Pharmacology & Pharmacy
Vol.06 No.11(2015), Article ID:61402,16 pages
10.4236/pp.2015.611053
Inhibition of Hepatitis C Virus Genotype 1a Non-Structural Proteins by Small Interference RNA in Human Hepatoma Cell Lines
Imran Shahid1,2*, Waleed Hassan AlMalki2, Shaia Saleh R. Almalki3, Ismail Muhammad AlTurkestany4, Hassan Ali AlGhamdi5, Saleh Ali AlMenshawi6
1Applied and Functional Genomics Laboratory, Center of Excellence in Molecular Biology, University of the Punjab, Lahore, Pakistan
2Department of Pharmacology and Toxicology, College of Pharmacy, Umm Al Qura University, Makkah, Saudi Arabia
3Laboratory Medicine, Faculty of Applied Medical Sciences, Albaha University, Albaha, Saudi Arabia
4Infection Control, Pilgrims City Hospital, Madina, Saudi Arabia
5Toxicology Center, Jeddah, Saudi Arabia
6Toxiology Research Center, Ministry of Interior, Jeddah, Saudi Arabia

Copyright © 2015 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).


Received 6 September 2015; accepted 21 November 2015; published 24 November 2015

ABSTRACT
Hepatitis C virus (HCV) infection and associated liver diseases are still challenging and represent a significant health care burden around the world. Although, the treatment strategies have been improved by the development of novel direct-acting antivirals, but such therapeutic options are still expensive and beyond the financial range of the most infected individuals in developing or even in resource replete countries. It demands an urgent need to search novel and improved alternate treatment strategies to treat the infection. The present study was aimed to develop an in vitro stable cell culture system, persistently expressing HCV genotype 1a non-structural genes and to characterize the inhibitory effects of synthetic siRNAs (short interference RNA) directed against the most conserved regions of nonstructural genes in an in vitro cell culture model. The continuous expression of nonstructural genes for more than 30 days post transfection was detected by reverse transcription polymerase chain reaction (RT-PCR) and Western blot analysis in stable human hepatoma cell line (Huh-7). The gene expression studies revealed significantly reduced gene expression of HCV nonstructural genes (i.e., NS2, NS4A and NS5A) both at mRNA and protein levels when treated against genome specific synthetic siRNAs in stable cell lines (51%, 47% and 54% respectively, p < 0.05). Similarly, a vivid decrease in HCV viral titer was exhibited by synthetic siRNAs in an in vitro viral replicate cell culture model (58%, 48% and 50%, respectively, p < 0.05) determined by quantitative Real-Time PCR (qPCR). Our data indicate that siRNA mediated gene silencing may be considered a promising alternate treatment strategy against HCV in combination with other effective therapeutic regimens in future.
Keywords:
Hepatitis C Virus, Non-Structural Proteins, Stable Cell Line, Anti-HCV Drugs, Short Interference RNA

1. Introduction
Afflicting around 170 million people worldwide, Hepatitis C virus (HCV) infection and associated liver diseases (hepatic cirrhosis, fibrosis and hepatocellular carcinoma) are significant health concerns in developing world and even in developed countries [1] . HCV infection results in more than 350,000 deaths every year predominantly due to hepatic cirrhosis and hepatocellular carcinoma [2] . HCV is a small, enveloped, positive sense single-stranded RNA (ssRNA) virus of approximately 9.6 kb long [3] . The viral genome consists of a single long open reading frame (ORF) encoding a single polypeptide of approximately 3100 amino acids [4] . The viral single polyprotein is proteolytically cleaved by host and viral encoded proteases into four mature structural and six nonstructural proteins [5] . The structural proteins are actively involved in virion formation while the nonstructural proteins are essential for viral replication, translation and polyprotein processing [6] . The proteins that significantly contribute to viral replication and translation (i.e., NS5B and NS3) are also the potential active target sites for the development of novel direct-acting antivirals and genome-based therapeutics [7] . Efforts to understand HCV life cycle and to identify effective anti-HCV agents have hampered by the lack of an efficient cell culture system [8] [9] . However, subgenomic replicons, HCV pseudo-particles, infection based cell culture systems and small animal models are currently available to study HCV replication and for the development of anti-HCV treatment strategies [10] [11] .
RNA interference (RNAi) is a gene-silencing mechanism by which a small double-stranded RNA (dsRNA) does sequential degradation of ssRNA (i.e., targeted mRNA) [12] . This degradation process of targeted mRNA completes in two discrete steps [13] . First, a long dsRNA is processed into 21 - 23 nucleotide siRNAs by Dicer (i.e., an RNase-III nuclease) [14] . Second, siRNA is incorporated into a multi-protein complex known as RISC (RNA-induced silencing complex) [15] . This siRNA-RISC complex leads to degradation of targeted mRNA recognized by the anti-sense strand of siRNA [15] . In-vitro and animal studies point out that RNAi therapeutics may be useful for future clinical use as the treatment of certain infectious diseases, cancer and certain metabolic disorders [16] -[18] . Similarly, the phenomenon is highly effective at low dosage which makes it an excellent candidate for future clinical use [19] . Consequently, RNAi as a drug class may have the potential to exert a transformational effect on modern molecular medicine.
The siRNA may be evaluated for their therapeutic activity against HCV, because HCV mRNA acts as a template both for viral translation and replication [20] [21] . For this reason, destruction of mRNA blocks viral replication and also inhibits protein synthesis. Several studies reported that siRNA mediated gene silencing blocked HCV replication in Huh-7 derived cell lines [22] [23] . However, viral escape mutants and off-target effects of RNA interference create significant problem to develop useful RNAi-based antiviral therapies [24] . This problem may be solved by searching highly conserved genome target sites while designing and constructing synthetic siRNAs.
The present study was aimed to characterize the inhibitory effects of siRNA against HCV genotype 1a nonstructural proteins in an in vitro stable Huh-7 cell culture system. We aimed to develop stable Huh-7 cell culture systems that persistently express HCV genotype 1a nonstructural proteins more than one month. Such in vitro cell line models would be helpful to understand the molecular pathogenesis of the infection and also to establish novel drug screening assays for anti-HCV compounds. We demonstrated a significant decrease in mRNA and protein expression of HCV 1a genotype nonstructural proteins (NS2, NS4A & NS5A) at mRNA and protein levels in stable cell lines. The specificity of siRNAs to inhibit HCV genome replication was also established in serum-infected Huh-7 cells.
2. Materials and Methods
2.1. PCR Amplification of HCV Nonstructural Genes
HCV non-structural genes (NS2, NS4A & NS5A) were amplified by using 200 ng HFL plasmid that expresses the complete HCV 1a genotype genome. Gene-specific primers with restriction enzyme sites (forward primers with EcoRV and reverse primers with Xba 1 restriction sites) were used to amplify the amplicons (Table 1). The restriction enzyme sites were further helpful to clone nonstructural genes into a mammalian expression plasmid pCR3.1/FlagTAG and to minimize complementarities with each other in order to avoid primer-dimer formations. PCR amplifications were performed by using 2X PCR master mix (Fermentas, Maryland USA) in a thermal cycler under optimized PCR conditions. The amplified product size was confirmed by resolving on 1.2% TAE (tris-acetate EDTA) agarose gel, stained DNA with ethidium bromide and analyzed under UV light in gel documentation apparatus.
2.2. Construction of Mammalian Expression Plasmid Expressing Nonstructural Genes
The mammalian expression plasmids (i.e., pCR3.1/FlagTAG/nonstructural genes) were constructed by cloning first FlagTAG into a pCR3.1 vector by following the manufacturer’s protocols and by using standard cloning
Table 1. Sequences of primers for PCR amplification with restriction sites and RT-PCR analysis of HCV genotype 1a nonstructural genes.
methods (Table 1). After that, NS2, NS4A and NS5A amplicons were cloned into the pCR3.1/FlagTAG plasmid. The cloning of nonstructural genes was confirmed by PCR and restriction digestion reactions by using standard PCR amplification and restriction enzyme digestion kit’s protocol (Fermentas, Maryland, USA).
