulose and oil per unit weight of peanut (Table 1) despite the wide differences in the applied mineral ions also described in part the biochemical adaptation of photosynthesis to light [45]. It is not known why it has not been possible to double photosynthesis per unit weight of plant tissue through physiological, genetic engineering and plant breeding approaches [11,46,47]. The evolutionary normalization of metabolism to produce similar yields of oil and cellulose per unit weight of seeds might account for part of the inability of plant breeding and related approaches to double the rate of photosynthesis per unit area of leaf.
Another step in the molecular adaptation of metabolism to mineral ions assured the accumulation of excess quantities of fatty acids etc (yield) per unit area of land. P + K-treated peanuts produced vegetable oil that were double that produced by the negative and positive control peanuts per unit land area [20]. The molecular adaptation was probably conferred in part by the inability of the GDH of PK-treated peanut to synthesize RNAs that are homologous to the mRNAs encoding ACCase and Pi translocator (Figure 3, Table 2). The cultivated peanut (Arachis hypogaea L.) possesses higher oil and protein contents than other legumes [10]. The two halves of the metabolic adaptation to mineral ions shared reciprocal relationship because mineral ion regimens (NS, NPKS, PN, and PS) that produced high percentage peanut oil weight inversely produced low NDF cellulosic biomass weight per hectare (Table 1). Whereas, those mineral regimens (PK, Pi) that produced low-median percentage peanut oil weights inversely produced the highest cellulose weights per hectare. Therefore, Arachis hypogaea L. was domesticated for its high oil content per unit area of cultivated land [20]. The molecular adaptations of peanut’s primary metabolic pathways that assure oil production irrespective of wide changes in mineral ion contents are not amenable to archeological and/or genetic logics of studies as were the cases in seed germination and harvesting, grain size enlargement, and enzyme encoded genes of domestication and diversification [5]. Furthermore, genetic code-based probes and primers may be equivocal for analyzing metabolic variations [29]. The molecular adaptations of metabolic pathways to mineral ion conditions could permit the formulation of mineral ion compositions and concentrations for optimizing and tripling of peanut yield per unit area of land possibly for bioenergy manufacture, without increasing energy inputs and costs in the agricultural processes.
REFERENCES
- J. R. Harlan, J. M. De Wet and E. G. Price, “Comparative Evolution of Cereals,” Evolution, Vol. 27, No. 2, 1973, pp. 311-325. doi:10.2307/2406971
- D. Zohary and M. Hopf, “Domestication of Plants in the Old World,” Oxford University Press, Oxford, 2000.
- M. E. Theodorou and W. C. Plaxton, “Metabolic Adaptations of Plant Respiration to Nutritional Phosphate Deprivation,” Plant Physiology, Vol. 101, No. 2, 1993, pp. 339-344.
- H. J. Bohnert and E. Sheveleva, “Plant Stress Adaptation: Making Metabolism Move,” Current Opinion in Plant Biology, Vol. 1, No. 3, 1998, pp. 267-274. doi:10.1016/S1369-5266(98)80115-5
- M. D. Purugganan and D. Q. Fuller, “The Nature of Selection during Plant Domestication,” Nature, Vol. 457, 2009, pp. 843-848. doi:10.1038/nature07895
- R. L. Meyer and N. Rask, “Major Food and Energy Crops: Trends and Prospects,” In: P. V. Ammirato, D. A. Evans, W. R. Sharp and Y. Yamada, Eds., Handbook of Plant Cell Culture Crop Species, Macmillan, New York, Vol. 2, 1984, pp. 19-47.
- A. Krapovickas, “Evolution of the Genus Arachis,” In: R. Moav, Ed., Agricultural Genetics, Wiley, New York, 1973, pp. 135-151.
- W. C. Gregory and M. P. Gregory, “Groundnut,” In: N. W. Simmonds, Ed., Evolution of Crop Plants, Longman, London and New York, 1976, pp. 151-154.
