American Journal of Plant Sciences
Vol.3 No.2(2012), Article ID:17540,12 pages DOI:10.4236/ajps.2012.32023
Gene Expression Profiling during Wilting in Chickpea Caused by Fusarium oxysporum f. sp. Ciceri
![]()
Plant Molecular Biology Unit, Division of Biochemical Sciences, National Chemical Laboratory, Pune, India.
Email: *vs.gupta@ncl.res.in
Received August 10th, 2011; revised September 20th, 2011; accepted November 3rd, 2011
Keywords: Cicer Arietinum; Fusarium oxysporum; Cdna-RAPD; Semi-Quantitative RT-PCR
ABSTRACT
Fusarium oxysporum f. sp. ciceri (Foc), one of the most important fungal pathogen of chickpea, is a constant threat to this crop plant. In the present study gene expression analysis of chickpea roots during Foc infection was performed using various approaches. cDNAs derived from total mRNA during infection process of susceptible (JG62) and resistant (Digvijay) cultivars, were amplified using random oligonucleotides. Sequence characterization of differentially expressed transcripts revealed their homology with many plant genes essential for various metabolic functions including defense. Further, expression patterns of specific candidate gene transcripts were analyzed in the Foc inoculated and uninoculated resistant and susceptible chickpea cultivars, on day 6 of infection. Semiquantitative RT-PCR analysis of defense related genes was performed using gene specific oligonucleotides in resistant and susceptible chickpea cultivars. The expression of fungal pathogenesis related genes and their race specific response was determined throughout the course of chickpea-Foc interaction. Temporal expression and race specific response of plant defense related and fungal virulence genes were studied in the resistant and susceptible cultivars of chickpea inoculated with three races of Foc highlighting the host-pathogen interactions. Few genes, involved in chickpea defense against Fusarium wilt which were not reported previously were unveiled in this study.
1. Introduction
Chickpea (Cicer arietinum L.) is the most abundantly grown legume in India, which contributes to 64% of the world production and serves as an important source of dietary protein for the Indian vegetarian population [1]. Chickpea plants are affected by various biotic and abiotic stresses. Fungal diseases, especially Fusarium wilt and Ascochyta blight are the important biotic factors for major yield loss of this crop worldwide. The causative agent of Fusarium wilt, Fusarium oxysporum is a soil borne, facultative, vascular wilt fungus that provokes economically important losses in approximately 80 crops including chickpea [2]. Individual pathogenic strains within the species; having an ability to infect a particular host range have been assigned to intraspecific groups called forma speciales [3]. Some of the forma speciales are further divided into races based on differential virulence to a set of cultivars e.g. F. oxysporum f. sp. ciceri (Foc) infecting chickpea. Similar to other soil borne diseases, various strategies have been employed for controlling Fusarium wilt, such as, use of chemical fungicides, biological control, etc., but most of these strategies have proven ineffective or have hazardous effect. Widely accepted and cost effective strategy is to develop and use wilt resistant cultivars [4].
A complex interaction between plant and its fungal pathogen is an outcome of expression of both, plant defense genes as well as fungal pathogenesis related genes. The result of such a relationship is projected as either resistance or disease development in the plant. There are multiple events involved that lead to successful plant defense during pathogen attack. Further, these defense mechanisms are governed by an array of genes, which are either singly or synergistically, involved in plant resistance traits. Many defense related genes have been cloned and characterized in an attempt to elucidate the mechanism of defense upon F. oxysporum attack in various plant species, including chickpea. In our earlier studies, enzymes like glucanases, chitinases and proteases have been shown to be probably involved in chickpea defense against Foc infection [5]. Further, various up-regulated transcript derived fragments like 14-3-3, WRKY and NBS-LRR type sequences as well as transposable elements, have been identified using cDNA-RAPD and cDNA-AFLP techniques [6]. However, exact molecular mechanisms involved in chickpea resistance are still unexplored.
Understanding the pathogenicity mechanism of fungi, on the other hand, demands knowledge of the virulence factors, which are active in the host environment. Till date, many pathogens have been studied in context to their virulence and various genes with a prime role in fungal pathogenesis have been identified. Various G protein subunits have been reported to be necessary for fungal morphogenesis, development as well as virulence [7]. A few genes specific for the virulence of Fusarium spp. have been identified. Chitin synthase genes [8] and transcription factors like Ftf1 [9] are essential for virulence of Fusarium. Since then, contribution of few more genes in pathogenesis of F. oxysporum has been established. Specifically, in F. oxysporum f. sp. lycopersici, genes like Fow1 and Fow2 [10,11]; Six1, Six2, and SSH1 [12] have been found to be involved in its virulence. However, indepth search of mechanisms and the genes involved in pathogenicity is essential in wilt causing root pathogens.
Race specificity is an important criterion in fungal pathogenesis, and plant defense response is dependent on it. In this study, we have analyzed the race specific expression of various plant and fungal genes and identified various up-regulated transcripts using several approaches. Crop plants are constantly exposed to their pathogens in the field which has directed us to consider a wide timescale (2 - 16 dai) for the expression analysis of plant and fungal genes involved in the chickpea-Foc interaction. An attempt has also been made to analyze the expression pattern of four important fungal genes previously reported to be essential for growth and pathogenesis, namely Fgb1 (G protein subunit), Gas1 (glucanosyltransferase), chs7 (chitin synthase chaperonin) and Fow1 (mitochondrial carrier protein). The host-pathogen interaction using chickpea-Foc system has been unveiled in this study.
2. Materials and Methods
2.1. Fungal Cultures
Standard races of Foc namely,1(NRRL 32153), 2 (NRRL 32154) and 4 (NRRL 32156) were obtained from the International Crops Research Institute for Semi-Arid Tropics (ICRISAT), Patancheru, Andhra Pradesh, India, where they were already characterized and classified for their race specificity using conventional method of race identification. Molecular diversity and race specificity of these cultures were established in our laboratory [13]. The cultures were maintained on Potato Dextrose Agar (PDA) slants with timely sub-culturing and infection to a susceptible cultivar, JG62.
2.2. Plant Material
C. arietinum seeds of cultivars, JG62 and Digvijay, used in this study were obtained from Mahatma Phule Krishi Vidyapeeth (MPKV), Rahuri, Maharashtra, India. Cultivar JG62 (pedigree-selection from germplasm) is susceptible while Digvijay (pedigree-Phule G-91028 × Bheema) is resistant to Fusarium wilt.
2.3. Seed Germination and Inoculation
Seeds were wrapped in wet sterile muslin cloth and stored at room temperature (24˚C to 26˚C) till sprouting. The sprouted seeds were transferred to trays containing sterile water with macroand micro-nutrients (half strength Hoagland’s nutrient medium) [6] and kept at 22˚C and 60% relative humidity under normal day conditions (14 h light/10 h dark). Freshly prepared spore suspension (1 × 106 spores/mL) of Foc races 1, 2 and 4 was added individually to the sterile hydroponic trays containing seven days old chickpea plants. Seedlings grown in similar trays with no pathogen (un-inoculated plants) served as control. The pathogenicity assays were conducted in triplicates.
2.4. Sample Collection and RNA Extraction
Total RNA was extracted from chickpea roots collected at various time intervals such as 2, 6, 9, 13 and 16 days after inoculation (DAI) using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) as described by Nimbalkar et al. (2006) [6]. The DNase treated RNA was used for the first strand cDNA sysnthesis using RT-PCR kit (Promega, Madison, WI, USA). Undiluted or 1:100 diluted cDNA was used for PCR using RAPD oligonucleotides (OPAD and OPAE series, Operon technologies, Huntsville, AL, USA) or gene specific oligonucleotides.
2.5. Cloning of Amplified cDNA Fragments and Their Sequence Characterization
cDNAs isolated from Foc inoculated and uninoculated JG62 and Digvijay plant roots were used as templates for amplification with RAPD oligonucleotides (OPAD and OPAE series, Operon technologies, USA). The amplified product from cDNA of resistant inoculated cultivar, Digvijay was cloned in pGEM-t easy vector (Promega, USA). Cloned cDNA fragments were sequenced by dideoxy termination method and the sequences were determined using an automated DNA sequencer (MegaBACE1000, Amersham Biosciences, NJ, USA). The nucleotide sequences were compared with the reported nucleotide sequences in Genbank nonredundant database using the BLAST sequence alignment program [14]. Oligonucleotides designed from amplified sequences were used for expression analysis of transcripts in inoculated and un-inoculated cDNA pool of Digvijay and JG62 root tissues. Plant specific cDNA pool was normalized using 18s rRNA oligonucleotides while fungal specific cDNA pool was normalized using ITS oligonucleotides.
2.6. Expression Analysis of Known Specific Genes
Gene specific oligonucleotides were designed from the conserved regions of plant defense related genes and fungal virulence related genes using sequences available in the NCBI Genbank database (database-fungi and databaseFabaceae) (http://www.ncbi.nlm.nih.gov) (Table 1).