2.3. Cell Culturing and Viable Cell Count
Huh-7 cell line, was grown in complete cell culture medium containing DMEM (Dulbecco’s modified Eagle medium), 10% FBS (Fetal bovine serum; Sigma-Aldrich, USA), and 1000 μg/ml antibiotics (i.e., penicillin and streptomycin) in tissue culture flasks under humidified atmosphere at 37˚C. The old cell culture medium was replaced with the new one after 72 h, and the cells were passaged every 4 th - 5th days. For viable cell count and transfection of expression plasmids, the cells were splitted and plated at a density of 3 × 105 cells/well in 6-well tissue culture plate. For this purpose, the old cell culture media was removed from tissue culture flask, washed the cells with 5 ml of 1X PBS (Phosphate?buffered saline) and incubated the cells with 3 ml of trypsin/EDTA (Invitrogen Life Technologies, USA) to dislodge the cells at 37˚C for 3 - 5 minutes. Then 5 ml of fresh cell culture media was added to neutralize trypsin/EDTA (Ethylenediaminetetraacetic acid), and the cell suspension was transferred to a falcon tube for centrifugation at 3000 rpm for 2 minutes. The supernatant was removed, and the cell pellet was resuspended in 5 ml fresh media as a uniform single cell suspension. The cells were counted in a hemocytometer by placing a cover slide on it with 10 μl of dispensing cell suspension under an inverted light microscope. The trypan blue dye (Sigma-Aldrich, USA) explosive method was used to count the viable Huh-7 cells by following the manufacturer’s protocol. Briefly, prepared a 1:1 of the cell suspension to trypan blue dye and dispensed 10 μl on a glass slide and counted the cells by using hemocytometer under an inverted light microscope. The cells were grown at a density of 3 × 105 cells/well in 6-well tissue culture plates and incubated at 37˚C for 24 h prior to transfection.
2.4. Transfection of Huh-7 Cells with Mammalian Expression Plasmid
After 24 h, when the cells were 60% - 70% confluent in 6-well tissue culture plates, removed the old cell culture media and washed the cell monolayers 3 - 4 times with 1X PBS. After that, the cells were transfected with constructed plasmids (pCR3.1/FlagTAG/non-structural genes) in a dose-dependent manner (0.5 μg, 1 μg, and 2 μg) at different time intervals (24 h, 36 h and 48 h) by using Lipofectamine™ 2000 (Invitrogen life technologies, CA) as a transfection reagent by following the manufacturer’s instructions. The empty vector (i.e., PCR3.1/ FlagTAG) was used as mock transfection (i.e., negative control). After 6 - 8 h, the cell culture media was replaced to complete cell culture medium, and incubated the cells under humidified atmosphere at 37˚C until harvested at 24, 48 and 72 h for mRNA and protein expression analysis.
2.5. Total RNA Extraction and RT-PCR for mRNA Expression
The total cellular RNA was extracted by using the Trizol reagent (Invitrogen Life sciences, CA) according to the manufacturer’s instructions, and isolated RNA was stored at −80˚C prior use. First-strand cDNA was synthesized using 1 μg of extracted RNA by the First strand cDNA synthesis kit (Fermentas, Maryland, USA) according to the kit’s protocol. The cDNA was amplified by regular PCR using forward and reverse primers of cellular gene GAPDH (Glyceraldehyde phosphate dehydrogenase) and non-structural genes-specific RT-PCR primers under the optimized PCR conditions (Table 1). The amplified PCR products were subjected to 1.2% TAE agarose gel electrophoresis to confirm the mRNA expression of transfected genes.
2.6. Protein Extraction and Western Blot Analysis
After 24 and 48 h post transfection, the transfected Huh-7 cells were lysed with cell lysis buffer (proteoJET mammalian cell lysis reagent, Fermentas, CA) and total protein was extracted for protein expression analysis by Western blot. For stable cell clones, the cells were harvested with cell lysis buffer after 10th, 20th and 30th days post transfection. Before protein extraction, removed the cell culture medium and washed the cell monolayers with 1X PBS. After that, added 100 μl cell lysis buffer, scraped the cells and incubated the cell pellet in an Eppendorf tube on ice for 10 minutes. The cells suspension was centrifuged at 13,000 rpm at 4˚C for 25 minutes. The supernatant containing protein was transferred to an Eppendorf tube and stored at −20˚C prior to use for protein estimation and Western blotting. The protein concentration of each sample was determined by Bradford assay® (Bio Red, Germany) and was subjected to electrophoresis on 12% SDS-PAGE (Sodium dodecyl sulphate-Poly acryl amide gel electrophoresis) gels. After electrophoresis, the gel protein samples were transferred to a blot (i.e., nitrocellulose membrane) by following the manufacturer’s instructions (Bio-Red, CA). After that, the blot was blocked with 5% skimmed milk for 1 h at room temperature or 4˚C overnight to remove non-specific binding sites. Then the membrane was washed twice with 1X TBST (Tris-Buffered Saline Tween 20) solution and incubated with anti-mouse monoclonal antibodies (i.e., Primary antibodies) specific to non-structural proteins (NS2, NS4A & NS5A; Santa Cruz Biotechnology Inc, USA), and GAPDH (Santa Cruz Biotechnology Inc, USA) in 2.5% skim milk while suspending the membrane in 1X TBST solution and incubated for 1-1/2h at room temperature. The membrane was washed twice with IX TBST solution with mild agitation to remove primary antibody and incubated with secondary horseradish peroxidase-conjugated antibody (Sigma-Aldrich, USA) for 1 h at 25˚C. The membrane was washed again with 1X TBST solution to remove secondary antibody in the same way as the primary one. The protein expression was determined by using an enhanced chemiluminescence detection kit by following the kit’s protocol (Sigma-Aldrich, St. Louis, USA) and imaged by using X-ray film (Fuji film, Japan).
2.7. Huh-7 Stable Cell Line Generation Expressing NS2, NS4A and NS5A Proteins
Huh-7 cell clones persistently expressing NS2, NS4A and NS5A proteins after 30 days post-transfection were produced under the selection of antibiotic Gentamycin (G 418). Huh-7 cells with transfected plasmid were replated in 60 mm tissue culture plate after 48 h post transfection. Immediately, replaced the old cell growth media with new one adding 1000 μg/ml Gentamycin (G 418) and incubated the cells at 37˚C with 5% CO2 and 95% air. The cell colonies viable/resistant to 1000 μg/ml Gentamycin (G 418) appeared after some days, were picked carefully and cultured in the complete cell growth medium. The selection of stable cell clones expressing nonstructural genes occurred after 30 days in the presence of 750 μg/ml Gentamycin (G418). Nonstructural proteins expression both at mRNA and protein level was confirmed by RT-PCR and Western blot analysis.
2.8. siRNA Designing
The genome sequences of NS2, NS4A and NS5B genes were used to find targets sequences for siRNA designing and these sequences were found by using a web-based tool “siRNA Target Finder” (Ambion) followed by generation of siRNAs using the Silencer siRNA Construction Kit (Ambion, USA). siRNAs with 21nt sequences in the target gene sequence started with AA nucleotides at the 5’ end followed by 19nt and 30% - 50% GC content was selected (Table 2).
In order to minimize the complementary effects of siRNAs against human genomes, the homology to coding sequences in humans was checked by BLAST, available on the NCBI server at (www.ncbi.nlm.nih.gov/BLAST). A scrambled siRNA (used as internal control) was also designed on the same pattern as the experimental ones
Table 2. Sequences of siRNA oligonucleotides directed against nonstructural genes of HCV 1a genotype.
that lack significant sequence homology to HCV and human genomes.