- B. Jayashree, F. Morgan, I. Dan, D. Jeff and H. C. Jonathan, “Analysis of Genomic Sequences from Peanut,” Electronic Journal of Biotechnology, Vol. 8, No. 3, 2005, pp. 226-237.
- Y. P. S. Bajaj, “Peanut,” In: P. V. Ammirato, D. A. Evans, W. R. Sharp and Y. Yamada, Eds., Handbook of Plant Cell Culture Crop Species, Macmillan, New York, Vol. 13, 1984, pp. 193-326.
- Z. Rengel, “Mineral Nutrition of Crops,” Food Products Press, New York, London, Oxford, 1999.
- J. F. Doebley, B. S. Gaunt and B. D. Smith, “The Molecular Genetics of Crop Domestication,” Cell, Vol. 127, No. 7, 2006, pp. 1309-1329. doi:10.1016/j.cell.2006.12.006
- M. R. Broadley and P. J. White, “Plant Nutritional Genomics,” Blackwell Publishing and CRC Press, Oxford, 2005.
- C. Colijn, A. Brandes, J. Zucker, D. S. Lun, B. Weiner, M. R. Farhat, T. Chen, D. B. Moody, M. Murray and J. E. Galagan, “Interpreting Expression Data with Metabolic Flux Models: Predicting Mycobacterium Tuberculosis Mycolic Acid Production,” PLoS Computational Biology, Vol. 5, No. 8, 2009, e1000489 doi:10.1371/journal.pcbi.1000489
- J. Hay and J. Schwender, “Computational Analysis of Storage Synthesis in Developing Brassica Napus L. (OilSeed Rape) Embryos: Flux Variability Analysis in Relation to 13C Metabolic Flux Analysis,” The Plant Journal, Vol. 67, No. 3, 2011, pp. 513-525. doi:10.1111/j.1365-313X.2011.04611.x
- J. A. Morgan and D. Rhodes, “Mathematical Modeling of Plant Metabolic Pathways,” Metabolic Engineering, Vol. 4, No. 1, 2002, pp. 80-89. doi:10.1006/mben.2001.0211
- G. O. Osuji and W. C. Madu, “Ammonium Ion-Dependent Isomerization of Glutamate Dehydrogenase in Relation to Glutamate Synthesis in Maize,” Phytochemistry, Vol. 39, No. 3, 1995, pp. 495-503. doi:10.1016/0031-9422(94)00976-Z
- G. O. Osuji and W. C. Madu, “Regulation of Peanut Glutamate Dehydrogenase by Methionine Sulphoximine,” Phytochemistry, Vol. 46, No. 5, 1997, pp. 817-825. doi:10.1016/S0031-9422(97)00395-6
- G. O. Osuji, A. S. Mangaroo, J. Reyes, A. Bulgin and V. Wright, “Biomass Enhancement in Maize and Soybean in Response to Glutamate Dehydrogenase Isomerization,” Biologia Plantarum, Vol. 47, 2003, pp. 45-52. doi:10.1023/A:1027324713682
- G. O. Osuji, T. K. Brown, S. M. South, J. C. Duncan and D. Johnson, “Doubling of Crop Yield through Permutation of Metabolic Pathways,” Advances in Biosciences and Biotechnology, Vol. 2, No. 5, 2011, pp. 364-379. doi:10.4236/abb.2011.25054
- G. O. Osuji, T. K. Brown and S. M. South, “Optimized Fat and Cellulosic Biomass Accumulation in Peanut Through Biotechnology,” International Journal Biotechnology & Biochemistry, Vol. 6, 2010, pp. 455-476.
- A. J. Norden, O. D. Smith and D. W. Gorbet, “Breeding of the Cultivated Peanut,” In: H. E. Pattee and C. T. Young, Eds., Peanut Science and Technology, American Peanut Research and Education Society, Inc., Yoakum, 1982, pp. 95-123.
