Advances in Lung Cancer
Vol.3 No.2(2014), Article
ID:47050,7
pages
DOI:10.4236/alc.2014.32007
Prognostic Role of miR-205 in Early-Stage (T1N0) Non-Small Cell Lung Cancer
Laura Boldrini1*#, Mirella Giordano1*, Adele Servadio1, Greta Alì2, Alice Cocco1, Marco Lucchi3, Pietro Bertoglio1, Franca Melfi3, Alfredo Mussi1, Gabriella Fontanini1
1Department of Surgical, Medical, Molecular Pathology and Critical Area, University of Pisa, Pisa, Italy
2Unit of Pathological Anatomy III, Azienda Ospedaliera Universitaria Pisana (A.O.U.P.), Pisa, Italy
3Unit of Thoracic Surgery, Azienda Ospedaliera Universitaria Pisana (A.O.U.P.), Pisa, Italy
Email: #laura.boldrini@med.unipi.it
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 20 March 2014; revised 20 April 2014; accepted 16 May 2014
ABSTRACT
Background: Published data have shown that microRNAs (miRNAs) could play a potential role as diagnostic and prognostic indicators in cancers. Data for the predictive value of miRNA let-7, miR-21, and miR-205 are inconclusive. The aim of the present analysis was therefore to evaluate the expression and the prognostic role of the above mentioned miRNAs in early-stage (T1N0) NSCLC patients. Methods: Quantification of let-7g, miR-21, and miR-205 expression was carried out into 105 early-stage NSCLC by quantitative Real Time-PCR (qRT-PCR). Results: a significant association between the low miR-205 expression and ADC histotype (p ( 0.0001) compared to SCC was found; moreover, survival analysis showed that tumors with a high miR-205 expression had a significantly shorter mean PFS and OS compared to the patients with a low expression of this miRNA (p = 0.02 and p = 0.03, respectively). No other statistically significant correlations were observed between the analysed miRNAs and the main clinico-pathological characteristics of the NSCLC patients. Conclusion: The results indicated that miR-205 could represent a useful marker in the prognostic management of the early-stage (T1N0) NSCLC patients.
Keywords
Non-Small Cell Lung Cancer, miR-21, let-7g, miR-205, Prognosis

1. Introduction
MicroRNAs (miRNAs) are short, non-coding RNAs of approximately 19 - 24 nucleotides in length, emerged for their ability to bind to the 3’-untranslated regions (UTRs) of messenger RNAs, where they interfere with translation, and thereby regulate cell differentiation, apoptosis, and tumorigenesis. Since their initial discovery in 1993 [1] , several published data found that some of these miRNAs act as oncogenes, whereas others exhibit tumor suppressor properties [2] -[4] . Altered miRNA expression has been implicated in several carcinomas, including non-small cell lung cancer (NSCLC); moreover, aberrant expression of miRNAs is significantly related to tumor stage, suggesting that they could be potential markers for cancer prognosis [5] [6] .
Similar to breast cancer, let-7 is down regulated in NSCLC [7] -[9] . Several investigators have shown that let-7 harbors tumor suppressive properties both in vitro and in vivo [10] -[13] .
MiR-21 has been identified as the best hit in a number of high-through screen experiments designed for the detection of cancer related miRNAs [14] . The exact molecular mechanism of miR-21 elevation and its role in cancer development and progression is currently unclear. Interestingly, several miRNAs, including miR-205, seem to be differently involved in the pathogenesis of the two major NSCLC histologies, adenocarcinomas (ADCs) and squamous cell carcinomas (SCCs).
Moreover, recent published studies [15] -[18] have correlated miRNA expression with prognosis in lung cancer, arising the possibility of using miRNA expression profiles to predict outcomes in NSCLC, especially in early stage lung cancer patients in order to identify those at high risk of relapse after surgical resection [19] .
The aim of this study was to evaluate let-7g, miR-21 and miR-205 expression in a series of 105 early-stage (T1N0) NSCLC patients to establish their potential prognostic value.
2. Patients and Methods
2.1. Patients Selection
One hundred and five NSCLC cases were retrospectively selected from patients operated at the Unit of Thoracic Surgery of the A.O.U.P. between 2000 and 2010. Histological diagnoses were independently formulated by two pathologists (G.F. and G.A.) according to the World Health Organization classification [20] -[22] . Clinico-pathological characteristics and clinical data were collected for all the patients. All the patients gave their informed consent to the molecular analyses.