The semi-quantitative RT-PCR conditions were as follows: initial denaturation at 94˚C for 5 min, followed by 20 to 25 cycles of 94˚C for 1 min denaturation, 55˚C

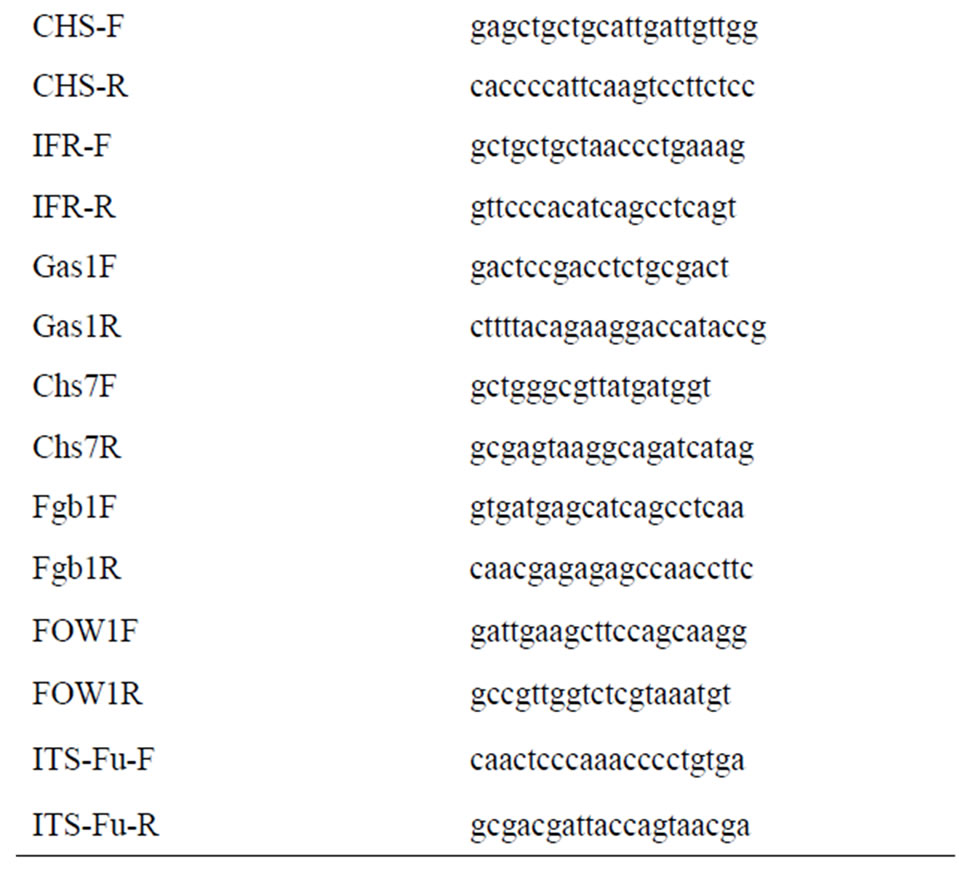
Table 1. Primers used for expression analysis of chickpeaFoc transcriptome.
for 1 min annealing and 72˚C for 1 min extension. Final extension was carried out at 72˚C for 7 min. The annealing temperature was varied according to the sequence of the oligonucleotide used. PCR products were visualized on 1.5% agarose gel by electrophoresis in TAE buffer system. Template used for this analysis was the cDNA from Digvijay and JG62 uninoculated and inoculated with Foc race 1, 2 and 4.
2.7. Genbank Accession Numbers
Nucleotide sequence of chickpea defense related genes namely, GroES2 (Genbank acc. no. GW342895), gene for 60s ribosomal protein (Genbank acc. no. GW342896), BetvI (Genbank acc. no. GW342897), CHS (Genbank acc. no. GW342898) and IFR (Genbank acc. no. GW342899) as well as Fusarium pathogenesis related genes namely, Fgb1 (Genbank acc. no. GW342900), Gas1 (Genbank acc. no. GW342901), Chs7 (Genbank acc. no. GW342902) and Fow1 (Genbank acc. no. GW342903) were deposited in NCBI database.
3. Results and Discussion
3.1. Pathogenecity Assays
Seven days old chickpea plants were inoculated with fungal spores of Foc races individually and uninoculated plants served as control. In general, Digvijay cultivar was found to grow slower as compared to JG62 cultivar. Upon infection JG62 showed wilting symptoms by 10 dai, while no wilting was observed in Digvijay even by 25 dai (till the plants were allowed to grow hydroponically). Within Foc infected JG62; race 1 infected JG62 showed wilting symptoms earlier than race 4 followed by race 2. Figure 1 shows the chickpea cultivars, JG62 and Digvijay, uninoculated (control) and race 1, 2 and 4 inoculated (showing wilting symptoms). Root tissues of both the cultivars (uninoculated and inoculated with Foc races) were further used for gene expression studies from several biological as well as technical replicates. For RAPD analysis Foc1 infected tissue at 6 dai, was used while for candidate gene expression studies Foc1, 2 and 4 inoculated tissues were analyzed at different time points from 2 - 16 dai.
3.2. cDNA-RAPD of Inoculated Resistant and Susceptible Chickpea Cultivars
Previously, Nimbalkar et al. (2006) [6] reported cDNARAPD and cDNA-AFLP analysis with Foc1 inoculated susceptible (JG62) and resistant (Vijay) chickpea varieties from our lab. In this study, the expression of transcription factors such as WRKY proteins, 14-3-3 proteins, and NBS-LRR-type gene sequences and other genes such as gamma-glutamyl-cysteine synthetase during chickpeaFoc1 interaction was analyzed.
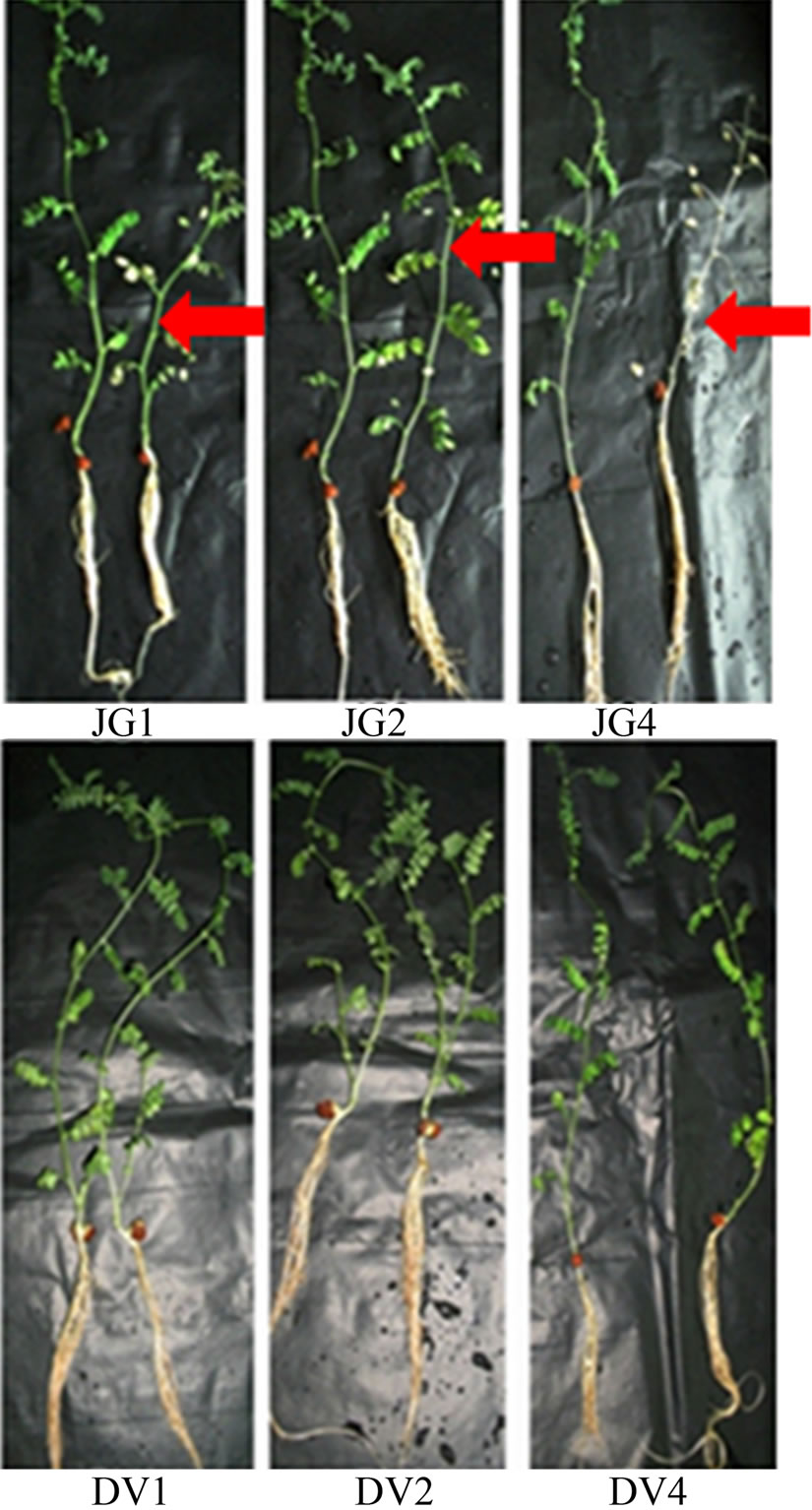
Figure 1. Foc race infected and uninfected JG62 and Digvijay chickpea cultivars. JG1, JG2 and JG4 represent Foc race 1, 2 and 4 inoculated JG62 plants (right), respectively in comparison to control plants (left). DV1, DV2 and DV4 represent Foc race 1, 2 and 4 inoculated Digvijay plants (right), respectively in comparison to control plants (left). Red arrows indicate wilted chickpea plants.