- G. O. Osuji, J. Konan and G. M’Mbijjewe, “RNA Synthetic Activity of Glutamate Dehydrogenase,” Applied Biochemistry and Biotechnology, Vol. 119, No. 3, 2004, pp. 209-228. doi:10.1007/s12010-004-0003-z
- D. Grierson, A. Slater, J. Speirs and G. A. Tucker, “The Appearance of Polygalacturonase mRNA in Tomatoes,” Planta, Vol. 163, No. 2, 1985, pp. 263-271. doi:10.1007/BF00393517
- G. O. Osuji, T. K. Brown and S. M. South, “Nucleotide-Dependent Reprogramming of mRNAs Encoding Acetyl Coenzyme A Carboxylase and Lipoxygenase in Relation to the Fat Contents of Peanut,” Journal of Botany, 2009. doi:10.1155/2009/278324
- G. O. Osuji and T. Brown, “Environment-Wide Reprogramming of mRNAs Encoding Phosphate Translocator and Glucosyltransferase in Relation to Cellulosic Biomass Accumulation in Peanut,” The ICFAI Journal Biotechnology, Vol. 1, No. 4, 2007, pp. 35-47.
- G. O. Osuji, A. S. Mangaroo and P. S. Roberts, “In Vitro Isomerization of Glutamate Dehydrogenase in Relation to Phytosequestration of Lead,” SAAS Bulletin, Biochemistry and Biotechnology, Vol. 14, 2001, pp. 60-72.
- D. Cammaerts and M. Jacobs, “A Study of the Polymorphism and the Genetic Control of the Glutamate Dehydrogenase Isoenzymes in Arabidopsis Thaliana,” Plant Science Letters, Vol. 31, No. 1, 1983, pp. 67-73. doi:10.1016/0304-4211(83)90130-X
- G. Osuji, T. K. Brown and S. M. South, “Discovery of RNA Synthetic Activity of Glutamate Dehydrogenase and its Application in Drug Metabolism Research,” The Open Drug Metabolism Journal, Vol. 2, 2008, pp. 1-13. doi:10.2174/1874073100802010001
- S. Knappe, U. Flugge and K. Fischer, “Analysis of the Plastidic Phosphate Translocator Gene Family in Arabidopsis and Identification of New Phosphate Translocator-Homologous Transporters, Classified by their Putative Substrate-Binding Site,” Plant Physiology, Vol. 131, No. 3, 2003, pp. 1178-1190. doi:10.1104/pp.016519
- M. G. James, K. Denyer and A. M. Myers, “Starch Synthesis in the Cereal Endosperm,” Current Opinion in Plant Biology, Vol. 6, No. 3, 2003, pp. 215-222. doi:10.1016/S1369-5266(03)00042-6
- E. J. Davis, I. J. Tetlow, C. G. Bowsher and M. J. Emes, “Molecular and Biochemical Characterization of Cytosolic Phosphoglucomutase in Wheat Endosperm,” Journal Experimental Botany, Vol. 54, No. 386, 2003, pp. 1351- 1360. doi:10.1093/jxb/erg151
- M. Swissa, Y. Aloni, H. Weinhouse and M. Benizman, “Intermediary Steps in Acetobacter Xylinum Cellulose Synthesis: Studies with Whole Cells and Cell-Free Preparations of the Wild Type and Celluloseless Mutant,” Journal of Bacteriology, Vol. 143, 1980, pp. 1142-1150.