2.2. RNA Isolation
Total RNA including miRNAs was isolated from 5 - 10 μm sections of formalin-fixed and paraffin-embedded (FFPE) tissues after manual tumor macrodissection using the miRNeasy FFPE Kit (Qiagen), according to the manufacturer’s instructions.
2.3. miRNA Expression
Quantification of let-7g, miR-21, miR-205, and RNU6B expression was carried out in triplicate into 105 NSCLC and into a pool of 27 non-cancerous lung tissues using specific TaqMan® MicroRNA Assays (Applied Biosystems), according to the manufacturer’s instructions. Briefly, 10 ng of RNA were retro-transcribed by the TaqMan® MicroRNA Reverse Transcription (RT) Kit (Applied Biosystems), and 1.3 μl of RT product were analysed by quantitative Real Time-PCR (qRT-PCR) on the Rotor-Gene 6000 (Corbett Research).
Threshold cycle (Ct) and baselines were determined by manual settings. miRNA expression was calculated by relative quantification and fold expression changes were determined by the 2−ΔΔCt method using the DataAssistTM software (Applied Biosystems).
2.4. Statistical Analysis
One-way analysis of variance and χ2 test were used to determine the association between miRNA expression and the different parameters, while survival analysis was performed by the Kaplan-Meier method. Statistical analyses were performed using the JMP10 software (SAS) and a two-tailed p value <0.05 was considered significant.
3. Results
3.1. Patient Characteristics
This study was conducted in 105 patients with early-stage NSCLC (T1N0), including 57 adenocarcinomas (ADCs), and 48 squamous cell carcinomas (SCCs). As regards histological classification, different histologic subtypes of adenocarcinoma were recognised: acinar (20/57, 35%), lepidic (18/57, 31.6%), mucinous (9/57, 15.8%), papillar (7/57, 12.3%), and solid (3/57, 5.3%) variants.
The median age at diagnosis was 67 years (range: 49 - 81) and the median follow up was 63 months (range: 6 - 161). Regarding grading, 5 tumors were G1, whereas 73 and 27 were G2 and G3, respectively. Disease progression and death from lung cancer were observed in 27 (25.7%) and in 21 (20%) of the 105 NSCLC patients, respectively. The median progression-free survival (PFS) and overall survival (OS) were 58 months (95% CI: 6 - 161) and 69 months (95% CI: 11 - 161), respectively. Regarding smoking habits, there were 15 non-smokers, 45 former smokers, and 45 current smokers.
3.2. let-7g, miR-21, and miR-205 Expression Profile and Clinico-Pathological Characteristics
We quantified the mature let-7g, miR-21, and miR-205 expression normalized to the RNU6B endogenous control in 105 NSCLC and in non-cancerous tissues. To determine whether miRNA profile was correlated with the main clinico-pathological characteristics, the NSCLC patients were divided into let-7g, miR-21, and miR-205 high and low expression groups based on median fold change values (0.451 for let-7g, 1.819 for miR-21, and 1.409 for miR-205). Except for a significant association between the low miR-205 expression and ADC histotype (p < 0.0001) compared to SCC, no other statistically significant associations were observed between the analysed miRNAs and the main clinico-pathological characteristics of the NSCLC patients (Table 1).
3.3. Survival Analysis
To evaluate the relationships of let-7g, miR-21, and miR-205 expression with prognosis of the NSCLC patients,
Table 1. Correlations between the expression of let-7g, miR-21, and miR-205 and the main clinico-pathological characteristics of the NSCLC patients.
we performed a survival analysis by the Kaplan-Meier method using the disease recurrence and the overall post-operative survival as endpoints. Interestingly, we found that the tumors with a high miR-205 expression showed a significantly shorter mean PFS and OS compared to the patients with a low expression of this miRNA (p = 0.02 and p = 0.03, respectively) (Figure 1). We did not observe any significant difference in PFS and OS of the NSCLC patients with a high let-7g or miR-21 expression compared to the patients with a low expression of these miRNAs.
4. Discussion
NSCLC accounts for approximately 70% of all lung cancer-related deaths worldwide [23] , reflecting the need for a better understanding of the mechanisms that underlie lung carcinogenesis. The chance of cure with surgery in early NSCLC is compromised by almost a 30% risk of developing distant metastases, and adjunctive prognostic parameters are necessary to select patients for adjuvant systemic treatments. Previously unknown markers, such as noncoding RNA gene, may also lend insight into the biology of lung cancer. MicroRNAs are a class of short noncoding RNAs that modulate gene expression by targeting messenger RNAs and triggering either the repression of translation or RNA degradation [24] . miRNAs are involved in tissue differentiation during both normal development and carcinogenesis, and miRNA expression profiles have been seen as a promising new class of markers for tumor diagnosis and prognosis [25] -[27] .