To explore these interactions further, transcript profiling of root cDNAs obtained from Foc1 inoculated JG62 and Digvijay (more resistant to Fusarium wilt than cultivar Vijay) cultivars at 6 dai, was accomplished, using RAPD oligonucleotides of OPAD and OPAE series (20 oligonucleotides each) in the present study. Differentially expressed and reproducible 134 transcripts exhibiting >200 bp size were selected for cloning and 117 clones thus obtained, were further sequence characterized. Based on homology search and the E-values, these clones represented 65% of plant (related to defense, shock, structural and DNA/protein synthesis) and 18% of fungal (related to metabolism, regulatory function and transposon) origin genes, respectively, which were not identified in our previous study (Table 2). About 17% of the transcripts were found to be novel in nature.
As exhibited in Table 2 one of these clones was homologous to C3HC4 type zinc finger protein of A. thaliana. Though the exact function of zinc finger protein in plant pathogen interactions is yet unknown; in a few hostpathogen interaction studies conducted till date, an upregulation of this gene has been consistently observed [15]. Three fungal genes represented in this study were homologous to Trichothecene 3-O-acetyltransferse (Tri), fungal transposase and fungal regulatory protein, respectively. Altogether, 27% of the clones were homologous to DNA/ protein synthesis related proteins such as histone proteins and ribosomal proteins; while 7% showed high homology to chaperonins, namely GroES2 and Hsp70. Further, a class of clones obtained from inoculated Digvijay roots revealed significant homologies to glycosyltransferase and methionine sulphoxide reductase (Msr) genes. A strong homology of one of the transcripts was observed to a ripening related protein of chickpea. This sequence information was used to design oligonucleotides, which were then deployed for expression analysis of the identified genes in inoculated and uninoculated, Digvijay and JG62 plants.
3.3. Involvement of Important Fungal and Plant Specific Genes during Foc Wilt
All the oligonucleotides designed from the sequence information obtained as above were used for semi-quantitative RT-PCR in case of JG62 and Digvijay inoculated with Foc race 1. However, only eight of these oligonucleotide pairs could reveal consistently reproducible and differential amplification profiles and were used for further analysis. Figure 2 shows the differential expression pattern of plant defense related and fungal specific transcripts of Digvijay and JG62 at 6 dai. Although resistant cultivar Digvijay showed no wilting symptoms upon Foc inoculation, fungal genes were found to be expressed; indicating the fungal growth and proliferation in the resistant cultivar. In this study, fungal Tri101 gene was found to be upregulated during Foc-chickpea interaction (Figure 2), although not all the species of the genus Fusarium express trichothecene. However, some species have been reported to harbour both, functional and nonfunctional copies of Tri genes [16]. Semi quantitative RT-PCR revealed the expression of transposase gene during fungal invasion of chickpea roots (Figure 2). Transcription of fungal transposons is known to occur during carbon or nitrogen starvetion condition; which mimics the pathogenesis conditions in fungi [17].
Apart from the above mentioned fungal genes, several plant genes were found to express in Digvijay upon Foc inoculation. Histone proteins are involved in chromosome duplication of eukaryotic genome and are shown to be overexpressed in plants during pathogen attack [18].
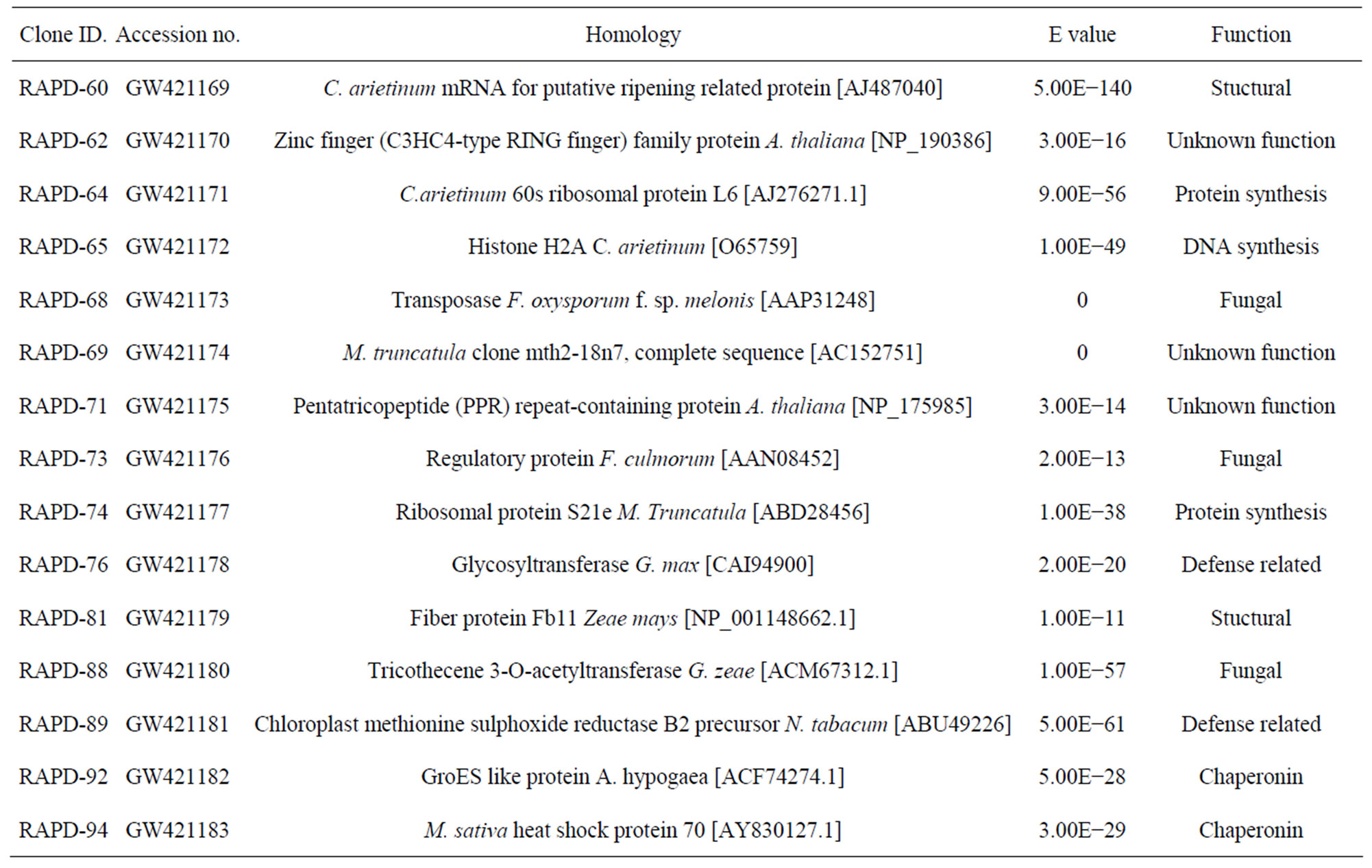
Table 2. Homology of cDNA clones generated using random primers with reported sequences in NCBI database.