2.9. Determination of siRNA Cellular Toxicity
MTT cell proliferation assay was used to evaluate the cytotoxic effects of siRNAs in Huh-7 cells by using Lipofectamine™ 2000 (Invitrogen, USA) as transfection reagent in a dose-dependent manner. Huh-7 cells at a density of 2 × 103 cells/well were cultured in 96-well tissue culture plate. After 24 h incubation, the cells were transfected with corresponding genome specific siRNAs from 10 nM to 50 nM dose. The plate was sealed and incubated at 37˚C in humidified atmosphere for overnight. After overnight incubation, replaced the old cell growth media with fresh one (100 μl) and added 20 μl of the MTT (3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide) solution in all wells of the plate. Wrapped the plate in an aluminum foil and incubated at 37˚C for 3 - 4 hrs. Again, carefully removed the cell growth media and added 100 μl of MTT solvent (solubilized in DMSO) in all wells of the plate to dissolve formazan crystals. Formazan products in cells (viable Huh-7 cells) were determined by measuring absorbance at a test wavelength (570 nm) and reference wavelength (620 nm) by an ELISA (Enzyme-linked immunosorbent assay) plate reader.
2.10. Transfection of Stable Cell Lines with Gene Specific siRNAs
To determine the inhibitory effects of siRNAs against nonstructural genes in stable cell lines, the cells were cultured in 6-well plate at a density of 3 × 105 cells/well for 24 h before transfection. When the plates were 60% - 80% cell confluent, removed the cell growth medium and washed the cells with 1X PBS. After that, transfected the cells by transfection mixture prepared by adding 50 nM of synthetic siRNAs (NS2, NS4A and NS5A genes specific) and 2 μl of lipofectamine™ 2000 diluted in 1ml DMEM supplemented with antibiotics but without 10% FBS. The mixture was added to each well of 6-well plate dropwise and swirled to ensure the equal distribution. After 6 h, removed the old cell growth media, washed the cells with 1X PBS and added cell growth medium without antibiotics and incubated at 37˚C with 5% CO2 and 95% air for 24 and 48 h. The total mRNA and protein was extracted after 24 h and 48 h post transfection to evaluate the siRNA inhibitory effects on NS2, NS4A and NS5A gene expression by RT-PCR and Western blot analysis
2.11. siRNA Inhibition Analysis by Quantitative Real-Time PCR
The quantitative siRNA inhibition against NS2, NS4A and NS5A genes in stable cell line was determined by quantitative Real-Time PCR (qPCR) ABI 7500 (Applied Biosystem, USA) using SYBR Green mix (Fermentas, Maryland, USA) and genome-specific primers at optimized conditions according to manufacturer’s instructions. The quantification of cellular GAPDH gene was used as an internal control for the data normalization. SDS 3.1 software was used for relative gene expression analysis. Each trial was performed in triplicate. Mean, standard deviation, standard error and level of significance was also determined by SPSS software (version 16.0).
2.12. Serum Sample Collection
The local HCV genotype 1a patient’s serum used in this study were collected from King Fahad Medical hospital and research center (KFMHRC) Jeddah, Saudi Arabia after clinical diagnosis under the provision of Institutional Review Board (IRB) of the KFMHRC. The patients under 18 years or above 70 and pregnant females were excluded. The participating subjects gave informed consent to collect blood samples for the study. The diagnosis of chronic HCV was based on elevated serum SGPT (Serum glutamic pyruvic transaminase) and SGOT (Serum glutamic oxaloacetic transaminase) levels at least for six months, histological examination and detection of active HCV mRNA by standard qualitative and quantitative PCR reactions. KFMHRC viral diagnostic laboratory determined the viral genotype of collected serum samples. Viral load was quantified by Real-Time quantitative PCR (qPCR) before conducting the trials. The samples were stored at −40˚C prior to precede RNA extraction.
2.13. Viral Inoculation of Huh-7 Cells and Co-Transfection with siRNAs
For viral inoculation experiments, we used similar protocols as described by Khaliq et al., 2010 with slight modifications [25] . A high viral load of patient’s sera (>5 × 107 IU/ml) were used to infect Huh-7 cells in these trials. Huh-7 cells (i.e., 3 × 105 cells/well) were cultured in 6-well tissue culture plate in complete cell culture media. When the plate was 60% - 80% cell confluent, the cells were washed twice with 1X PBS, inoculated with 500 μl (5 × 107 IU/ml) serum and 500 μl FBS-free cell culture media. After 6 h, cells were washed thrice with 1X PBS and complete cell culture media containing 10% FBS was added. The cells were incubated at 37˚C in humidified atmosphere for overnight. Next day, the adherent cell monolayer was washed twice with 1X PBS to get rid of remaining infection serum; added 2 ml of fresh cell culture medium and cell incubation was continued. After 72 h of viral inoculation, the cell monolayer was washed twice with 1X PBS before harvesting and extracting viral mRNA. The viral mRNA in Huh-7 cells was quantified by qPCR.
To determine the inhibitory effects of siRNAs on viral replication by calculating a decrease in HCV viral titer, serum infected Huh-7 cells were cultured again into 6 well plates (3 × 105 cells/well) until the cells become 60% - 80% confluent in 2 ml cell culture medium. The cells were transfected with 50 nM/well synthetic siRNAs directed against NS2, NS4A and NS5A genes and scrambled siRNA (internal control) by using Lipofectamine 2000™ (Invitrogen, USA) as transfection reagent according to the manufacturer’s protocol. After 72 h incubation, the total RNA was extracted from the cells by using the Purescript® RNA Isolation kit (Gentra System Pennsylvania, USA) following the kit protocol.
2.14. Quantitative HCV RNA Detection in Huh-7 Cells by Quantitative Real-Time PCR
HCV Real-TM Quant SC kit (Cepheid Sunnyvale, USA), was used for the quantitative detection of HCV using fluorescent reporter dye probes specific in the SmartCycler® (Cepheid). Reagents were thawed, and 16 Smart Cycler reaction tubes (25 μl) were prepared. The reaction mixture was prepared by adding into the tube with DTT (Dithiothreitol) 300 μl of RT-PCR-mix-1 (containing Cye 3 dye), 200 ml of RT-PCR-mix-2, 20 ml of Hot Start Polymerase and 10 μl of MMLV (Moloney Murine Leukemia Virus) Revertase. Vortexed the tube and mixed thoroughly by brief centrifugation. 12.5 ml of reaction mix and 12.5 ml of extracted RNA samples were added to an appropriate Smart Cycler tube. For each run, 6 standards and 1 negative control were prepared. 12.5 ml of quantitation standards for HCV (QS1 HCV, QS2 HCV and QS3 HCV) were added to 3 labeled tubes, while 12.5 μl of TE-Buffer was added to the tube labeled as negative control. After brief centrifugation, the tubes were inserted in the thermal cycler according to the set parameters for Real-Time PCR reaction conditions.
The concentration of HCV RNA in each sample was calculated by the following formula. The concentration was calculated in HCV IU/ml.
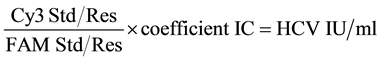 .
.
2.15. Statistical Analysis
SPSS software (version 16.0, SPSS Inc) was used to analyze the data. The data were presented as mean ± standard deviation (SD). p value < 0.05 was considered statistically significant.
3. Results
3.1. Construction of Mammalian Expression Plasmids Expressing HCV Nonstructural Proteins
The mammalian expression plasmids pCR3.1/FlagTAG, persistently expressing HCV non-structural proteins more than 30 days post-transfection were constructed to establish an in vitro stable Huh-7 cell culture system. First, we amplified NS2, NS4A and NS5A genes by using 200 ng HFL plasmid under optimized PCR conditions and confirmed the genes identity by rePCR, restriction digestion and sequencing PCR reactions (Figure 1).
Then the amplicons were cloned successfully into pCR3.1/FlagTAG expression plasmid that was confirmed by restriction digestion reactions and sequencing PCR analysis. The genome sequences of the cloned products were reported in a complete sequence homology with the parent HFL plasmid.