This study was aimed to evaluate let-7g, miR-21, and miR-205 expression profile in early stage NSCLC patients (T1N0), with particular reference to their role in prognosis. In our study, miR-205 evaluation has been shown to have a significant association with SCC histotype, as it has been previously reported [28] . Considering
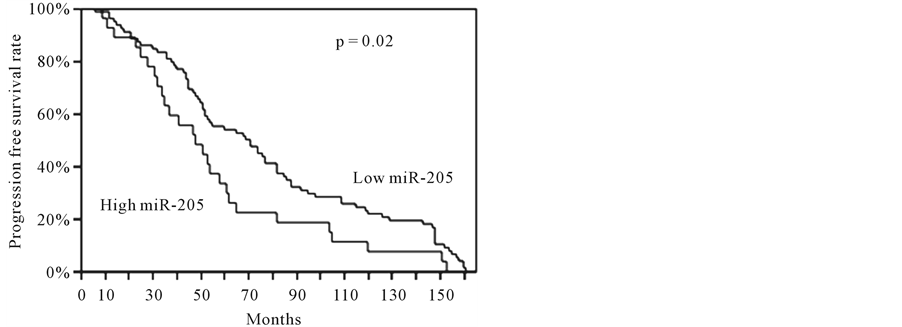 (a)
(a) (b)
(b)
Figure 1. Kaplan-Meier curves in the NSCLC patients with a different miR-205 expression.
that specific therapies for NSCLC patients are selected depending on the histologic diagnosis, a clinical diagnostic assay based on the expression levels of miR-205 should provide an alternative tool of differential diagnosis, when routinary immunohistochemistry fails. As far as is concerned the clinico-pathological characteristics, we did not observed any significant correlation between the miRNAs expression and the clinico-pathological features, including age, gender, hystotype, tumor grade, smoking habit, and recurrences. In our cases, KaplanMeier survival estimates showed that the early stage NSCLC patients with high miR-205 had a poorer survival than the patients with low expression, either for progression-free survival or overall survival, suggesting that miR-205 could represent a useful marker in the prognostic management of the early-stage (T1N0) NSCLC patients. No difference in the prognosis was found for let-7g and miR-21. Regarding the prognostic role of the analysed miRNAs, Takamizawa et al. [7] first reported low expression of let-7 family members was associated with shorter overall survival in NSCLC; on the contrary, the prognostic role of mature let-7 family members was further investigated, but there were no differences in overall survival between clusters of cases with varying let-7 expression [8] . Similarly, the association between miR-21 or miR-205 expression and NSCLC prognosis has been explored in several studies, but the results are controversial. While some studies showed that miR-21 or miR-205 overexpression was associated with poor NSCLC survival [14] [29] [30] , other studies indicated the converse or insignificant association [31] [32] . To explain discrepancies on the prognostic value of miRNAs in NSCLC, some issues should be addressed. Different kinds of samples are used to detect miRNAs including frozen tissues, formalin-fixed paraffin-embedded (FFPE) tissues, serum and sputum. Saito et al. reported that the results concerning the association of miR-21 expression and NSCLC prognosis from frozen tissues were in apparent contrast to those from FFPE tissues [33] . Therefore, the type of samples could influence the results, and an appropriate sample type should be determined to extract miRNAs. Moreover, published studies have used different cut-point for high vs. low miRNA expression, but a standard cut-off should be identified in order to correctly compare data. In view of these points, further investigations are necessary to have a systematic evaluation of miRNAs in the molecular characterization of the NSCLC patients.
5. Conclusion
Based on our results, miRNA 205 expression may represent a useful marker in prognostic management of the early-stage (T1N0) NSCLC patients.
Financial Disclosure
This work was supported by Grant PRIN 20092009LMEEEH_004 from the Italian Ministry for University and Scientific Research. The authors declared no conflict of interests.