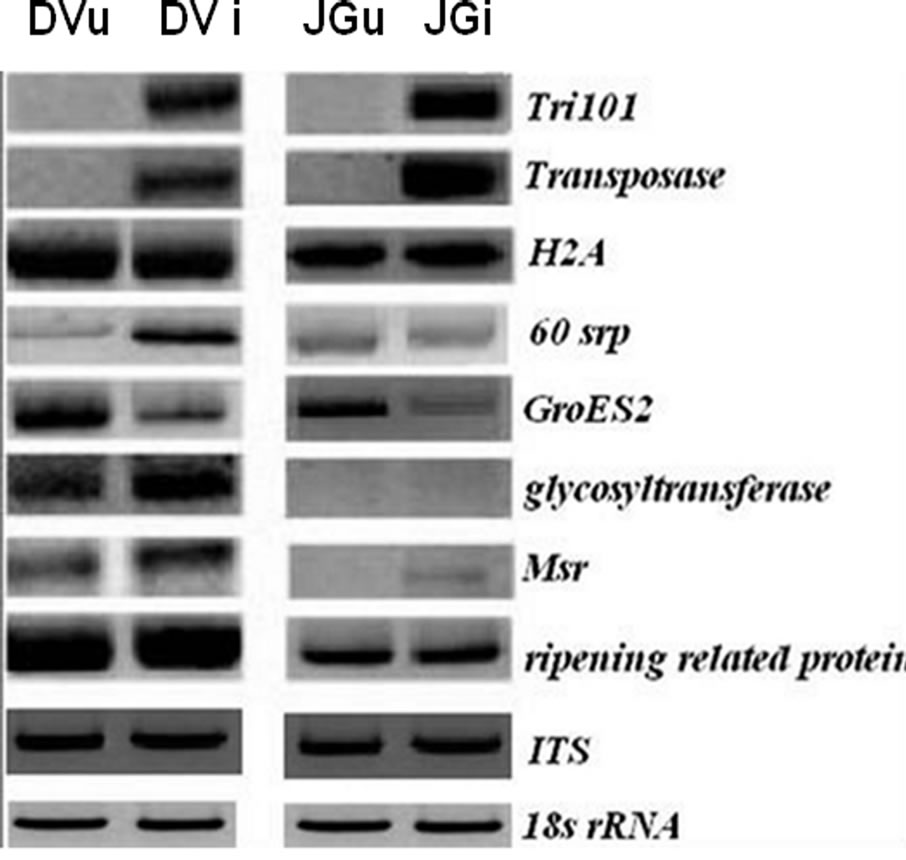
Figure 2. Expression of transcripts in non-inoculated and Foc1 inoculated Digvijay (DVu and DVi) and JG62 (JGu and JGi) chickpea cultivars at 6 dai. Tri101 (tricothecene 3-O-acetyltransferase), transposase, H2A (histone protein 2), 60srp (gene for 60s ribosomal protein), GroES2 (chaperonin), glycosyltransferase, Msr (methionine sulfoxide reductase), ripening related protein, ITS (Intertranscribed spacer region) and 18s rRNA (for cDNA normalization). Semi-quantitative RT-PCR was performed at 20 to 25 cycles depending upon the specific genes and their amplified products.
In the present study, expression of H2A histone protein was found to be unaffected upon infection (Figure 2), while previous reports showed an increase in histone protein level upon fungal infection in Petroselinum crispum [19]. On the other hand, gene for 60s ribosomal protein (60srp) was found to be abundantly expressed in the inoculated Digvijay plants, suggesting active changes in metabolism of inoculated chickpea roots (Figure 2). It is likely, since the higher need of protein synthesis arises during the plant defense [15]. Similar results were obtained by McFadden et al. (2004) [20], where they showed an increase in ribosomal protein transcript level during cotton root infection by F. oxysporum f. sp. vasinfectum.
Glycosyltransferase gene was upregulated in inoculated Digvijay plants as compared to its expression in healthy plants, suggesting its role in mounting defense against pathogen attack (Figure 2). This is concordant with the information available from tomato and tobacco plants wherein, glycosyltransferases have been shown to respond rapidly to signals from wounds and pathogen attack [21,22]. In inoculated roots of chickpea plants, upregulation in the expression of Msr gene was observed (Figure 2). In earlier reports also, Cauliflower mosaic virus (CMV) exposed Arabidopsis leaves showed a strong induction of the plastidic Msr gene (c-pmsr) after 2 to 3 weeks of chronic pathogen infection [23].
Ripening related proteins are involved in fruit development and ripening. However, these proteins are also thought to protect the developing fruit from pathogen attack and are expressed during symbiotic relationships in plants [24]. A slight increase in the transcripts of ripening related protein was observed, in the Foc inoculated chickpea cultivars as compared to uninoculated one, projecting its involvement in plant defense (Figure 2). However, the exact role of this protein during pathogen attack needs to be justified. Chaperonin molecules are reported to be essential to the plants during biotic and abiotic stress conditions [25]. However, in our studies GroES2 chaperonin was found to be downregulated in Foc inoculated chickpea plants as compared to the uninoculated plants at 6 dai (Figure 2).
3.4. Plant Defense Related Gene Expression in Chickpea upon Inoculation with Foc Races 1, 2 and 4
Variation in the expression pattern of specific genes observed in the above studies led us to perform detailed analysis of expression of candidate plant defense genes in order to understand the contribution of these genes in plant defense. For this various stages of disease development in the susceptible and resistant chickpea cultivars, JG62 and Digvijay, respectively, inoculated with all the three races of Foc, namely Foc1, 2 and 4 were used. As mentioned previously, chickpea root cDNA, control as well as Foc race 1, 2 and 4 inoculated, were normalized using 18s rRNA oligonucleotides. The normalized cDNA samples were then used for candidate gene study using plant defense related gene oligonucleotides. While evaluating the expression of various defense related genes in Foc1, 2 and 4 inoculated JG62 and Digvijay, the constitutive expression of these genes in uninoculated plants was negated from the inoculated plants, respectively (Figures 3 and 4).
3.4.1. GroEs2, 60srp and Betvi Expression in Chickpea upon Inoculation with Foc Races 1, 2 and 4
3.4.1.1. BetvI Gene
Pathogenesis related proteins (PR proteins) are reported to be extensively involved in chickpea defense against various pathogens [26]. In our study, PR2, PR5 and PR10 as well as BetvI which is known to show signifycant homologies to PR10 proteins and is a major pollen allergen of several plant species [27-30] were screened for their reproducible, race specific and prolonged expression (2 - 16 dai) in both the chickpea cultivars.
Except BetvI, all the other PR proteins could not meet the above mentioned selection criteria. In inoculated plants, BetvI expression was pronounced around early stages of infection i.e. 2 dai in both, the resistant and susceptible cultivars, and the expression level dropped in both the cases by 13 dai (Figure 3). This is in accordance with the previous studies conducted by Foster-Hartnett et al. (2007) [32] where, during infection by Erysiphe pisi, resistant and susceptible cultivars of Medicago truncatula showed transcript accumulation. By 6 dai only Foc1 inoculated JG62 showed higher expression as compared to Foc2 and 4 inoculated JG62. In Foc1, 2 and 4 inoculated resistant cultivar, Digvijay, higher gene expression was observed at 2 dai, i.e. in the earlier stage of infection,
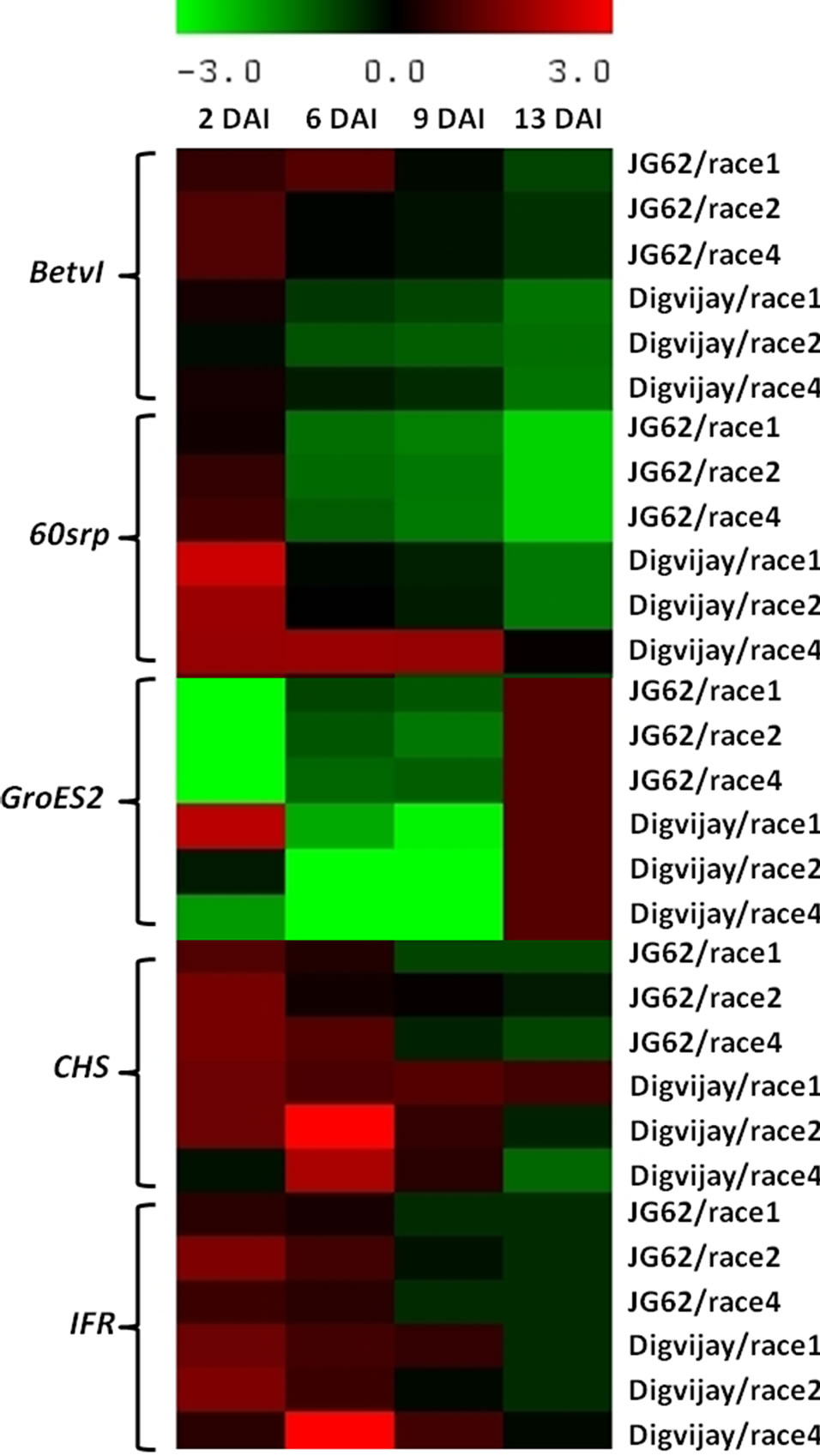
Figure 3. Heat map depicting the expression of plant defense related genes in Foc inoculated resistant and susceptible cultivars. While generating the heat map of various defense related genes in Foc1, 2 and 4 inoculated JG62 and Digvijay, the constitutive expression of these genes in control plants was negated from the inoculated plants, respecttively at all the time points of the study (Based on relative density index values). Heat map was generated using Multiexperiment viewer software [31].