3.2. Stable Cell Line Generation Expressing NS2, NS4A and NS5A Proteins in Huh-7 Cells
Stable Huh-7 cell clones persistently expressing (NS2, NS4A & NS5A proteins up to 30th days post-transfection was produced under the selection pressure of antibiotic Gentamycin (G418). The non-structural proteins expression both at mRNA and protein levels was confirmed by RT-PCR and Western blot analysis (Figure 2 and Figure 3).
Figure 1. PCR amplification of HCV non-structural genes. HCV non structural genes (NS2, NS4A and NS5A) were amplified as separate amplicons from HFL plasmid. The amplified product size was confirmed by resolving on 1.2% TAE agarose gel, stained DNA with ethidium bromide and analyzed under UV light in gel documentation apparatus. (a) Lane M: 500 bp DNA size marker, NS2 (651 bp); (b) Lane M: 100 bp DNA size marker, NS4A (165 bp); (c) Lane M: 1 kb plus DNA size marker, NS5A (1344 bp).
Figure 2. HCV genotype 1a non-structural gene expression at mRNA level in stable Huh-7 cell lines. (a) GAPDH mRNA expression was used as an internal control in stable Huh-7 cell lines and for equal loading of the samples during RT-PCR at day 10th, 20th and 30th respectively. Lane M: 100 bp DNA size marker, -ve: negative control, Lane Huh-7: Huh-7 cells without any gene expression plasmid, Lane 1 - 9: GAPDH mRNA expression of stable Huh-7 cells trasnfected with NS2, NS4A and NS5A mammalian expression plasmid at day 10th, 20th and 30th respectively; (b) mRNA expression of NS2 gene (383 bp) in stable Huh-7 cell line detected by semi-quantitative RT-PCR at day 10th, 20th and 30th respectively. Lane M: 50 bp DNA size marker, -ve: Huh-7 cells without NS2 expression plasmid, Lane 1 - 3: NS2 mRNA expression (383 bp) at day 10th, 20th and 30th respectively; (c) mRNA expression of NS5A gene (260 bp) in stable Huh-7 cell line detected by semi-quantitative RT-PCR at day 10th, 20th and 30th respectively. Lane M: 100 bp DNA size marker, -ve: Huh-7 cells without NS5A expression plasmid, Lane 1 - 3: NS5A mRNA expression (260 bp) at day 10th, 20th and 30th respectively; (d) mRNA expression of NS4A gene (140 bp) in stable Huh-7 cell line detected by semi-quantitative RT-PCR at day 10th, 20th and 30th respectively. Lane M: 50 bp DNA size marker, -ve: Huh-7 cells without NS4A expression plasmid, Lane 1 - 3: mRNA expression of NS4A (260 bp) gene at day 10th, 20th and 30th respectively. GAPDH: Glyceraldehyde phosphate dehydogenase, bp: base pair.
Hybridization with anti-HCV anti-mouse monoclonal antibodies clearly showed the persistent expression of nonstructural proteins at day 10th, 20th, and 30th respectively (Figure 3). GAPDH mRNA and protein expression was used as a housekeeping gene (i.e., internal control) both in stable cell clones and non-transfected Huh-7 cells (negative control) and for equal loading of mRNA and protein samples during RT-PCR and Western blot analysis (Figure 2 and Figure 3).
3.3. Silencing Effects of Genome Specific siRNAs in Stable Cell Clones
Before the screening of siRNA inhibitory effects against NS2, NS4A and NS5A genes in the stable cell line, we determined that the constructed siRNAs have no cellular toxicity to Huh-7 cells by the MTT cell proliferation assay. The results revealed that at 50 nM dose, siRNA had no cellular toxicity to Huh-7 cells that were determined by calculating the percentage viable cell count against specific siRNAs as compared to the scrambled siRNA (Figure 4).
After that, we transfected stable cell lines with 50 nM dose of genome specific siRNAs named accordingly to NS2, NS4A and NS5A genes and observed the inhibition of corresponding gene expression at different time intervals (i.e., 24 h and 48 h). Results showed significant inhibition of mRNA and protein expression of corresponding genes in a dose dependent manner as compared to the positive control (Figures 5-7). RT-PCR and Western blot analysis revealed a significant decrease in NS2 gene expression at 48 h post transfection as compared to 24 h with both siRNAs (NS2-is11 & NS2-is22) (Figures 5(a)-(c)).
We further validated the results of RT-PCR and Western blot by quantitative Real-Time PCR. The relative quantitative analysis by Real-Time PCR revealed that the mRNA transcript level of NS2 gene decreased 17% (p < 0.05) with NS2-is11 and 27% (p < 0.05) with NS2-is22 after 24 h post-transfection (Figure 5(d)). The mRNA inhibition was 38% (p < 0.05) to NS2-is11 and 51% (p < 0.05) with NS2-is22 after 48 h post-transfection as compared to scrambled siRNA with no significant change in mRNA levels of NS2 gene in controlled Huh-7 cells (Figure 5(d)). Comparatively, the siRNA NS2-is22 significantly decreased NS2 gene expression in stable cell clones by reaching a maximum inhibition of 51% after 48 h post transfection (Figure 5(d)).
NS4A gene is considered to be the smallest gene of HCV genome. Therefore, only one siRNA (NS4A-is 55) was designed to determine whether siRNA trigger RNA interference against NS4A gene into stable cell clones. RT-PCR and Western blot analysis showed a significant decrease in expression of NS4A gene after 24 h and 48 h post-transfection (Figures 6(a)-(c)).
The relative quantitative analysis by qPCR demonstrated that the mRNA levels of NS4A gene were decreased 47% (p < 0.05) after 24 h post-transfection (Figure 6(d)), while 46% (p < 0.05) inhibition was noticed after 48 h
Figure 3. HCV genotype 1a non-structural genes expression at protein level in stable Huh-7 cell lines. (a) GAPDH protein expression (34 kDa) was used as housekeeping gene for equal loading of the protein samples and detected by using specific GAPDH monoclonal antibodies at day 10th, 20th and 30th respectively in stable Huh-7 cells; (b) NS2 (23 kDa), NS4A (8 kDa) and NS5A (52 kDa) protein expression was detected by Western blot analysis using anti-NS2, NS4A and NS5A anti-mouse monoclonal antibodies at day 10th, 20th and 30th respectively in stable Huh-7 cells.
Figure 4. MTT cell proliferation assay to evaluate siRNAs cyto-toxicity. Each MTT trial was performed in triplicate. The absorbance of colored formazan product (in viable Huh-7 cells after siRNA transfection) was quantified by measuring at a sample wavelength (570 nm) and reference wavelength (620 nm) by an ELISA plate reader. The percentage viable cell count after siRNA transfection was determined in comparison to scrambled siRNA.
Figure 5. Inhibition of NS2 gene expression at mRNA and protein level by synthetic siRNAs in stable Huh-7 cell line. (a)-(b) Dose dependent (at 50 nM) mRNA inhibition of NS2 gene after 24hrs and 48hrs post transfection in stable Huh-7 cells. The expression levels of control (i.e., NS2 plasmid expression in stable cell clones), GAPDH (i.e., housekeeping gene) and scrambled siRNA was also depicted in the Figure 5(a) and Figure 5(b). Each trial was performed in triplicate. Lane L: 100 bp DNA size marker (GAPDH), 50 bp DNA size marker (NS2). M: Mock transfection (Huh-7 cells without NS2 expression plasmid), C: Control (i.e., NS2 mRNA expression in stable cell line). GAPDH: Glyceraldehyde phosphate dehydrogenase, bp: base pair; (c) Inhibition of NS2 protein expression by synthetic siRNAs after 24 hrs and 48 hrs post transfection in stable Huh-7 cells. The GAPDH protein expression was also shown in the figure for equal loading of the protein samples for Western blot analysis. Lane M: Mock transfection (Huh-7 cells without NS2 expression plasmid), C: Control (i.e., NS2 protein expression in stable Huh-7 cell line). kDa: kilo Dalton; (d) Relative mRNA inhibition of NS2 gene in stable Huh-7 cells was determined by Real-Time PCR using gene specific primers after 24 hrs and 48 hrs post transfection. Each trial was performed in triplicate. p-value < 0.05 was considered statistically significant.
post-transfection as compared to the scrambled siRNA (Figure 6(d)).