References
- Lee, R.C., Feinbaum, R.L. and Ambros, V. (1993) The C. elegans Heterochronic Gene Lin-4 Encodes Small RNAs with Antisense Complementarity to Lin-14. Cell, 75, 843-854. http://dx.doi.org/10.1016/0092-8674(93)90529-Y
- Ambros, V., Bartel, B., Bartel, D.P., Burge, C.B., Carrington, J.C., Chen, X., Dreyfuss, G., Eddy, S.R., Griffiths-Jones, S., Marshall, M., Matzke, M., Ruvkun, G. and Tuschl, T. (2003) A Uniform System for MicroRNA Annotation. RNA, 9, 277-279. http://dx.doi.org/10.1261/rna.2183803
- Chen, C.Z. (2005) MicroRNAs as Oncogenes and Tumor Suppressors. New England Journal Medicine, 353, 1768-1771. http://dx.doi.org/10.1056/NEJMp058190
- Calin, G.A. and Croce, C.M. (2006) MicroRNA-Cancer Connection: The Beginning of a New Tale. Cancer Research, 66, 7390-7394. http://dx.doi.org/10.1158/0008-5472.CAN-06-0800
- Ferracin, M., Veronese, A. and Negrini, M. (2010) Micromarkers: miRNAs in Cancer Diagnosis and Prognosis. Expert Review of Molecular Diagnostics, 10, 297-308. http://dx.doi.org/10.1586/erm.10.11
- Nana-Sinkam, S.P., Fabbri, M. and Croce, C.M. (2010) MicroRNAs in Cancer: Personalizing Diagnosis and Therapy. Annals of the New York Academy of Sciences, 1210, 25-33. http://dx.doi.org/10.1111/j.1749-6632.2010.05822.x
- Takamizawa, J., Konishi, H., Yanagisawa, K., Tomida, S., Osada, H., Endoh, H., Harano, T., Yatabe, Y., Nagino, M., Nimura, Y., Mitsudomi, T. and Takahashi, T. (2004) Reduced Expression of the Let-7 MicroRNAs in Human Lung Cancers in Association with Shortened Postoperative Survival. Cancer Research, 64, 3753-3756. http://dx.doi.org/10.1158/0008-5472.CAN-04-0637
- Inamura, K., Togashi, Y., Nomura, K., Ninomiya, H., Hiramatsu, M., Satoh, Y., Okumura, S., Nakagawa, K. and Ishikawa, Y. (2007) Let-7 MicroRNA Expression Is Reduced in Bronchioloalveolar Carcinoma, a Noninvasivecarcinoma, and Is Not Correlated with Prognosis. Lung Cancer, 58, 392-396. http://dx.doi.org/10.1016/j.lungcan.2007.07.013
- Nicoloso, M., Spizzo, R., Shimizu, M., Rossi, S. and Calin, G. (2009) MicroRNAs-Themicro Steering Wheel of Tumour Metastases. Nature Reviews Cancer, 9, 293-302. http://dx.doi.org/10.1038/nrc2619
- Johnson, C.D., Esquela-Kerscher, A., Stefani, G., Byrom, M., Kelnar, K., Ovcharenko, D., Wilson, M., Wang, X., Shelton, J., Shingara, J., Chin, L., Brown, D. and Slack, F.J. (2007) The Let-7 MicroRNA Represses Cell Proliferation Pathways in Human Cells. Cancer Research, 67, 7713-7722. http://dx.doi.org/10.1158/0008-5472.CAN-07-1083
- Kumar, M.S., Erkeland, S.J., Pester, R.E., Chen, C.Y., Ebert, M.S., Sharp, P.A. and Jacks, T. (2008) Suppression of Non-Small Cell Lung Tumor Development by the Let-7 MicroRNA Family. Proceedings of the National Academy of Sciences, 105, 3903-3908. http://dx.doi.org/10.1073/pnas.0712321105
- Trang, P., Medina, P., Wiggins, J., Ruffino, L., Kelnar, K., Omotola, M., Homer, R., Brown, D., Bader, A.G., Weidhaas, J.B. and Slack, F.J. (2009) Regression of Murine Lung Tumors by the Let-7 MicroRNA. Oncogene, 29, 1580-1587. http://dx.doi.org/10.1038/onc.2009.445
- Trang, P., Wiggins, J., Daige, C., Cho, C., Omotola, M., Brown, D., Weidhaas, J.B., Bader, A.G. and Slack, F.J. (2011) Systemic Delivery of Tumor Suppressor Microrna Mimics Using a Neutral Lipid Emulsion Inhibits Lungtumors in Mice. Molecular Therapy, 19, 1116-1122. http://dx.doi.org/10.1038/mt.2011.48