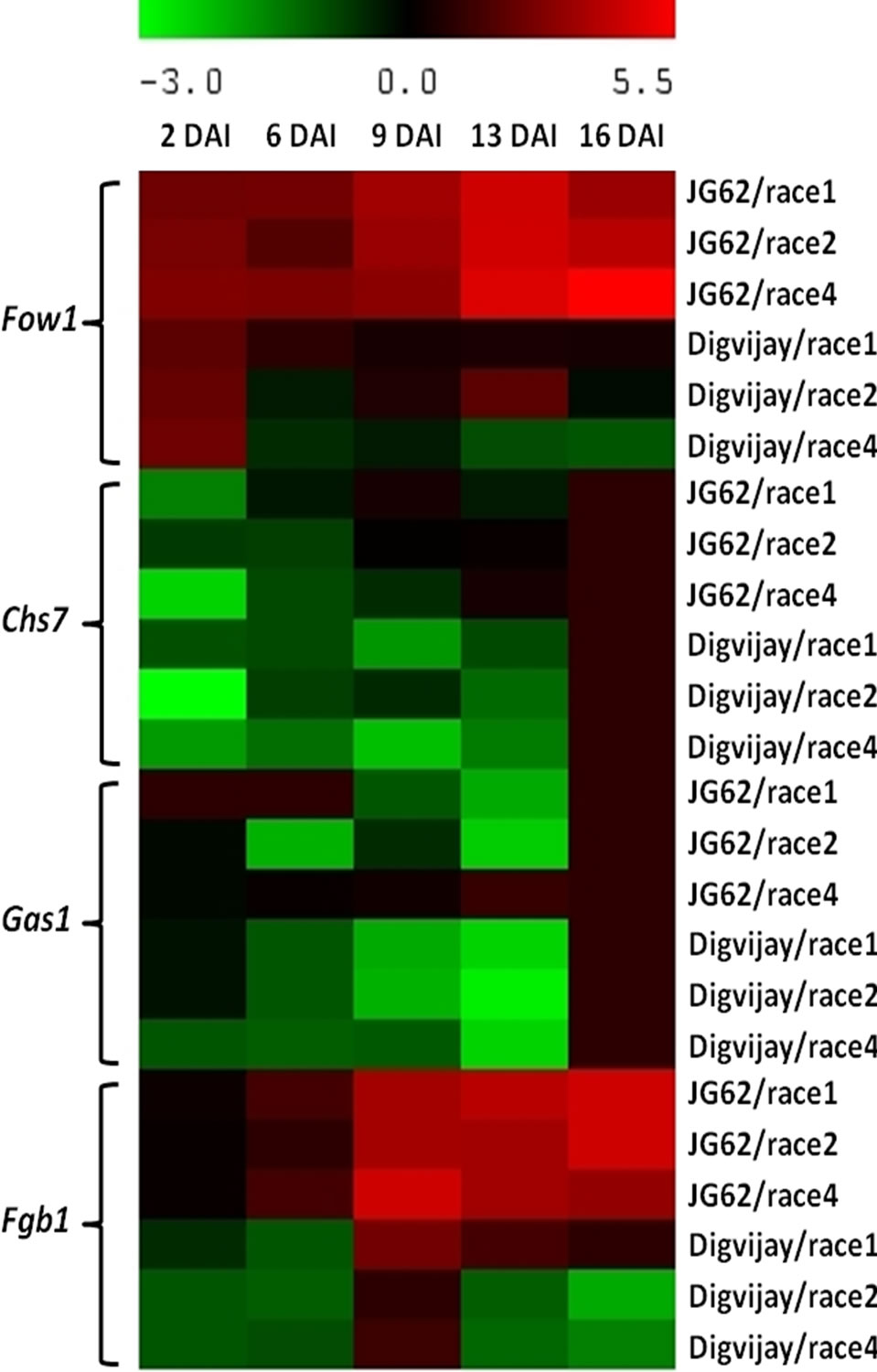
Figure 4. Heat map depicting the expression of fungal virulence related genes in Foc inoculated resistant and susceptible cultivars. While generating the heat map of various virulence related genes in Foc1, 2 and 4 inoculated JG62 and Digvijay, the constitutive expression of these genes in control plants was negated from the inoculated plants, respectively at all the time points of the study (Based on relative density index values). Heat map was generated using Multiexperiment viewer software [31].
which declined by 6 dai. Interestingly, BetvI showed the highest expression in the early stages of infection (2 dai) and that too in susceptible cultivar, JG62 inoculated with Foc races (Figure 3).
3.4.1.2. Gene for 60s Ribosomal Protein (60srp)
In Foc1, 2 and 4 inoculated chickpea expression of 60srp gene was the highest during early stage of infection followed by a gradual decline as the disease progressed. By 9 dai, 60srp gene expression was seen only in the resistant cultivar inoculated with Foc race 4 (Figure 3). In earlier reports also, consistent upregulation of 60srp gene transcripts was observed within mycorrhizal root tissues of tomato plants (at 10 dai) and during arbuscular mycorrhizal symbiosis of M. truncatula [33,34]. In comparative analysis, Foc challenged Digvijay showed an elevated expression of 60srp gene as compared to that in JG62 (Figure 3). However, Foc4 challenged Digvijay showed higher 60srp gene response as compared to Foc1 followed by Foc2.
3.4.1.3. GroES2 Gene
As mentioned previously, chaperonins such as GroES2 are essential for plant function during stress conditions including pathogen attack. When inoculated with Foc races chickpea GroES2 chaperonin gene was found to express at 2 dai in race 1 inoculated Digvijay. After this the transcripts’ level declined drastically till 9 dai, however, the expression was maximum at 13 dai (Figure 3). Similarly, in Foc inoculated JG62, GroES2 expression was initially weak but showed an increase at 6 - 9 dai and was the maximum at 13 dai (Figure 3).
3.4.2. Expression of Major Genes of Phenylpropanoid Pathway in Chickpea upon Inoculation with Foc Races 1, 2 and 4
Phenolic compounds like phytoanticipins and phytoalexins are important plant secondary metabolites; which have been suggested to play diverse roles in plant defense against phytopathogens [35,36]. Medicarpin and Maakianin are the major phytoalexins involved in chickpea defense and are produced through phenylpropanoid pathway [37, 38]. Chalcone synthase (CHS) and Isoflavone reductase (IFR) are the two key enzymes that are involved in isoflavonoid production against biotic stresses.
3.4.2.1. Chalcone Synthase (CHS) Gene
In this study, expression of both, CHS and IFR, was analyzed against Foc inoculation in chickpea. When JG62 and Digvijay inoculated with Foc1, 2 and 4 were compared for CHS expression; it was higher at 2 - 9 dai with a decline by 13 dai in the resistant cultivar, while the expression steadily decreased with the progression of disease in the susceptible cultivar. This decline was pronounced only in the case of race 2 and 4 inoculated Digvijay at 13 dai. However, interestingly Foc2 inoculated Digvijay showed the highest CHS gene expression at 6 dai when compared to the expression level of both, JG62 and Digvijay, inoculated with Foc races (Figure 3).
3.4.2.2. Isoflavone Reductase (IFR) Gene
A constitutive expression of IFR gene was detected in all the analyzed root tissues. In general, expression level was higher in the resistant cultivar as compared to that in the susceptible one, in the inoculated tissues. Previous results obtained by Arfaoui et al. (2007) [39] supported this data; where they also showed an increase in IFR transcripts in resistant line suggesting the role of fungicidal isoflavones in the restriction of pathogen infection.