To determine the siRNAs silencing effects against NS5A gene in stable Huh-7 cells, we transfected the cells with two synthetic siRNAs (NS5A-is66, NS5a-is77) at 50 nM dose for 24 and 48 h post transfection. The results exhibited that NS5A gene expression was significantly decreased by NS5A-is66 siRNA as compared to NS5A-is77 after 24 and 48 h post-transfection respectively (Figures 7(a)-(c)).
We further varified the RT-PCR and Western blot results by qPCR which demonstrated that mRNA levels of NS5A gene were decreased 52% (p < 0.05) with NS5A-is66 and 47% (p < 0.05) with NS5A-is77 after 24 h post-transfection (Figure 7(d)). Similarly, 54% (p < 0.05) decrease to mRNA expression was noticed with NS5A-is66 and 44% (p < 0.05) with NS5A-is77 after 48 h post-transfection (Figure 7(d)). Consequently, the siRNA NS5A-is66 was the most effective siRNA by reaching a maximum inhibition of 54% after 48 h post- transfection (Figure 7(d)).
3.4. Silencing Effects of siRNAs in Serum Infected Huh-7 Cells
We further elaborated the study while determining the silencing effects of siRNAs against HCV replication in serum infected Huh-7 cells by calculating a percentage decrease in HCV viral titer by Real Time PCR (Figure 8).
For this purpose, the cells were infected with high viral titer (>5 × 107 copies/μl) of HCV genotype 1a patient’s serum and incubated with gene-specific siRNAs. The total RNA was extracted from the cells, and relative quantitative Real-Time PCR was performed to determine a percentage decrease in HCV transcript levels (i.e., HCV viral titer) against gene specific siRNAs. The decrease in viral titer was much more significant with siRNA NS2-is22 i.e., 58% (p < 0.05) as compared to NS2-is11, which exhibited only 56% (p < 0.05) inhibition
Figure 6. Inhibition of NS4A gene expression at mRNA and protein level by synthetic siRNAs in stable Huh-7 cell line. (a)-(b) Dose dependent (at 50 nM) mRNA inhibition of NS2 gene after 24 hrs and 48 hrs post transfection in stable Huh-7 cells. The expression levels of control (i.e., NS4A plasmid expression in stable cell clones), GAPDH (i.e., housekeeping gene) and scrambled siRNA was also depicted in the Figure 6(a) and Figure 6(b). Each trial was performed in triplicate. Lane L: 100 bp DNA size marker (GAPDH), 50 bp DNA size marker (NS4A). M: Mock transfection (Huh-7 cells without NS4A expression plasmid), C: Control (i.e., NS4A mRNA expression in stable cell line). GAPDH: Glyceraldehyde phosphate dehydrogenase, bp: base pair; (c) Inhibition of NS4A protein expression by synthetic siRNAs after 24 hrs and 48 hrs post transfection in stable Huh-7 cells. The GAPDH protein expression was also shown in the figure for equal loading of the protein samples for Western blot analysis. Lane M: Mock transfection (Huh-7 cells without NS4A expression plasmid), C: Control (i.e., NS4A protein expression in stable Huh-7 cell line). kDa: kilo Dalton; (d) Relative mRNA inhibition of NS4A gene in stable Huh-7 cells was determined by Real-Time PCR using gene specific primers after 24 rs and 48 rs post transfection. Each trial was performed in triplicate. p-value < 0.05 was considered statistically significant.
(Figure 8(a)). The inhibition in viral load was 48% (p < 0.05) with NS4A genome specific siRNA (NS4A-is55) (Figure 8(b)). The siRNAs targeting NS5A region demonstrated 50% (p < 0.05) inhibition of viral load with siRNA NS5A-is66 and 44% (p < 0.05) with NS5A-is77 (Figure 8(c)). These findings suggested that the decrease in HCV viral titer by siRNAs may be helpful to block HCV replication. Consequently, the use of selective inhibitors like chemically synthesized siRNA might be considered as potential therapeutic approach against HCV.
4. Discussion
Hepatitis C virus is a serious human liver pathogen and considered to be a serious risk factor for associated hepatic diseases (hepatic fibrosis, cirrhosis, and hepatocellular carcinoma) in infected individuals [1] . The poor therapeutic response to current standard of care i.e., pegylated interferon (PEG-IFN) and ribavirin (RBV) and high therapy cost of the newer drugs demands urgently to improve the existing therapeutic approaches and to develop novel alternate anti-HCV treatment strategies [1] . The HCV nonstructural proteins play an essential role in viral replication, translation, polyprotein processing, interferon drug resistance and other regulatory activities [26] . Furthermore, these proteins are potential drug target sites for newly developed direct-acting antivirals and other anti-HCV agents [26] . Considering the importance of these proteins, we constructed the mammalian expression plasmids (i.e., pCR3.1/FlagTAG/HCV nonstructural genes) for siRNA inhibition studies against the functional residues of nonstructural proteins in stable Huh-7 cell lines. Huh-7 and their derivative cell lines are the most promising cell culture systems to study liver associated diseases and for the development of novel an-
Figure 7. Inhibition of NS5A gene expression at mRNA and protein level by synthetic siRNAs in stable Huh-7 cell line. (a)-(b) Dose dependent (at 50 nM) mRNA inhibition of NS5A gene after 24 hrs and 48 hrs post transfection in stable Huh-7 cells. The expression levels of control (i.e., NS5A plasmid expression in stable cell clones), GAPDH (i.e., housekeeping gene) and scrambled siRNA was also depicted in the Figure 7(a) and Figure 7(b). Each trial was performed in triplicate. Lane L: 100 bp DNA size marker for GAPDH and NS5A. M: Mock transfection (Huh-7 cells without NS5A expression plasmid), C: Control (i.e., NS5A mRNA expression in stable cell line). GAPDH: Glyceraldehyde phosphate dehydrogenase, bp: base pair; (c) Inhibition of NS5A protein expression by synthetic siRNAs after 24 hrs and 48 hrs post transfection in stable Huh-7 cells. The GAPDH protein expression was also shown in the figure for equal loading of the protein samples for Western blot analysis. Lane M: Mock transfection (Huh-7 cells without NS5A expression plasmid), C: Control (i.e., NS5A protein expression in stable Huh-7 cell line). kDa: kilo Dalton; (d) Relative mRNA inhibition of NS5A gene in stable Huh-7 cells was determined by Real-Time PCR using gene specific primers after 24 hrs and 48 hrs post transfection. Each trial was performed in triplicate. p-value < 0.05 was considered statistically significant.
Figure 8. siRNA Inhibitory effects on viral load in serum infected Huh-7 cells. A percentage decrease in viral titer upon transfection of synthetic siRNAs directed against NS2, NS4A and NS5A genes was determined by relative quantitative Real-Time PCR in serum infected Huh-7 cells. (a)-(c): The decrease in viral titer upon transfection of siRNAs directed against NS2, NS4A and NS5A genes. Each trial was performed in triplicate. p-value < 0.05 was considered statistically significant as compared to the S 1a. S 1a (i.e., serum of HCV genotype 1a patient; +ve control).
ti-HCV compounds. Several researchers have demonstrated HCV genotype 1 - 4 replication in primary human hepatocytes [10] . Some studies also reported the in vitro stable cell culture system persistently expressing HCV genotype 3a proteins circulating in the local populations [9] . In the current study, we reported an in vitro stable Huh-7 cell culture system, which allowed continuous, defined and reproducible expression of HCV genotype 1a nonstructural proteins (NS2, NS4A, and NS5A) over 30 days post transfection (Figure 2 and Figure 3). The ultimate purpose of this approach was to evaluate the inhibitory/silencing effects of siRNAs against the expression of nonstructural proteins in stable cell clones. Some studies have described that siRNAs inhibitory effects in transient gene expression were not so much significant and useful to evaluate their therapeutic activity [27] [28] , so we adopted this strategy. The stable expression of nonstructural genes at mRNA and protein level was confirmed by RT-PCR and Western blot analysis (Figure 2 and Figure 3). The results revealed reproducible mRNA and protein expression of NS2, NS4A and NS5B genes over time at day 10th, 20th, and 30th to be used for in vitro RNA interference studies.
