E-Health Telecommunication Systems and Networks
Vol.05 No.01(2016), Article ID:64772,12 pages
10.4236/etsn.2016.51003
Agent Petri Nets Framework for Modeling Staphylococcus epidermidis Biofilm Formation
Borhan Marzougui1, Kamel Barkaoui2, Mohamed Amine Makni3
1Information Technology Department, ECT, Abu Dhabi, United Arab Emirates
2CNAM, Paris, France
3National Bone-Marrow Transplantation Center of Tunis, Tunis, Tunisia

Copyright © 2016 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 19 December 2015; accepted 17 March 2016; published 21 March 2016
ABSTRACT
This Staphylococcus epidermidis has been discovered as the most frequent germ detected during indwelling medical devices infection. This fact is well attached with the ability of this bacterium to form structured layered population known as biofilm. Inside S. epidermidis biofilm, bacterial cells present more different behavior than in their planktonic counterpart. This paper describes the thriving application of Petri net theory for modeling of interaction between different regulations actors leading S. epidermidis to switch from Planctonik to Biofilm. Indeed this biologic system is very sensible and has dangerous effect. We propose Agent Petri Nets model to describe and analyze the process of formation of Biofilm molecule. This model presents a formal framework based on Multi Agents system characteristics.
Keywords:
Staphylococcus epidermidis, Biofilm, Petri Net, Modeling, Agent

1. Introduction
Inside biological system, cells are composed by thousands of components that interact in a myriad of ways. Despite this interconnection, it is necessary to classify these networks of cells according to their biological function.
The emerging of systems biology with multi-disciplinary field is involved to the study of the relationships between various parts of a biological system, and modeling method. They are vital role in the drive to present the processes of life. Advancements in experimental technologies in biology and medicine have generated an amount of biological data. Many different molecular cell processes interact and change their behavior quickly. So, we need develop methods for exploring this various data. Much formalism from the fields of biology, mathematics and the computer sciences is used to integrate, represent and analyze the vast amount of biological data.
To understand the functioning of complex biological systems, it is necessary to model the interactions that take place. In fact, the use of a formal method is crucial to prevent ambiguities, uncertainties and even contradictions to appear in dynamic biological systems. Petri Nets allow the analysis of qualitative structural to quantitative behavioral properties. PNs are effective for the modeling of molecular networks [1] . In fact, the mathematical formalism of Petri net theory is able to encompass many of these techniques. Various extensions to the original theory of Petri nets have been used for modeling molecular biology systems and metabolic [2] . Such systems permit to coordinate various molecules. We propose in this work to model this molecule as agent and biologic system as Multi agent system. The specification of biologic system is complex and each entity can interact and communicate in a dynamic environment. Indeed the complexity of the systems studied is increasing. The precision, reliability and the hardiness have become difficult factors to reach. Therefore, the integration of a mathematical tool offers an exact way, in presence of graphic tools, to succeed the conception of these systems, especially the multi agent systems.
This paper focuses on the theoretical foundations of modeling Biological Systems based on Agent Petri Nets. Section 2 introduces various preliminaries, including the advantages notion of Multi Agents System and Agent Petri nets. Section 3 discusses the specification of Biological systems, such the properties as Multi Agent System of this system is introduced. Section 4 shows how to create a general framework to model Biological systems based on Agent Petri nets. Related work is discussed in Section 6. Section 7 concludes the paper.
The formatter will need to create these components, incorporating the applicable criteria that need following.
2. Preliminaries
This section introduces basic concepts related to Multi Agents System and Petri nets. Moreover, we introduce the notion of Agent Petri Nets. This formalism will be used to descript and model Biological systems.
2.1. Multi Agents System
Multi agents system is used to model complex systems which can be decomposed into several interacting entities called agents.
An agent is defined as an autonomous entity capable of communicating with other agents to partially discern at least its environment and the objects that surround it, and to have correct or erroneous representations about the behaviors of a part or the set of the gents of the environment. So, contrary to the objects, an agent possesses an autonomous behavior. It is capable of taking some decisions and establishing plans of actions to accomplish complex activities. An intelligent agent resides in a dynamic environment and can realize autonomous actions in order to achieve its goals. In deeded, the most important reason to implement agent paradigm when designing a complex a system such Biologic system, is that agent has the potential and the competence to assure the reliability of the modeling process. The multi agent system is expected to be autonomous, adaptable robust and distributed. Multi agent systems can be involves two main concepts: agent and environment. The most important actions in MAS specification is that of communication and interaction among the agents.
Several researches treated the concept of formal descriptions of multi agent system. Formal descriptions aim to assess proprieties and to provide formal specifications of this complex system.
2.2. Petri Nets
Petri Nets may serve as convenient formalism integrating quantitative and qualitative modeling and analysis techniques. Petri Nets are often used in the context of Biological systems. Various models employ Petri Nets as the internal representation used for process analyzing Biological system.
Definition 1 (Petri Net): A Petri Net is a Tuple 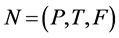 consists of two finite, nonempty, and disjoint sets of Places P and set of Transitions T, a flow relation
consists of two finite, nonempty, and disjoint sets of Places P and set of Transitions T, a flow relation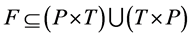 ,
, 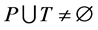 and
and . N can be define as
. N can be define as 
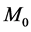 presents the initial marking. Places and transitions are collectively called nodes. For a node
presents the initial marking. Places and transitions are collectively called nodes. For a node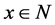 , we
, we
define its pre-nodes by 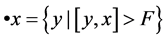 and its pre-nodes by
and its pre-nodes by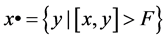 .
.
Definition 2 (Behavior): Transition  is enabled in marking
is enabled in marking 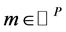 iff, for all
iff, for all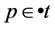 ,
, . We denote this by
. We denote this by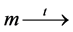 . If t is enabled, t can fire in m, yielding marking
. If t is enabled, t can fire in m, yielding marking 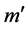 where, for all
where, for all