Foc1 inoculated Digvijay showed higher level of IFR gene expression than JG62 upto 9 dai while by 13 dai no expression difference was observed in Digvijay and JG62. Foc2 inoculated Digvijay and JG62 showed similar IFR gene expression profile at 2 - 13 dai. In case of Foc4 inoculated Digvijay, expression was maximum at 6 dai, while it declined by 13 dai (Figure 3).
3.5. Highlighting Chickpea Defense Response to Foc Races
It was observed that race 1 inoculated, resistant cultivar, Digvijay showed higher expression of defense related genes namely, GroES2, 60srp, CHS and IFR, except BetvI, as compared to the susceptible cultivar JG62, throughout the study (Figure 3). In case of the Foc inoculated plants, expression of CHS and IFR genes was higher in Digvijay as compared to JG62 throughout the course of disease development, indicating its importance in defense against Foc. However, CHS showed less expression initially at 2 dai in Foc4 inoculated Digvijay as compared to JG62, which enhanced at 6 and 9 dai where the expression of these two genes was higher in the resistant cultivar as compared to the susceptible cultivar JG62. In general, except for BetvI all the other genes namely, GroES2, 60srp, CHS and IFR showed higher expression in Digvijay as compared to JG62 almost till 13 dai in case of race 1, 2 and 4 indicating that these are involved in mounting defense against Foc attack.
3.6. Expression Analysis of Fungal Pathogenesis Related Genes from Foc
An array of fungal pathogenesis related genes have been reported till date which are involved in cell signaling, adhesion and appressoria formation, production of cell wall degrading enzymes (CWDEs) and toxins, etc. [40]. In the present study, expression of many of these genes during Foc infection to chickpea (both resistant and susceptible cultivars), was attempted, however, only a few genes could be successfully analyzed throughout the course of infection (2 to 16 dai). For example, genes encoding for CWDEs did not show a prolonged expression, while genes involved in appressoria formation could not be amplified in Foc. Thus, a few important genes essential for Foc infection were extensively studied, at various time points, throughout the course of infection. Foc1, 2 and 4 inoculated JG62 and Digvijay root cDNAs were normalized prior to candidate gene studies with fungal specific ITS oligonucleotides. All the cDNA showed uniform amplification with ITS oligonucleotdes. The oligonucleotides being specific to F. oxysporum, they did not show amplification in uninoculated chickpea cultivars. While evaluating the expression of various virulence related genes in Foc1, 2 and 4 inoculated JG62 and Digvijay, the expression of ITS gene in these plants was negated from expression of virulence genes in inoculated JG62 and Digvijay, at all the stages, respectively.
3.6.1. Fow1 Gene
Mitochondrial carrier protein Fow1, which is responsible for the transfer of tricarboxylates [41], has been reported to be essential for pathogenesis of F. oxysporum [10]. In the present study, Fow1 gene expression was initially strong in Foc1, 2 and 4 infecting both, JG62 and Digvijay. Gradually, with the disease progression; the expression level enhanced in case of Foc infecting susceptible cultivar and diminished in resistant cultivar by 16 dai (Figure 4).
In comparative analysis, Fow1 gene expression was found to be higher in case of race 4 infecting JG62, especially at 9 to 16 dai. In case of Foc infected Digvijay, though the expression was initially high at 2 dai, it declined sharply by 6 dai after which it was gained again by 9 dai and was maximum only in case of Foc2 infecting Digvijay at 13 dai (Figure 4).
3.6.2. Chs7 Gene
Chitin synthases (CSs) are integral membrane bound proteins that participate in the biosynthesis of chitin and are important for cell wall synthesis, hyphal growth and differentiation. Fungal CSs occur in specialized vesicles in cytosole called chitosomes [42] which are responsible for their transport from ER to the cell surface. Chitin synthase VII (Chs7) is a chaperonin like ER protein involved in export of CS from ER to cell surface.
In the present study Chs7 gene expression was initially weak, however, its prolonged expression was observed in case of Foc inoculated JG62 than in case of Foc inoculated Digvijay (Figure 4). In general, Chs7 gene expression was the highest at 16 dai in case of both, the resis tant and the susceptible cultivars.
3.6.3. Gas1 Gene
Glucanosyltransferases are essential for growth and morphogenesis of fungi and thus, are involved in fungal pathogenesis as well, when the fungus comes in contact with the host tissue [43]. The temporal expression of Gas1 revealed involvement of this gene only in the early stage of disease development and establishment. Gas1 was found to be expressing in Foc1 infecting JG62 at 2 to 6 dai and in Foc4 at 13 to 16 dai while Gas1 expression was observed in Foc inoculated Digvijay only at 16 dai. In case of race 1 inoculated JG62, Gas1 gene expression was observed at 16 dai as well (Figure 4).
3.6.4. Fgb1 Gene
F. oxysporum G protein subunit (Fgb1) has been reported to be essential for cell signaling, hyphal growth, conidiation as well as virulence. In this study race specific response of Fgb1 was observed in Foc race 1, 2 and 4 inoculated resistant and susceptible cultivars (Figure 4). Foc1 and Foc2 showed progressively intense expression of Fgb1 gene till 16 dai as compared to Foc4 infecting JG62, which showed maximum expression at 9 dai. A reduced gene expression response was observed in case of Digvijay, especially where no expression of Fgb1 gene was observed at 2 to 6 dai in case of Foc1, 2 and 4 infecting Digvijay; while race 1 inoculated Digvijay showed expression from 9 to 16 dai, though it was manifold low as compared to JG62. A slight expression was detected at 9 dai in case of Foc inoculated Digvijay which diminished at 13 dai. Comparative analysis of expression of Fgb1 in case of susceptible (JG62) and resistant (Digvijay) cultivars of chickpea with Foc races 1, 2 and 4 indicated that in general Fgb1 gene expression was higher in case of Foc inoculated JG62 as compared to Digvijay (Figure 4).
3.7. Temporal Expression of Disease Resistance Genes (in Chickpea) in Response to Foc Inoculation
This study was performed to understand chickpea-Fusarium interactions at transcriptional level for both, plant defense as well as fungal virulence related genes. Not many studies have been attempted to determine the defense related gene expression in chickpea against biotic stresses like Fusarium wilt. A few studies which have been previously conducted specify the defense transcript accumulation only during the first few hours of plant infection process, when the pathogen has merely entered the host plant and has started to establish itself. Analysis of defense related genes such as basic glucanase, ascorbate peroxidase, glutathione reductase [44], phenylalanine ammonium lyase, CHS and IFR [39] revealed only slight increase in the expression of these genes between resistant and susceptible accessions. It indicated that no significant differential expression of these genes correlates with Fusarium wilt resistance. Comparatively, in our studies imperative variations were observed which could be due to the time scale chosen for gene expression analysis. Also, continuous exposure to the pathogen load (mimicking the field conditions) could be a reason for such dramatic differences. We included a stretched time scale (2 to 16 dai), throughout the course of fungal infection and disease development; for determining the defense and virulence gene expression. Earlier, individual studies using Foc race 0 and race 1 have been conducted for profiling defense gene expression in chickpea [39,44], however; this is the first report of a comparative study exploring the gene specific transcript accumulation against three different Foc races causing wilt in the Indian subcontinent. In previous studies conducted using Foc inoculated cultivars, the expression level of defense related genes was found to be much higher in moderately resistant accession than the susceptible accession [38]. Similar results were reported by others [45] who showed that the level of accumulation of transcripts could be correlated with the differences in resistance/susceptibility of the host plant. Overall, in the present study, an enhanced expression of plant defense related genes was observed in case of resistant inoculated cultivar as compared to susceptible inoculated cultivar. Additionally, genes like glycosyltransferase, Msr, BetvI and GroES2 which have not been previously harnessed for their roles in plant defense; especially in chickpea, were found to express upon pathogen attack.
Fungal virulence genes like Fgb1, Gas1, Chs7 and Fow1 which have been previously reported to be essential for various cellular functions including fungal pathogenesis; showed an elevated expression during 9 to 16 dai indicating the window period of disease progression. Earlier studies conducted for the above mentioned virulence genes established their role in fungal pathogenesis and development in several systems [10,46], while in our study the temporal expression and race specific behavior of the suite of these virulence genes against both, the resistant and the susceptible cultivars, has been analyzed.
3.8. Race Specific Interaction of Chickpea—Fusarium oxysporum
Interestingly, in the present study, the response of plant defense genes has been observed to be specific to Foc races. All the genes except BetvI, namely GroES2, 60srp, CHS and IFR, showed higher expression in resistant chickpea cultivar, Digvijay vis-à-vis susceptible cultivar JG62, when exposed to race 1. This clearly indicated that, to establish resistance against Foc race 1 collective response of all the four genes under present study was essential whereas for remaining two races (Foc 2 and Foc 4) differential upregulation of some of these genes was sufficient to give complete resistance in Digvijay.