As a gene silencing mechanism, RNA interference (RNAi) is one of the key methods used in applied and functional genomics to study the knockout functions of some genes involved in the pathogenesis of certain viral and infectious diseases. Because of its effectiveness and specificity in gene silencing, RNAi is applicable to be used for hard-to-cure diseases like HIV infection, cancers and certain genetic disorders [16] -[18] . A number of investigators have demonstrated that antiviral siRNAs interfere with the replication of animal viruses like HIV-I (Human immunodeficiency virus), flock house virus (FVH) and poliovirus [24] . Similarly, many researchers have also reported that both siRNAs and shRNAs (short hairpin RNA) remarkably inhibits viral replication by targeting HCV genotype 1a and 1b genome in replicon cells [29] . In this study, we targeted the highly conserved regions of HCV nonstructural proteins (NS2, NS4A and NS5A) with synthetic siRNAs according to the well established rules and protocols. We purposed that RNAi-based anti-HCV strategy could be used to target NS2, NS4A and NS5A region of HCV 1a genotype in an in-vitro stable cell line model. Chemically synthesized siRNAs can be introduced into stable Huh-7 cell lines by transfecting with some lipophilic reagents. We were able to show that the transfection of synthetic siRNAs significantly decreased NS2, NS4A and NS5A gene expression in stable Huh-7 lines. RNA interference activities by measuring the mRNA and protein expression of the gene of interest in stable Huh-7 cells may helpful to identify potent siRNA duplexes which selectively suppress HCV gene expression. To achieve optimized efficacy and to determine significantly reduced gene expression, sequence-specific siRNAs directed against NS2, NS4A, and NS5A genes were induced into stable Huh-7 cells in a dose-dependent manner. Our findings are according to the study of Liu et al. and Kim et al. who described significantly decreased expression of HCV genotype 1a and 1b structural and nonstructural genes against the several siRNAs in a dose-dependent manner [30] [31] .
NS2 is an essential protein for the completion of HCV replication in the cytoplasm of host cells. In 1993, Grakoui et al. described that residue H143, E163 and C184 (H952, E972 and C993 when numbered in polyprotein) constitute a catalytic triad of NS2/NS3 protease and are conserved among all HCV genotypes [32] . Therefore, two siRNAs (NS2-is11 & NS2-is22) were designed against the targeted regions which encode H143, E163 and C184 residues. NS2-is22 demonstrated 51% (p < 0.05) mRNA inhibition at 48 h post transfection as measured by quantitative Real-Time PCR (Figure 5(d)). The decrease in mRNA expression by NS2-is11 after 24 and 48 h post transfection and NS2-is22 after 24 h were not much significant (Figure 5(d)). Consequently, NS2-is22 was considered the most effective siRNA to decrease mRNA transcript level of NS2 gene at a significant level in stable Huh-7 cell line.
NS4A is a cofactor and an activating subunit of NS3-4A serine protease complex. Bartenschlager et al. and Tanji et al. in 1995 showed that central region of the protein (residue 21 to 34) is essential and sufficient for cofactor function of the protein [33] [34] . Some studies revealed that NS4A is essential for the optimal activity of NS3 serine protease. In its absence, only one cleavage site (i.e., NS5A/NS5B) is partially processed by NS3 protease alone. It is a small size protein, so only one siRNA NS4A-is55 was designed from the central region of the gene and evaluated for RNAi activity against NS4A in a stable cell line. The qPCR revealed a vivid decrease in the mRNA transcript level of NS4A gene up to 47% (p < 0.05) after 24 h and 48 h post-transfection (Figure 6(d)).
NS5A is a phosphoprotein that plays an essential role in HCV replication, modulation of cell signaling and confers resistance to interferon therapy [35] [36] . Initially, NS5A grabbed the attention due to its role in modulating the interferon therapy response [37] . It contains a domain that confers resistance to interferon treatment in HCV-infected patients known as interferon-α sensitivity determining region (ISDR) [38] . The inhibition of NS5A may impair the resistance mechanism against interferon by reinstating the innate immune responses. Two siRNAs (NS5A-is66 and NS5A-is77) were designed to NS5A gene, and mRNA transcript level was decreased up to 44% - 54% (p < 0.05) in stable cell line after 24 h and 48 h post-transfection (Figure 7(d)).
The researchers have also been reported HCV replication in serum-infected liver cell lines which presumably resemble to naturally occurring HCV infection in humans [10] [39] [40] . Similarly in 2005, Guha et al. demonstrated that in vitro Huh-7 cell culture models are the best to determine viral infectivity and to evaluate anti-viral agents against HCV infection [41] . To rationalize our study in this prospectus, we further validated the therapeutic induction of siRNAs against NS2, NS4A and NS5A genes in an in vitro viral replicate cell culture model (i.e., serum infected Huh-7 cells). For this purpose, first Huh-7 cells were infected with HCV genotype-1a patient’s serum and after that transfected with sequence specific siRNAs. A decrease in viral titer was determined by the detection of 5’ UTR of viral copies using qPCR after 3rd day post-infection [25] and compared to the positive control (serum of HCV 1a genotype patients) which remained unaltered. The viral titer was decreased up to 58% (p < 0.05) by NS2 specific siRNAs in serum infected Huh-7 cells (Figure 8(a)). NS4A genome specific siRNA decreased viral replication only 48% (p < 0.05) (Figure 8(b)), while the siRNAs directed against NS5A gene decreased HCV viral titer about 50% (p < 0.05) (Figure 8(c)). These findings are in accordance to the studies demonstrated by Zekri et al. in 2009 who described inhibition of HCV replication in serum infected Huh-7 cells by siRNAs directed against 5’ UTR of HCV genotype 4 [23] . Furthermore, some researchers have also reported the inhibition of HCV replication in serum infected Huh-7 cells by targeting NS3, NS5A and NS5B genes [42] -[44] . We presume that a significant decrease in HCV viral titer with siRNAs in serum infected Huh-7 cells may be an adjuvant therapy to inhibit HCV replication with other combination drugs. Therefore, siRNA mediated gene silencing may be considered a potential candidate for an alternate therapy to treat the chronic HCV infection. Pan et al. demonstrated one supportive study in 2009 while reporting that RNAi in combination with interferon-α might significantly increase their individual antiviral effects [45] [46] .
5. Conclusion
In conclusion, siRNA inhibitory effects against HCV nonstructural proteins in an in vitro stable cell line model and in serum infected Huh-7 cells may indicate that there is an enormous potential for RNAi-based therapeutics against HCV infection. Thus, if the issue with efficient siRNA delivery, off-target effects, viral escape mutants and safety concerns are overcome, it would be trivial to apply this technology to many different hepatotropic diseases.
Acknowledgements
The authors highly acknowledge Mrs. Sidrah Hafeez, Department of English, Pakistan international school Jeddah, Saudi Arabia, for her kind help in proofreading the manuscript.
Declaration of Interest
The authors declare that they have no potential conflict of interests by any means to any institution, organization while publishing this study.