- L. Peng, K. Yasushi, H. Pat and D. Deborah, “Sitosterol-β-Gluciside as Primer for Cellulose Synthesis in Plants,” Science, Vol. 295, No. 5552, 2002, pp. 147-150. doi:10.1126/science.1064281
- I. M. Saxena and R. M. Brown, “Identification of Cellulose Synthase(s) in Higher Plants: Sequence Analysis of Processive β-Glycosyltransferases with the Common Motif ‘D, D, D35Q(R,Q)XRW’,’’ Cellulose, Vol. 4, No. 1, 1977, pp. 33-49. doi:10.1023/A:1018411101036
- W. B. Parker, L. C. Marshall, J. D. Burton, D. A. Somers, D. L. Wyse, J. W. Gronwald and B. G. Gengenbach, “Dominant Mutations Causing Alterations in AcetylCoenzyme a Carboxylase Confer Tolerance to Cyclohexanedione and Aryloxyphenoxypropionate Herbicides in Maize,” Proceedings National Academy Science, Vol. 87, No. 18, 1990, pp. 7175-7179. doi:10.1073/pnas.87.18.7175
- P. M. C. Smith and C. A. Atkins, “Purine Biosynthesis: Big in Cell Division, Even Bigger in Nitrogen Assimilation,” Plant Physiology, Vol. 128, No. 3, 2002, pp. 793- 802. doi:10.1104/pp.010912
- K. M. Schnorr, P. Nygaard and M. Laloue, “Molecular Characterization of Arabidopsis thaliana cDNAs Encoding three Purine Biosynthetic Enzymes,” The Plant Journal, Vol. 6, No. 1, 1999, pp. 113-121. doi:10.1046/j.1365-313X.1994.6010113.x
- A. Kleinhofs, R. L. Warner, J. M. Lawrence, J. M. Melzer and D. A. Kudrna, “Molecular Genetics of Nitrate Reductase in Barley,” In: J. L. Wray and J. R. Kinghorn, Eds., Molecular and Genetic Aspects of Nitrate Assimilation, Oxford Science Publications, Oxford, 1989, pp. 197- 211.
- C. P. Vance, S. S. Miller, R. G. Gregerson, D. A. Samac, D. L. Robinson and J. S. Gantt, “Alfalfa NADH-Dependent Glutamate Synthase: Structure of the Gene and Importance in Symbiotic N2 Fixation,” Plant Journal, Vol. 8, No. 3, 1995, pp. 345-358. doi:10.1046/j.1365-313X.1995.08030345.x
- L. Husted,” Cytological Studies on the Peanut, Arachis II Chromosome Number, Morphology and Behavior and Their Application to the Origin of Cultivated Forms,” Cytologia, Vol. 7, No. 3, 1936, pp. 396-423. doi:10.1508/cytologia.7.396
- U. Klahre, S. A. Leuenberger, V. A. Iglesias and F. Meins, “High Molecular Weight RNAs and Small Interfering RNAs Induce Posttranscriptional Gene Silencing in Plants,” Proceedings of the National Academy of Sciences of the USA, Vol. 10, 2002, pp. 1973-1078.
- C. A. Perez-Novo, C. Claeys, F. Speleman, P. V. Cauwenberge, C. Bachert and J. Vandesompele, “Impact of RNA Stability on Reference Gene Expression Stability,” Biotechniques, Vol. 39, No. 1, 2005, pp. 52-56. doi:10.2144/05391BM05
- D. Q. Fuller, “Contrasting Patterns in Crop Domestication and Domestication Rates: Recent Archaeobotanical Insights from the Old World,” Annals of Botany (London), Vol. 100, No. 51, 2007, pp. 903-924. doi:10.1093/aob/mcm048
- K. O. Burkey, F. L. Booker W. A. Pursley and A. S. Heagle, “Elevated Carbon Dioxide and Ozone Effects on Peanut: II. Seed Yield and Quality,” Crop Science, Vol. 47, No. 4, 2007, pp. 1488-1497. doi:10.2135/cropsci2006.08.0538
- P. Horton, “Prospects for Crop Improvement Through the Genetic Manipulation of Photosynthesis: Morphological and Biochemical Aspects of Light Capture,” Journal of Experimental Botany, Vol. 51, No. 1, 2000, pp. 475-485. doi:10.1093/jexbot/51.suppl_1.475
- R. A. Richards, “Selectable Traits to Increase Crop Photosynthesis and Yield of Grain,” Journal of Experimental Botany, Vol. 51, No. 1, 2000, pp. 447-458. doi:10.1093/jexbot/51.suppl_1.447
NOTES
*Corresponding author.