Similarly out of four fungal genes assessed in the present study, all the four genes revealed higher expression in the susceptible chickpea cultivar, JG62 than in the resistant chickpea cultivar, Digvijay when inoculated with race 1. While only Fgb1 and Fow1 genes were upregulated in Foc inoculated JG62 at 2 to 6 dai. The study indicated that amongst these four genes, Fow1 and Fgb1 are the most essential genes for prolonged virulence in Foc during its infection to chickpea; followed by Gas1 and then Chs7, confirming race specific involvement of these genes in establishing pathogenicity of Foc in chickpea.
Thus, it is noteworthy that, though a higher level of fungal virulence gene expression is essential for disease development in chickpea; comparatively a low level of plant defense related gene expression is sufficient to allow complete disease resistance in resistant chickpea cultivarDigvijay. Protecting Digvijay from pathogen attack can be accredited to the enhanced expression of the defense related genes; however, there is a need to further explore the exact role of these genes and their interaction with others during defense, for the confirmation of this hypothesis.
4. Acknowledgements
GSG acknowledges the fellowship from the Council of Scientific and Industrial Research, India. The work was supported by a grant from the McKnight Foundation, USA. Authors acknowledge MPKV, Rahuri, India for providing the seed material.
REFERENCES
- FAO, 2007. http://faostat.fao.org/
- C. H. Beckman, “The Nature of Wilt Diseases of Plants,” American Phytopathological Society, St. Paul, 1987.
- G. M. Armstrong and J. K. Armstrong, “Formae Speciales and Races of Fusarium oxysporum Causing Wilt Disease,” In: P. E. Nelson, T. A. Toussoun and R. J. Cook, Eds., Fusarium: Disease, Biology, and Taxonomy, Pennsylvania State University, University Park, 1981, pp. 391-399.
- M. P. Haware and Y. L. Nene, “Races of Fusarium oxysporum f. sp. Ciceri,” Plant Disease, Vol. 66, 1982, pp 809-810. doi:10.1094/PD-66-809
- A. P. Giri, A. M. Harsulkar, A. G. Patankar, V. S. Gupta, M. N. Sainani and V. V. Deshpande, “Association of Induction of Protease and Chitinase in Chickpea Roots with Resistance to Fusarium oxysporum f. sp. Ciceri,” Plant Pathology, Vol. 47, No. 6, 1998, pp 693-699. doi:10.1046/j.1365-3059.1998.00299.x
- S. B. Nimbalkar, A. M. Harsulkar, A. P. Giri, M. N. Sainani, V. Franceschi and V. S. Gupta, “Differentially Expressed Gene Transcripts in Roots of Resistant and Susceptible Chickpea Plant (Cicer arietinum L.) upon Fusarium oxysporum Infection,” Physiologicaland Molecular Plant Pathology, Vol. 68, No. 4-6, 2006, pp. 176- 188. doi:10.1016/j.pmpp.2006.10.003
- J. A. Seo, K. H. Han and J. H. Yu, “Multiple Roles of a Heterotrimeric G Protein Gamma Subunit in Governing Growth and Development of Aspergillus nidulans,” Genetics, Vol. 171, No. 1, 2005, pp. 81-89. doi:10.1534/genetics.105.042796
- M. P. Madrid, A. Di Pietro and M. I. G. Roncero, “Class V Chitin Synthase Determines Pathogenesis in the Vascular Wilt Fungus Fusarium oxysporum and Mediates Resistance to Plant Defence Compounds,” Molecular Microbiology, Vol. 47, No. 1, 2003, pp. 257-266. doi:10.1046/j.1365-2958.2003.03299.x
- B. Ramos, F. M. Alves-Santos, M. A. García-Sánchez, N. Martín-Rodrigues, A. P. Eslava and J. M. Díaz-Mínguez, “The Gene Coding for a New Transcription Factor (ftf1) of Fusarium oxysporum is Only Expressed during Infection of Common Bean,” Fungal Genetics and Biology, Vol. 44, No. 9, 2007, pp. 864-876. doi:10.1016/j.fgb.2007.03.003
- I. Iori, N. Fumio and T. Takashi, “Plant Colonization by the Vascular Wilt Fungus Fusarium oxysporum Requires Fow1, a Gene Encoding a Mitochondrial Protein,” Plant Cell, Vol. 14, No. 8, 2002, pp. 1869-1883. doi:10.1105/tpc.002576
- I. Iori, K. Makoto, I. Yuichiro and T. Takashi, “Fow2, a Zn(II)2Cys6-Type Transcription Regulator, Controls Plant Infection of the Vascular Wilt Fungus Fusarium oxysporum,” Molecular Microbiology, Vol. 63, No. 3, 2007, pp 737-753.
- H. C. van der Does, B. Lievens, L. Claes, P. M. Houterman, B. J. Cornelissen and M. Rep, “The Presence of a Virulence Locus Discriminates Fusarium oxysporum Isolates Causing Tomato Wilt from Other Isolates,” Environmental Microbiology, Vol. 10, No. 6, 2008, pp. 1475- 1485. doi:10.1111/j.1462-2920.2007.01561.x
- G. S. Gurjar, M. P. Barve, A. P. Giri and V. S. Gupta, “Identification of Indian Pathogenic Races of Fusarium oxysporum f. sp. ciceris with Gene Specific, ITS and Random Markers,” Mycologia, Vol. 101, 2009, pp. 484- 495. doi:10.3852/08-085
- S. F. Altschul, W. Gish, W. Miller, E. W. Myers and D. J. Lipman, “Basic Local Alignment Search Tool,” Journal of Molecular Biology, Vol. 215, No. 3, 1990, pp. 403-410.
- W. Chen, N. J. Provart, J. Glazebrook, F. Katagiri, H. Chang, T. T. Eulgem, F. Mauch, S. Luan, G. Zou, S. A. Whitham, P. R. Budworth, Y. Tao, Z. Xie, X. Chen, S. Lam, J. A. Kreps, J. F. Harper, A. Si-Ammour, B. Mauch-Mani, M. Heinlein, K. Kobayashi, T. Hohn, J. L. Dangl, X. Wang and T. Zhu, “Expression Profile Matrix of Arabidopsis Transcription Factor Genes Suggests Their Putative Functions in Response to Environmental Stresses,” Plant Cell, Vol. 14, No. 3, 2002, pp. 559-574. doi:10.1105/tpc.010410
- T. Tokai, M. Fujimura, H. Inoue, T. Aoki, K. Ohta, T. Shibata, I. Yamaguchi and M. Kimura, “Concordant Evolution of Trichothecene 3-O-Acetyltransferase and an rDNA Species Phylogeny of Trichothecene-Producing and Nonproducing Fusaria and Other Ascomycetous Fungi,” Microbiology, Vol. 151, 2005, pp. 509-519. doi:10.1099/mic.0.27435-0
- M. Rep, H. C. van der Does and B. J. Cornelissen, “Drifter, a Novel, Low Copy hAT-Like Transposon in Fusarium oxysporum is Activated during Starvation,” Fungal Genetics and Biology, Vol. 42, No. 6, 2005, pp. 546- 553. doi:10.1016/j.fgb.2005.03.007
- R. Jeong, W. Lim, S. Kwon and K. Kim, “Identification of Glycine max Genes Expressed in Response to Soybean Mosaic Virus Infection,” Journal of Plant Pathology, Vol. 21, No. 1, 2005, pp. 47-54. doi:10.5423/PPJ.2005.21.1.047
- E. Logemann, S. C. Wu, J. Schröder, E. Schmelzer, I. E. Somssich and K. Hahlbrock, “Gene Activation by UV Light, Fungal Elicitor or Fungal Infection in Petroselium crispum is Correlated with Repression of Cell CycleRelated Genes,” Plant Journal, Vol. 8, 1995, pp. 865-876. doi:10.1046/j.1365-313X.1995.8060865.x
- H. G. McFadden, I. W. Wilson, R. M. Chapple and C. Dowd, “Fusarium Wilt (Fusarium oxysporum f. sp. vasinfectum) Genes Expressed during Infection of Cotton (Gossypium hirsutum),” Molecular Plant Pathology, Vol. 7, No. 2, 2004, pp. 87-101. doi:10.1111/j.1364-3703.2006.00327.x
- P. J. O’Donnell, M. R. Truesdale, C. M. Calvert, A. Dorans, M. R. Roberts and D. J. Bowles, “A Novel Tomato Gene That Rapidly Responds to Woundand Pathogen-Related Signals,” Plant Journal, Vol. 14, No. 1, 1998, pp 137-142. doi:10.1046/j.1365-313X.1998.00110.x
- M. R. Roberts, S. A. J. Warner, R. Darby, E. K. Lim, J. Draper and D. J. Bowles, “Differential Regulation of a Glucosyl Transferase Gene Homologue during Defense Responses in Tobacco,” Journal of Experimental Botany, Vol. 50, 1999, pp. 407-410
 . doi:10.1093/jexbot/50.332.405
. doi:10.1093/jexbot/50.332.405 - A. Sadanandom, Z. Poghosyan, D. J. Fairbairn and D. J. Murphy, “Differential Regulation of Plastidial and Cytosolic Isoforms of Peptide Methionine Sulfoxide Reductase in Arabidopsis,” Plant Physiology, Vol. 123, No. 1, 2000, pp. 255-263. doi:10.1104/pp.123.1.255
- A. De Beer and M. A. Vivier, “Vv-AMP1, a Ripening Induced Peptide from Vitis vinifera Shows Strong Antifungal Activity,” BMC Plant Biology, Vol. 8, No. 75, 2008, pp. 75-90. doi:10.1186/1471-2229-8-75
- S. L. Rutherford, “Between Genotype and Phenotype: Protein Chaperones and Evolvability,” Nature Reviews in Genetics, Vol. 4, No. 4, 2003, pp. 263-274. doi:10.1038/nrg1041
- R. Saikia, B. P. Singh, R. Kumar and D. K. Arora, “Detection of Pathogenesis-Related Proteins-Chitinase and -1,3-Glucanase in Induced Chickpea,” Current Science, Vol. 89, 2005, pp. 659-663.