- Gao, W., Yu, Y., Cao, H., Shen, H., Li, X., Pan, S. and Shu, Y. (2010) Deregulated Expression of miR-21, miR-143 and miR-181a in Non Small Cell Lung Cancer Is Related to Clinicopathologic Characteristics or Patient Prognosis. Biomedicine Pharmacotherapy, 64, 399-408. http://dx.doi.org/10.1016/j.biopha.2010.01.018
- Yanaihara, N., Caplen, N., Bowman, E., Seike, M., Kumamoto, K., Yi, M., Stephens, R.M., Okamoto, A., Yokota, J., Tanaka, T., Calin, G.A., Liu, C.G., Croce, C.M. and Harris, C.C. (2006) Unique MicroRNA Molecular Profiles in Lung Cancer Diagnosis and Prognosis. Cancer Cell, 9, 189-198. http://dx.doi.org/10.1016/j.ccr.2006.01.025
- Raponi, M., Dossey, L., Jatkoe, T., Wu, X., Chen, G., Fan, H. and Beer, D.G. (2009) MicroRNA Classifiers for Predicting Prognosis of Squamous Cell Lung Cancer. Cancer Research, 69, 5776-5783. http://dx.doi.org/10.1158/0008-5472.CAN-09-0587
- Landi, M.T., Zhao, Y., Rotunno, M., Koshiol, J., Liu, H., Bergen, A.W., Rubagotti, M., Goldstein, A.M., Linnoila, I., Marincola, F.M., Tucker, M.A., Bertazzi, P.A., Pesatori, A.C., Caporaso, N.E., McShane, L.M. and Wang, E. (2010) MicroRNA Expression Differentiates Histology and Predicts Survival of Lung Cancer. Clinical Cancer Research, 16, 430-441. http://dx.doi.org/10.1158/1078-0432.CCR-09-1736
- Patnaik, S.K., Kannisto, E., Knudsen, S. and Yendamuri, S. (2010) Evaluation of microRNA Expression Profiles that May Predict Recurrence of Localized Stage I Non-Small Cell Lung Cancer after Surgical Resection. Cancer Research, 70, 36-45. http://dx.doi.org/10.1158/0008-5472.CAN-09-3153
- Lu, Y., Govindan, R., Wang, L., Liu, P.Y., Goodgame, B., Wen, W., Sezhiyan, A., Pfeifer, J., Li, Y.F., Hua, X., Wang, Y., Yang, P. and You, M. (2012) MicroRNA Profiling and Prediction of Recurrence/Relapse-Free Survival in Stage I Lung Cancer. Carcinogenesis, 33, 1046-1054. http://dx.doi.org/10.1093/carcin/bgs100
- Travis, W.D., Brambilla, E., Muller-Hemerlink, H.K. and Harris, C.C. (2004) World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of the Lung, Pleura, Thymus and Heart. IARC Press, Lyon.
- Travis, W.D., Brambilla, E., Noguchi, M., Nicholson, A.G., Geisinger, K.R., Yatabe, Y., Beer, D.G., Powell, C.A., Riely, G.J., Van Schil, P.E., Garg, K., Austin, J.H., Asamura, H., Rusch, V.W., Hirsch, F.R., Scagliotti, G., Mitsudomi, T., Huber, R.M., Ishikawa, Y., Jett, J., Sanchez-Cespedes, M., Sculier, J.P., Takahashi, T., Tsuboi, M., Vansteenkiste, J., Wistuba, I., Yang, P.C., Aberle, D., Brambilla, C., Flieder, D., Franklin, W., Gazdar, A., Gould, M., Hasleton, P., Henderson, D., Johnson, B., Johnson, D., Kerr, K., Kuriyama, K., Lee, J.S., Miller, V.A., Petersen, I., Roggli, V., Rosell, R., Saijo, N., Thunnissen, E., Tsao, M. and Yankelewitz, D. (2011) International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society International Multidisciplinary Classification of Lung Adenocarcinoma. Journal of Thoracic Oncology, 6, 244-285. http://dx.doi.org/10.1097/JTO.0b013e318206a221
- Travis, W.D., Brambilla, E., Noguchi, M., Nicholson, A., Geisinger, K., Yatabe, Y., Ishikawa, Y., Wistuba, I., Flieder, D.B., Franklin, W., Gazdar, A., Hasleton, P.S., Henderson, D.W., Kerr, K.M., Petersen, I., Roggli, V., Thunnissen, E. and Tsao, M. (2013) Diagnosis of Lung Cancer in Small Biopsies and Cytology: Implications of the 2011 International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society Classification. Archives of Pathology and Laboratory Medicine, 137, 668-684. http://dx.doi.org/10.5858/arpa.2012-0263-RA