2.3. Agent Petri Nets [3]
Most of the result presented in the paper, can be adapted for various Multi Agent System. However, we use Agent Petri Nets to formalize the main framework of Biological Systems and to prove their correctness.
Agent Petri nets was introduced in [3] - [5] . An Agent Petri Nets is defined as being a directed bipartisan graph that has two types of nodes (place and transition). Every transition carries the functions that manipulate the internal state and the behavior of an Agent (Token) in its environment. The distribution of the tokens in the places at a given moment is called marking of the Agent Petri nets. A marking gives the state of the system that depends on the interaction between the entities that compose it. The change in internal state or the behavior of every Agent or of the set of system is assured by Agents functions.
We present a description of most functionality of APN by the following definitions:
In a formal manner, Agent Petri Nets defined by the 9-uplet:

where:
P: a non-empty finished set of places;
T: a non-empty finished set of transitions;
A: a non-empty finished set of agents;
Meadow: 
Post: 
Prj: pre condition of firing;
F (Ai, Aj): agent relation function presenting the condition of firing;
Ft: function agent that uses 3 variables:
Envk: Environment of work that describes Multi Agent System;
Definition 4: Function of Adherence (Relative to an Agent).
This function gives rise to a relation between an agent and its environment.
In a formal manner, the adherence function of an agent Ai, in an environment Envk noted Apai is defined by:
where:
b: constraint = Prj (b = 0 or b = 1): the engagement of Ai in Envk.

Definition 5: Function Agent Ft.
The function agent describes the relation between two agents. It modifies the values descended directly of an agent. These define the capacity to discern and to react to the modifications occurred in its environment. Generally, it is written as follows:
Definition 6: Cardinality.
The cardinality of an elementary (Agent) in a group of elements Env (all environments) describes the membership of this element, or a subgroup. We must ensure that:
We define a constraint on an Agent by the Boolean function: Cont (Ai, K, j).
Cont (Ai, K, j) is defined as a pre firing condition from a Transition T to a place P. In a formal way, we define a constraint on completion of a place P. According to the theory of parts:
Let K and I both sets. One can verify that K is à subset of I:

With this basic description of Agent Petri Nets, we introduce in this paper other extension of models. We create a general framework for MAS, special for Staphylococcus epidermidis Biofilm Formation.
3. Molecular Basis of Staphylococcus epidermidis Biofilm Formation
Inside S. epidermidis biofilm, bacterial cells present different behavior than in their planktonic counterpart. Much knowledge is gathered concerning molecular mechanism and cells behavior inside biofilm toward external environment. Biofilm formation in S. epidermidis is a four-step process, it begins with initial cell attachment to native or conditioned a biotic surface, the second step, known as accumulation step, is marked with active cell multiplication and multi-layers population forming, the third step is the biofilm maturation during which biofilm micro-colonies takes a mushroom like form owing to metrical components distribution. The last step is the detachment of cells which regains their planktonic statute [6] . Different genes involved in biofilm formation in S. Epidermidis are under complex regulation in time and in space, indeed, an important genes network interacting together and with different targets involved directly or not in biofilm formation are behind the chronologically organized growth phases as well as the defined structure of S. epiermidis biofilm.
IcaR, the fifth gene of icaoper on, is located upstream to icaADBC genes. This gene is divergently transcribed from the other ica genes [7] .
The Teicoplan in Associated Regulator “TcaR” was reported as a negative regulator of ica transcription since inactivation of this gene enhanced the transcriptional level of icaADBC [8] .
Rbf (regulator of biofilm) is a member of AraC/XylS transcriptional regulators family. This protein was reported to play an important role in biofilm formation in S. epidermidis [9] .
In S. epidermidis, sigma B (σB) alternative factor plays a key role in the relationship of bacterial cell to its external environment (Figure 1). Indeed this factor is activated by numerous environmental stresses including high temperature, high osmolarity, antibiotics, or extreme pH [10] .
Sar proteins were classified in three sub-families basing on their structural properties. The first subfamily contains proteins acting as homodimers and binds DNA with a single DNA binding domain [12] . As conclusion, SarA enables S. epidermidis to switch between mechanisms of biofilm formation, ensuring the adaptation of this bacteriumo hostile environment [13] .
The accessory gene regulator (agr) quorum sensing system is a chromosomal oper on encoding two divergently transcribed transcripts, RNAII and RNAIII [14] (Figure 2).
Figure 1. Sigma B factor molecular pathway [11] .
Figure 2. Agr quorum sensing system molecular pathway [14] .
Figure 3. GlobalInteraction leading S. epidermidis to switch from Planctonik to Biofilm.
4. APN Model for Biofilm Formation
In formal manner this reaction is defined by:
where:




We can deduce that:
The set of tasks TS can be performed by a group of Genes A and can obtain a new set of performed tasks noted. Tsj. This change can infect the behaviour and the structure of the set of Genes. The Gene Ai employed to perform a task tsj leading to achieve some Goals. This Gene undergoes behavioural and structural changes.
In formal manner this reaction is defined by:
where:


Figure 4. Performed Reaction without Change of Gene.



where:
N: New relation between genes;
D: Destruction of gene;
S: Substitution: base is replaced by one of the other three bases;
Dl: Deletion: block of one or more DNA pairs are lost;
Is: Insertion: block of one or more DNA pairs are added;
Iv: Inversion: 180˚ rotation of DNA piece;
R: Reciprocal translocation: parts of no homologous chromosomes change places;
C: Chromosomal rearrangements: affect many genes at one time.
After perform a Reaction there are a behavioural and a structural changes of Gene noted by: performed/TG. We obtain new instance of Gene Ai (Figure 5):
In this case:
Definition1 presents a formal description of performed task (reaction). After firing the transition Performed there are new result (.tsj). This action can’t infect the gene:
But, in Definition 2, after firing the transition 

In most of Biologic system reaction, there are major changes in behaviour and in structure of gene. So Performed is a particular case of Performed/TG.
We define the function given the set of result of reaction Ts performed by a set of Gene Ai related to the transition Tk by: Perfect (Tk) =
In Petri Nets model, Perfect (tk) presents a condition of firing the transition Tk.
Figure 6 presents an example of Performed reaction achieved by set of Gene.
Figure 5. Performed Reaction with behavioural and structural changes of Gene.
Figure 6. Example of APN performed reaction.
We transform the APN model to a Matrix of Gene Transformation. This matrix mention in each line the transformation achieved for the Gene Ai when there are perform of reaction tsj.
For example, 


Any reaction tsj can be performed by a set of Gene A. We define
Using the same APN model presented in Figure 7, we can deduce that:





A goal of set of Gene is define as the achievement of all reaction planned for the total of Biologic system (Figure 7): Goal (Gp) =
In APN model, we use Inhibitor arc to firing the transition T2. In this state all reaction tsj are performed successfully.
We define the migration model of mobile Gene by a following Sub Petri Nets:
where:
P: a non-empty finished set of Places;





n: number of Places;
k: number of place in Env1;
n-k: number of Places in Env2;





Figure 7. Achievement of all reaction in biologic system.
5. APN Model for SigmaB Factor Molecular Pathway
In this section we present an APN model for modeling Staphylococcus epidermidis Biofilm Formation (Figure 8). We define all reaction that can be performing by all Genes in the reaction system.
Figure 8. APN Model for modeling Staphylococcus epidermidis Biofilm Formation.
Table 1. Method for modelling Biological system [2] .
6. Related Work
Much formalism has been used to model Biological Systems, in part due to the various phenomena that occur in those systems. The second part due to multi-disciplinarily of research groups. Formal Method for Biologists may be more familiar with mathematical modelling and computer scientists. Several works has been discussed the dichotomy between mathematical and computational models elsewhere such in [14] . Indeed, using mathematical models of cellular metabolism, it is possible to automatism test of generate sub-optimal phenotypes for specific applications [9] . Although different formal approaches has been questioned if there is such Petri Nets models. [17] proposes colored Petri Nets to simulate enzymatic reaction process: token is a pair encompassing the name and the concentration of the related substrate. Transitions present a kinetic function. [13] proves that with standard PNs we can modeling the essential components in biochemical pathways, and that PN models can be used to perform a qualitative analysis. In those models, places represent reactants, products or enzymes. Whereas transitions represent reactions [15] , it was elaborated a Physicochemical modeling of cell signaling pathways. They address the model design process, as well as, model verification, interpretation validation, calibration and publication of models. Another recent review on the modeling of signaling networks can be found in [8] . Recently an excellent review was elaborated by [18] for Modeling Signaling Networks with Different Formalisms. In Table 1, [2] summarizes some of the literature references reviewed herein, classified by type of intracellular process implemented.
7. Conclusions
We present in this paper a formal framework based on Multi agent System and Petri Nets to model Staphylococcus epidermidis Biofilm Formation. We provided a high-level description for this formalization, with a semantics given using the Agent Petri Nets. All process was present rigorously and clearly with APN. This method can be used to describe the reaction among molecules, their interaction and their transformation. With this framework it’s easy to present the behavior and the structure of each entity or the total of biologic system. Such system has a dynamic behavior, quickly generation of result and incomprehensible reaction. The objective of the dynamic model consists in proposing a formal method to understand the functioning of the Staphylococcus epidermidis Biofilm Formation and it is possible to perform formal analyses on environments thus described.
The ability to predict system behavior with an APN helps evaluate model completeness as well as improve our understanding of the function of biological systems. In fact, the meaning facets framework establishes a new methodology for computer-aided collaborative modeling in Systems Biology.
Our meaning facets are also a way for structuring and clarifying our understanding of bio-models. Due to the graphical visualization of biologic system by Petri nets, a bioscientist can intuitively understand the modeled process.
Cite this paper
BorhanMarzougui,KamelBarkaoui,Mohamed AmineMakni, (2016) Agent Petri Nets Framework for Modeling Staphylococcus epidermidis Biofilm Formation. E-Health Telecommunication Systems and Networks,05,19-30. doi: 10.4236/etsn.2016.51003