Cite this paper
ImranShahid,Waleed HassanAlMalki,Shaia SalehR. Almalki,Ismail MuhammadAlTurkestany,Hassan AliAlGhamdi,Saleh AliAlMenshawi, (2015) Inhibition of Hepatitis C Virus Genotype 1a Non-Structural Proteins by Small Interference RNA in Human Hepatoma Cell Lines. Pharmacology & Pharmacy,06,502-517. doi: 10.4236/pp.2015.611053
References
- 1. Shahid, I., AlMalki, W., Hafeez, M. and Hassan, S. (2014) Hepatitis C Virus Infection Treatment: An Era of Game Changer Direct Acting Antivirals and Novel Treatment Strategies. Critical Reviews in Microbiology, 6, 1-13.
http://dx.doi.org/10.3109/1040841X.2014.970123 - 2. Shahid, I., Ahmad, W., Ijaz, B., Javed, F.T., Gull, S., Kausar, H., Sarwar, M.T., Asad, S., Sumrin, A., Khaliq, S., Jahan, S., Pervaiz, A. and Hassan, S. (2013) Stable Huh-7 Cell Line Development Expressing HCV Non-Structural Proteins of Genotype 1a. Journal of Virological Methods, 4, 4.
- 3. Asad, S., Ijaz, B., Ahmad, W., Kausar, H., Sarwar, T., Gull, S., Shahid, I., Khan, M. and Hassan S. (2012) Development of Persistent HCV Genotype 3a Infection Cell Culture Model in Huh-7 Cell. Virology Journal, 9, 11.
http://dx.doi.org/10.1186/1743-422X-9-11 - 4. Ansar, M., Ashfaq, U.A., Shahid, I., Sarwar, T., Javed, T., Rehman, S., Hassan, S. and Riazuddin S. (2011) Inhibition of Full Length Hepatitis C Virus Particles of 1a Genotype through Small Interference RNA. Virology Journal, 8, 203.
http://dx.doi.org/10.1186/1743-422X-8-203 - 5. Ahmad, W., Ijaz, B., Javed, F.T., Jahan, S., Shahid, I., Khan, M. and Hassan S. (2010) HCV Genotype Distribution and Possible Transmission Risks in Lahore, Pakistan. World Journal of Gastroenterology, 16, 4321-4328.
http://dx.doi.org/10.3748/wjg.v16.i34.4321 - 6. Ahmad, W., Shabbiri, K., Ijaz, B., Asad, S., Sarwar, M.T., Gull, S., Kausar, H., Fouzia, K., Shahid, I. and Hassan S. (2011) Claudin-1 Required for HCV Virus Entry Has High Potential for Phosphorylation and O-Glycosylation. Virology Journal, 8, 229.
http://dx.doi.org/10.1186/1743-422X-8-229 - 7. Sarwar, M.T., Kausar, H., Ijaz, B., Ahmad, W., Ansar, M., Sumrin, A., Ashfaq, U.A., Asad, S., Gull, S., Shahid, I. and Hassan S. (2011) NS4A Protein as a Marker of HCV History Suggests that Different HCV Genotypes Originally Evolved from Genotype 1b. Virology Journal, 8, 317.
http://dx.doi.org/10.1186/1743-422X-8-317 - 8. Lazaro, A., Chang, M., Tang, W., Campbell, J., Sullivan, G., Gretch, R., Corey, L., Coombs, W. and Fausto N. (2007) Hepatitis C Virus Replication in Transfected and Serum-Infected Cultured Human Fetal Hepatocytes. American Journal of Pathology, 170, 478-489.
http://dx.doi.org/10.2353/ajpath.2007.060789 - 9. Butt, S., Idrees, M., Rehman, U., Ali, L., Hussain, A., Ali, M., Ahmed, N., Saleem, S. and Fayyaz M. (2011) Establishment of Stable Huh-7 Cell Lines Expressing Various Hepatitis C Virus Genotype 3a Protein: An In-Vitro Testing System for Novel Anti-HCV Drugs. Genetics Vaccines Therapy, 9, 12-17.
http://dx.doi.org/10.1186/1479-0556-9-12 - 10. Buck, M. (2008) Direct Infection and Replication of Naturally Occurring Hepatitis C Virus Genotypes 1, 2, 3 and 4 in Normal Human Hepatocyte Cultures. PLoS One, 3, e2660.
http://dx.doi.org/10.1371/journal.pone.0002660 - 11. Molina, S., Castet, V., Pichard-Garcia, L., Wychowski, C., Meurs, E., Pascussi, M., Sureau, C., Fabre, M., Sacunha, A. and Larrey, D. (2008) Serum-Derived Hepatitis C Virus Infection of Primary Human Hepatocytes Is Tetraspanin CD81 Dependent. Journal of Virology, 82, 569-574.
http://dx.doi.org/10.1128/JVI.01443-07 - 12. Lisowski, L., Elazar, M., Chu, K., Glenn, J. and Kay, M. (2013) The Anti-Genomic (Negative) Strand of Hepatitis C Virus Is Not Targetable by shRNA. Nucleic Acids Research, 41, 3688-3698.
http://dx.doi.org/10.1093/nar/gkt068 - 13. Zheng, D., Giljohann, A. and Chen, L. (2012) Topical Delivery of siRNA-Based Spherical Nucleic Acid Nanoparticle Conjugates for Gene Regulation. Proceedings of the National Academy of Sciences of the United States of America, 109, 11975-11980.
http://dx.doi.org/10.1073/pnas.1118425109 - 14. Gao, K., and Huang, L. (2009) Nonviral Methods for siRNA Delivery. Molecular Pharmacology, 6, 651-658.
http://dx.doi.org/10.1021/mp800134q - 15. Zintchenko, A., Philipp, A., Dehshahri, A. and Wagner, E. (2008) Simple Modifications of Branched PEI Lead to Highly Efficient siRNA Carriers with Low Toxicity. Bioconjugation Chemistry, 19, 1448-1455.
http://dx.doi.org/10.1021/bc800065f - 16. Caplen, J., Parrish, S., Imani, F., Fire, A. and Morgan, A. (2001) Specific Inhibition of Gene Expression by Small Double-Stranded RNAs in Invertebrate and Vertebrate Systems. Proceedings of the National Academy of Sciences of the United States of America, 98, 9742-9747.
http://dx.doi.org/10.1073/pnas.171251798 - 17. Coburn, A. and Cullen, R. (2002) Potent and Specific Inhibition of Human Immunodeficiency Virus Type 1 Replication by RNA Interference. Journal of Virology, 76, 9225-9231.
http://dx.doi.org/10.1128/JVI.76.18.9225-9231.2002 - 18. Brummelkamp, R., Bernards, R. and Agami, R. (2002) Stable Suppression of Tumorigenicity by Virus-Mediated RNA Interference. Cancer Cell, 2, 243-247.
http://dx.doi.org/10.1016/S1535-6108(02)00122-8 - 19. Hannon, J. (2002) RNA Interference. Nature, 418, 244-251.
http://dx.doi.org/10.1038/418244a - 20. Korf, M., Meyer, A., Jarczak, D., Beger, C., Manns, P. and Kruger, M. (2007) Inhibition of HCV Subgenomic Replicons by siRNAs Derived from Plasmids with Opposing U6 and H1 Promoters. Journal of Viral Hepatology, 14, 122-132.
http://dx.doi.org/10.1111/j.1365-2893.2006.00793.x - 21. Seo, Y., Abrignani, S., Houghton, M. and Han, H. (2003) Small Interfering RNA-Mediated Inhibition of Hepatitis C Virus Replication in the Human Hepatoma Cell Line Huh-7. Journal of Virology, 77, 810-812.
http://dx.doi.org/10.1128/JVI.77.1.810-812.2003 - 22. Khaliq, S., Jahan, S., Ijaz, B., Ahmad, W., Asad, S. and Hassan, S. (2011) Inhibition of Hepatitis C Virus Genotype 3a by siRNAs Targeting Envelope Genes. Archives of Virology, 156, 433-442.