- Siegel, R., Naishadham, D. and Jemal, A. (2012) Cancer Statistics, 2012. CA: A Cancer Journal for Clinicians, 62, 10-29. http://dx.doi.org/10.3322/caac.20138
- Bartel, D.P. (2004) MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell, 116, 281-297. http://dx.doi.org/10.1016/S0092-8674(04)00045-5
- Mascaux, C., Laes, J.F., Anthoine, G., Haller, A., Ninane, V., Burny, A. and Sculier, J.P. (2009) Evolution of Micro-RNA Expression during Human Bronchial Squamous Carcinogenesis. European Respiratory Journal, 33, 352-359. http://dx.doi.org/10.1183/09031936.00084108
- Iorio, M.V. and Croce, C.M. (2009) MicroRNAs in Cancer: Small Molecules with a Huge Impact. Journal of Clinical Oncology, 27, 5848-5856. http://dx.doi.org/10.1200/JCO.2009.24.0317
- Lu, J., Getz, G., Miska, E.A., Alvarez-Saavedra, E., Lamb, J., Peck, D., Sweet-Cordero, A., Ebert, B.L., Mak, R.H., Ferrando, A.A., Downing, J.R., Jacks, T., Horvitz, H.R. and Golub, T.R. (2005) MicroRNA Expression Profiles Classify Human Cancers. Nature, 435, 834-838. http://dx.doi.org/10.1038/nature03702
- Lebanony, D., Benjamin, H., Gilad, S., Ezagouri, M., Dov, A., Ashkenazi, K., Gefen, N., Izraeli, S., Rechavi, G., Pass, H., Nonaka, D., Li, J., Spector, Y., Rosenfeld, N., Chajut, A., Cohen, D., Aharonov, R. and Mansukhani, M. (2009) Diagnostic Assay Based on hsa-miR-205 Expression Distinguishes Squamous from Nonsquamous Non-Small-Cell Lung Carcinoma. Journal of Clinical Oncology, 27, 2030-2037. http://dx.doi.org/10.1200/JCO.2008.19.4134
- Liu, X.G., Zhu, W.Y., Huang, Y.Y., Ma, L.N., Zhou, S.Q., Wang, Y.K., Zeng, F., Zhou, J.H. and Zhang, Y.K. (2012) High Expression of Serum miR-21 and Tumor miR-200c Associated with Poor Prognosis in Patients with Lung Cancer. Medical Oncology, 29, 618-626. http://dx.doi.org/10.1007/s12032-011-9923-y
- Gao, W., Shen, H., Lui, L., Xu, J., Xu, J. and Shu, Y. (2011) miR-21 Overexpression in Human Primary Squamous Cell Lung Carcinoma Is Associated with Poor Patient Prognosis. Journal of Cancer Research Clinical Oncology, 137, 557-566. http://dx.doi.org/10.1007/s00432-010-0918-4
- Markou, A., Tsaroucha, E.G., Kaklamanis, L., Kaklamanis, L., Fotinou, M., Georgoulias, V. and Lianidou, E.S. (2008) Prognostic Value of Mature MicroRNA-21 and MicroRNA-205 Overexpression in Non-Small Cell Lung Cancer by Quantitative Real-Time RT-PCR. Clinical Chemistry, 54, 1696-1704. http://dx.doi.org/10.1373/clinchem.2007.101741
- Voortman, J., Goto, A., Mendiboure, J., Sohn, J.J., Schetter, A.J., Saito, M., Dunant, A., Pham, T.C., Petrini, I., Lee, A., Khan, M.A., Hainaut, P., Pignon, J.P., Brambilla, E., Popper, H.H., Filipits, M., Harris, C.C. and Giaccone, G. (2010) MicroRNA Expression and Clinical Outcomes in Patients Treated with Adjuvant Chemotherapy after Complete Resection of Non-Small Cell Lung Carcinoma. Cancer Research, 70, 8288-8298. http://dx.doi.org/10.1158/0008-5472.CAN-10-1348
- Saito, M., Schetter, A.J., Mollerup, S., Kohno, T., Skaug, V., Bowman, E.D., Mathé, E.A., Takenoshita, S., Yokota, J., Haugen, A. and Harris, C.C. (2011) The Association of microRNA Expression with Prognosis and Progression in Early-Stage, Non-Small Celllung Adenocarcinoma: A Retrospective Analysis of Three Cohorts. Clinical Cancer Research, 17, 1875-1882. http://dx.doi.org/10.1158/1078-0432.CCR-10-2961
NOTES

*These authors equally contributed.
#Corresponding author.