References
- 1. Claudine, C. (2007) Petri Net Modelling of Biological Networks. Briefings in Bioinformatics, 8, 210-219.
http://dx.doi.org/10.1093/bib/bbm029 - 2. Daniel, M., Rafael, S., Miguel, R., Eugenio, C., Bruce, T. and Isabel, R. (2011) Modeling Formalisms in Systems Biology. AMB Express, 1, 45.
- 3. Marzougui, B., Hassinek, K. and Barkaoui, K. (2010) A New Formalism for Modeling a Multi Agent Systems: Agent Petri Nets. Journal of Software Engineering and Applications (JSEA), 3, 1118-1124.
http://dx.doi.org/10.4236/jsea.2010.312130 - 4. Marzougui, B., Hassinek, K. and Barkaoui, K. (2013) Modeling Migration of Mobile Agents. Lecture Notes in Business Information Processing, 132, 530-540.
- 5. Marzougui, B., Barkaoui, K. and Alouane, N.H. (2013) APN Model for Specification of the Communication Protocols in Multi-Agent System. Journal of Software Engineering and Applications, 6, 14-22.
- 6. Boles, B.R. and Horswill, A.R. (2011) Staphylococcal Biofilm Disassembly. Trends in Microbiology, 19, 449-455.
http://dx.doi.org/10.1016/j.tim.2011.06.004 - 7. Jefferson, K.K., Cramton, S.E., Gotz, F. and Pier, G.B. (2003) Identification of a 5-Nucleotide Sequence that Controls Expression of the ica Locus in Staphylococcus aureus and Characterization of the DNA-Binding Properties of IcaR. Molecular Microbiology, 48, 889-899.
http://dx.doi.org/10.1046/j.1365-2958.2003.03482.x - 8. Jefferson, K., Pier, D.B., Goldmann, D.A. and Pier, G.B. (2004) The Teicoplanin-Associated Locus Regulator (TcaR) and the Intercellular Adhesin Locus Regulator (IcaR) Are Transcriptional Inhibitors of the ica Locus in Staphylococcus aureus. Journal of Bacteriology, 186, 2449-2456.
http://dx.doi.org/10.1128/JB.186.8.2449-2456.2004 - 9. Rowe, S., Mahon, V., Smith, G. and Gara, J.P.O. (2011) A Novel Role for SarX in Staphylococcus epidermidis Biofilm Regulation. Microbiology, 157, 1042-1049.
http://dx.doi.org/10.1099/mic.0.046581-0 - 10. Lauderdale, K.J., Boles, B.R., Cheung, A.L. and Horswill, A.R. (2009) Interconnections between Sigma B, agr, and Proteolytic Activity in Staphylococcus aureus Biofilm Maturation. Infection and Immunity, 77, 1623-1635.
http://dx.doi.org/10.1128/IAI.01036-08 - 11. Knobloch, J., Jager, S., Horstkotte, M., Rohde, H. and Mack, D. (2004) RsbU-Dependent Regulation of Staphylococcus epidermidis Biofilm Formation Is Mediated via the Alternative Sigma Factor σB by Repression of the Negative Regulator Gene icaR. Infection and Immunity, 72, 3838-3848.
http://dx.doi.org/10.1128/IAI.72.7.3838-3848.2004 - 12. Burgard A, Pharkya P, Maranas C (2003) Optknock: A Bilevel Programming Framework for Identifying Gene Knockout Strategies for Microbial Strain Optimization. Biotechnology and Bioengineering, 84, 647-657.
http://dx.doi.org/10.1002/bit.10803 - 13. Patil, K., Rocha, I., Forster, J. and Nielsen, J. (2005) Evolutionary Programming as a Platform for in Silico Metabolic Engineering. BMC Bioinformatics, 6, 308.
http://dx.doi.org/10.1186/1471-2105-6-308 - 14. Hunt, C., Ropella, G., Park, S. and Engelberg, J. (2008) Dichotomies between Computational and Mathematical Models. Nature Biotechnology, 26, 737-738.
http://dx.doi.org/10.1038/nbt0708-737 - 15. Pammi, M., Liang, R., Hicks, J., Mistretta, T.A. and Versalovic, J. (2013) Biofilm Extracellular DNA Enhances Mixed Species Biofilms of Staphylococcus epidermidis and Candida albicans. BMC Microbiology, 13, 257.
- 16. Kong, K。, Vuong, C. and Otto, M. (2006) Staphylococcus Quorum Sensing in Biofilm Formation and Infection. International Journal of Medical Microbiology: IJMM, 296, 133-139.
http://dx.doi.org/10.1016/j.ijmm.2006.01.042 - 17. Genrich, H., Kuffner, R. and Voss, K. (2001) Executable Petri Net Models for the Analysis of Metabolic Pathways. International Journal on Software Tools for Technology Transfer, 3, 394-404.
- 18. Geisinger, E., Adhikari, R.P., Jin, R., Ross, H.F. and Novick, R.P. (2006) Inhibition of Rot Translation by RNAIII, a Key Feature of agr Function. Molecular Microbiology, 61, 1038-1048.
http://dx.doi.org/10.1111/j.1365-2958.2006.05292.x






