http://dx.doi.org/10.1007/s00705-010-0887-6 - 23. Zekri, R., Bahnassy, A., El-Din, M. and Salama, M. (2009) Consensus siRNA for Inhibition of HCV Genotype-4 Replication. Virology Journal, 6, 13.
http://dx.doi.org/10.1186/1743-422X-6-13 - 24. Shin, D., Lee, H., Kim, I., Yoon, Y. and Kim M. (2009) Optimization of Linear Double-Stranded RNA for the Production of Multiple siRNAs Targeting Hepatitis C Virus. RNA, 15, 898-910.
http://dx.doi.org/10.1261/rna.1268209 - 25. Khaliq, S., Jahan, S., Ijaz, B., Ahmad, W., Asad, S., Pervaiz, A., Samreen, B., Khan, M. and Hassan, S. (2010) Inhibition of Core Gene of HCV 3a Genotype Using Synthetic and Vector Derived siRNAs. Virology Journal, 7, 318.
http://dx.doi.org/10.1186/1743-422X-7-318 - 26. Ahmad, W., Ijaz, B., Javed, T., Kausar, H., Sarwar, M.T., Gull, S., Asad, S., Shahid, I. and Hassan, S. (2011) HCV Genotype-Specific Correlation with Serum Markers: Higher Predictability for Genotype 4a. Virology Journal, 8, 293.
http://dx.doi.org/10.1186/1743-422X-8-293 - 27. Grimm, S. (2004) The Art and Design of Genetic Screens: Mammalian Culture Cells. Nature Reviews in Genetics, 5, 179-189.
http://dx.doi.org/10.1038/nrg1291 - 28. Wurm, M. (2004) Production of Recombinant Protein Therapeutics in Cultivated Mammalian Cells. Nature Biotechnology, 22, 1393-1398.
http://dx.doi.org/10.1038/nbt1026 - 29. Jarczak, D., Korf, M., Beger, C., Manns, P. and Kruger, M. (2005) Hairpin Ribozymes in Combination with siRNAs against Highly Conserved Hepatitis C Virus Sequence Inhibit RNA Replication and Protein Translation from Hepatitis C Virus Subgenomic Replicons. FEBS Journal, 272, 5910-5922.
http://dx.doi.org/10.1111/j.1742-4658.2005.04986.x - 30. Kim, M., Shin, D., Kim, S. and Park, M. (2006) Inhibition of Hepatitis C Virus Gene Expression by Small Interfering RNAs Using a Tri-Cistronic Full-Length Viral Replicon and a Transient Mouse Model. Virus Research, 122, 1-10.
http://dx.doi.org/10.1016/j.virusres.2006.05.003 - 31. Liu, M., Ding, H., Zhou, P., Qin, L., Gao, J., Cao, M., Luan, J., Wu, B. and Qi, T. (2006) RNA Interference Effectively Inhibits mRNA Accumulation and Protein Expression of Hepatitis C Virus Core and E2 Genes in Human Cells. Bioscience Biotechnology and Biochemistry, 70, 2049-2055.
http://dx.doi.org/10.1271/bbb.60001 - 32. Grakoui, A., McCourt, W., Wychowski, C., Feinstone, M. and Rice, M. (1993) Characterization of the Hepatitis C Virus-Encoded Serine Proteinase: Determination of Proteinase-Dependent Polyprotein Cleavage Sites. Journal of Virology, 67, 2832-2843.
- 33. Bartenschlager, R., Lohmann, V., Wilkinson, T. and Koch, O. (1995) Complex Formation between the NS3 Serine-Type Proteinase of the Hepatitis C Virus and NS4A and Its Importance for Polyprotein Maturation. Journal of Virology, 69, 7519-7528.
- 34. Tanji, Y., Hijikata, M., Satoh, S., Kaneko, T. and Shimotohno, K. (1995) Hepatitis C Virus Encoded Non-Structural Protein NS4A Has Versatile Functions in Viral Protein Processing. Journal of Virology, 69, 1575-1581.
- 35. Macdonald, A., Crowder, K., Street, A., McCormick, C. and Harris, M. (2004) The Hepatitis C Virus NS5A Protein Binds to Members of the Src Family of Tyrosine Kinases and Regulates Kinase Activity. Journal of General Virology, 85, 721-729.
http://dx.doi.org/10.1099/vir.0.19691-0 - 36. Reed, E., Xu, J. and Rice, M. (1997) Phosphorylation of the Hepatitis C Virus NS5A Protein in Vitro and in Vivo: Properties of the NS5A-Associated Kinase. Journal of Virology, 71, 7187-7197.
- 37. Tan, L. and Katze, G. 92001) How Hepatitis C Virus Counteracts the Interferon Response: The Jury Is Still Out on NS5A. Virology, 284, 1-12.
http://dx.doi.org/10.1006/viro.2001.0885 - 38. Gale, J., Korth, M.J., Tang, M., Tan, L., Hopkins, A., Dever, E., Polyak, J., Gretch, R. and Katze, G. (1997) Evidence That Hepatitis C Virus Resistance to Interferon Is Mediated through Repression of the PKR Protein Kinase by the Non-Structural 5A Protein. Virology, 230, 217-227.
http://dx.doi.org/10.1006/viro.1997.8493 - 39. El-Awardy, K., Tabll, A., El-Abd, S., Bahgat, M., Shoeb, A., Youssef, S., El-Din, B., Redwan, M., El-Demellawy, M., Omran, H., El-Garf, T. and Goueli, A. (2006) HepG2 Cells Support Viral Replication and Gene Expression of Hepatitis C Virus Genotype 4 in Vitro. World Journal of Gastroenterology, 12, 4836-4842.
- 40. Lazaro, A., Chang, M., Tang, W., Campbell, J., Sullivan, G., Gretch, D., Corey, L., Coombs, W. and Fausto, N. (2007) Hepatitis C Virus Replication in Transfected and Serum-Infected Cultured Human Fetal Hepatocytes. American Journal of Pathology, 170, 478-489.
http://dx.doi.org/10.2353/ajpath.2007.060789 - 41. Guha, C., Lee, W., Chowdhury, R. and Chowdhury, J. (2005) Cell Culture Models and Animal Models of Viral Hepatitis. Part II: Hepatitis C. Laboratory Animals, 34, 39-47.
http://dx.doi.org/10.1038/laban0205-39 - 42. Kanda, T., Steele, R., Ray, R. and Ray, B. (2007) Small Interfering RNA Targeted to Hepatitis C Virus 5’ Nontranslated Region Exerts a Potent Antiviral Effect. Journal of Virology, 81, 669-676.
http://dx.doi.org/10.1128/JVI.01496-06 - 43. Kapadia, B., Brideau-Andersen, A. and Chisari, V. (2003) Interference of Hepatitis C Virus RNA Replication by Short Interfering RNAs. Proceedings of the National Academy of Sciences of the United States of America, 100, 2014-2018.
http://dx.doi.org/10.1073/pnas.252783999 - 44. McCaffrey, P., Mause, L., Pham, T., Conklin, S., Hannon, J. and Kay, A. (2002) RNA Interference in Adult Mice. Nature, 418, 38-39.
http://dx.doi.org/10.1038/418038a - 45. Pan, Q., Henry, D., Metselaar, J., Scholte, B., Kwekkeboom, J., Tilanus, W., Janssen, L. and van der Laan, J. (2009) Combined Antiviral Activity of Interferon-Alpha and RNA Interference Directed against Hepatitis C without Affecting Vector Delivery and Gene Silencing. Journal of Molecular Medicine, 87, 713-722.
http://dx.doi.org/10.1007/s00109-009-0470-3 - 46. Smith, M., Smolic, R., Volarevic, M. and Wu, Y. (2007) Positional Effects and Strand Preference of RNA Interference against Hepatitis C Virus Target Sequences. Journal of Viral Hepatitis, 14, 194-212.
http://dx.doi.org/10.1111/j.1365-2893.2006.00794.x
NOTES

*Corresponding author.