- S. M. Borch, K. Sletten and A. M. Hagen, 16th Nordic Congress on Allergology, Tromsoe, Abstract VI-19, June 1987, p. 65,.
- D. Lüttkopf, U. Müller, P. S. Skov, B. K. Ballmer-Weber, B. Wüthrich, K. S. Hansen, L. K. Poulsen, M. Kästner, D. Haustein and S. Vieths, “Comparison of Four Variants of a Major Allergen in Hazelnut (Corylus avellana) Cor a 1.04 with the Major Hazel Pollen Allergen Cor a 1.01,” Molecular Immunology, Vol. 38, 2002, pp. 515-525. doi:10.1016/S0161-5890(01)00087-6
- B. Bohle, A. Radakovics, B. Jahn-Schmid, K. Hoffmann-Sommergruber, G. F. Fischer and C. Ebner, “Bet v 1, the Major Birch Pollen Allergen, Initiates Sensitization to Api g 1, the Major Allergen in Celery: Evidence at the T Cell Level,” European Journal of Immunology, Vol. 33, 2003, pp. 3303-3310. doi:10.1002/eji.200324321
- P. Neudecker, K. Lehmann, J. Nerkamp, T. Haase, A. Wangorsch, K. Fötisch, S. Hoffmann, P. Rösch, S. Vieths and S. Scheurer, “Mutational Epitope Analysis of Pru av 1 and Api g 1, the Major Allergens of Cherry (Prunus avium) and Celery (Apium graveolens): Correlating IgE Reactivity with Three-Dimensional Structure,” Journal of Biochemistry, Vol. 376, No. 1, 2003, pp. 97-107. doi:10.1042/BJ20031057
- A. I. Saeed, V. Sharov, J. White, J. Li, W. Liang and N. Bhagabati, “TM4: A Free, Open-Source System for Microarray Data Management and Analysis,” Biotechniques, Vol. 34, No. 2, 2003, pp. 374-378.
- D. Foster-Hartnett, D. Danesh, S. Penuela, N. Sharapova, G. Endre, K. A. Vandenbosch, N. D. Young and D. A. Samac, “Molecular and Cytological Responses of Medicago truncatula to Erysiphe pisi,” Molecular Plant Pathology, Vol. 8, No. 3, 2007, pp. 307-319. doi:10.1111/j.1364-3703.2007.00395.x
- J. Taylor and L. A. Harrier, “Expression Studies of Plant Genes Differentially Expressed in Leaf and Root Tissues of Tomato Colonised by the Arbuscular Mycorrhizal Fungus Glomus mosseae,” Plant Molecular Biology, Vol. 51, No. 4, 2003, pp. 619-629. doi:10.1023/A:1022341422133
- S. Weidmann, L. Sanchez, J. Descombin, O. Chatagnier, S. Gianinazzi and V. Gianinazzi-Pearson, “Fungal Elicitation of Signal Transduction-Related Plant Genes Precedes Mycorrhiza Establishment and Requires the dmi3 Gene in Medicago truncatula,” Molecular Plant-Microbe Interaction, Vol. 17, No. 12, 2004, pp. 1385-1393. doi:10.1094/MPMI.2004.17.12.1385
- F. Daayf, A. Schmitt and R. R. Bélanger, “Evidence of Phytoalexins in Cucumber Leaves Infected with Powdery Mildew Following Treatment with Leaf Extracts of Reynoutria sacchalinensis,” Plant Physiolögy, Vol. 113, 1997, pp. 719-727.
- A. C. Ramos-Valdivia, R. Heijden and R. Verpoorte, “Elicitor Mediated Induction of Anthraquinone Biosynthesis and Regulation of Isopentenyl Diphosphate Isomerase and Farnesyl Diphosphate Synthase Activities in Cell Suspension Cultures of Cinchona robusta How,” Planta, Vol. 203, 1997, pp. 155-161. doi:10.1007/s004250050177
- H. Kessmann and W. Barz, “Accumulation of Isoflavones and Pterocarpan Phytoalexins in Cell Suspension Cultures of Different Cultivars of Chickpea (Cicer arietinum L.),” Plant Cell Reports, Vol. 6, 1987, pp. 55-59. doi:10.1007/BF00269739
- H. Kessmann, S. Daniel and W. Barz, “Elicitation of Pterocarpan Phytoalexins in Cell Suspension Cultures of Different Chickpea (Cicer arietinum L.) Cultivars by an Elicitor from the Fungus Ascochyta rabiei,” Z. Naturforsch, Vol. 43, 1988, pp. 529-535.
- A. Arfaoui, A. El Hadrami, Y. Mabrouk, B. Sifi, A. Boudabous, I. El Hadrami, F. Daayf and M. Cherif, “Treatment of Chickpea with Rhizobium Isolates Enhances the Expression of Phenylpropanoid Defense-Related Genes in Response to Infection by Fusarium oxysporum f. sp. Ciceris,” Plant Physiology and Biochemistry, Vol. 45, No. 6-7, 2007, pp. 470-479. doi:10.1016/j.plaphy.2007.04.004
- P. Tudzynski and A. Sharon, “Fungal Pathogenicity Genes,” In: D. K. Arora and G. G. Khachatourians, Eds., Applied Mycology and Biotechnology, (3) Fungal Genomics, Elsevier Science, Amsterdam, 2003, pp. 187-212.
- J. A. Mayer, D. Kakhniashvili, D. A. Gremse, C. Campbell, R. Krämer, A. Schroers and R. S. Kaplan, “Bacterial Overexpression of Putative Yeast Mitochondrial Transport Proteins,” Journal of Bioenergy and Biomembrane, Vol. 29, 1997, pp, 541-547.
- S. Bartnicki-Garcia and Ch. E. Bracker, “Unique Properties of Chitosomes,” In: C. Nombela, Ed., Microbial Cell Wall Synthesis and Autolysis, Elsevier Science, Amsterdam, 1984, pp. 101-112.
- I. Mouyna, W. Morelle, M. Vai, M. Monod, B. Léchenne, T. Fontaine, A. Beauvais, J. Sarfati, M. C. Prévost, C. Henry and J. P. Latgé, “Deletion of GEL2 Encoding for a Beta (1-3) Glucanosyltransferase Affects Morphogenesis and Virulence in Aspergillus fumigates,” Molecular Microbiology, Vol. 56, 2005, pp. 1675-1688. doi:10.1111/j.1365-2958.2005.04654.x
- S. Cho and F. J. Muehlbauer, “Genetic Effect of Differentially Regulated Fungal Response Genes on Resistance to Necrotrophic Fungal Pathogens in Chickpea (Cicer arietinum L.),” Physiological and Molecular Plant Pathology, Vol. 64, No. 2, 2004, pp. 57-66. doi:10.1016/j.pmpp.2004.07.003
- X. Wang, A. El Hadrami, L. R. Adam and F. Daayf, “Local and Distal Gene Expression of pr-1 and pr-5 in Potato Leaves Inoculated with Isolates from the Old (US-1) and the New (US-8) Genotypes of Phytophthora infestans (Mont.) de Bary”. Environmental and Experimental Botany, Vol. 57, No. 1-2, 2006, pp. 70-79. doi:10.1016/j.envexpbot.2005.04.006
- J. Delgado-Jarana, A. L. Martínez-Rocha, R. RoldánRodriguez, M. I. G. Roncero and A. Di Pietro, “Fusarium oxysporum G-Protein β Subunit Fgb1 Regulates Hyphal Growth, Development, and Virulence through Multiple Signalling Pathways,” Fungal Genetics and Biology, Vol. 42, No. 1, 2004, pp. 61-72. doi:10.1016/j.fgb.2004.10.001
NOTES
*Corresponding author.

