Advances in Microbiology
Vol. 2 No. 2 (2012) , Article ID: 19666 , 7 pages DOI:10.4236/aim.2012.22009
Molecular Assessment of 16S-23S rDNA Internal Transcribed Spacer Length Polymorphism of Aeromonas hydrophila
1Programme d’Epigenomique, Institute of Systems and Synthetic Biology Genopole, French National Center for Scientific Research (CNRS), Evry, France
2National Bureau of Fish Genetic Resources, Lucknow, India
3Department of Biochemistry, Faculty of Science, Banaras Hindu University, Varanasi, India
Email: *vijaisingh15@gmail.com
Received January 20, 2012; revised February 8, 2012; accepted March 12, 2012
Keywords: Aeromonas hydrophila; diseases; ITS; tRNA-Glu; molecular marker
ABSTRACT
Aeromonas hydrophila is an important bacterial pathogen which causes the hemorrhagic septicemia in fishes, amphibians and humans. Genetic relationships of diverse isolates of A. hydrophila were recovered from fish and water sources. These isolates were investigated by flanked region of 16S and 23S Ribosomal DNA (rDNA) internal transcribed spacer (ITS). Here we analyzed polymorphism of PCR-amplified 16S-23S rDNA ITS and their revealed band pattern consisting of one to four DNA fragments. The fragment size ranged from 500 to 1000 bp. The DNA band patterns revealed a considerable genetic diversity in interaspecies. The 750 bp size of band was common in all isolates of A. hydrophila except one Isolates AH21. The tRNA-Glu sequences were identified from 750 bp size of ITS region that could be used as strain level potential genotypic markers.
1. Introduction
An epidemic of Aeromonas hydrophila infection with a high rate of mortality (95%) in turtles (Pseudemis scripta) has been reported in Italy [1]. The disease source due to A. hydrophila has caused extensive losses among cultured fresh water fishes in mainland China [2,3]. Infection of A. hydrophila occurs in all developmental stages of fish. Several important virulence factors are involved in the pathogenesis, which reduce the immunity and thereby damage cellular activity in fish that allows the susceptibility to disease outbreaks and high rate of mortality. Aeromonas has been isolated from diarrheic children, fish, milk and ice creams; and 57% isolates produced the enterotoxin [4]. Moreover A. hydrophila has also been isolated from blood and bilious fluid of women with liver cirrhosis [5]. A. hydrophila causes several diseases in humans like diarrhea, soft tissue infections, meningitis, endocarditis, peritonitis, hemolytic-uremic syndrome and septicemia in immunocompromised [6].
A. hydrophila is a significant bacterial pathogen of a wide variety of hosts, which is associated with hemorrhagic septicemia in fishes, reptiles and amphibians [7].
In fishes, they cause well known diseases like hemorrhagic septicemia, fin as well as tail rot and result in the high mortality in commercial aquaculture system [8]. Freshwater reservoirs are decreasing due to growing populations, increased human consumption, urbanization and the lack of cost-effective sewage water treatment systems [9,10]. Several species of Aeromonas isolates were recovered from cultured fish and was characterized by biochemical as well as 16S rDNA sequences. The pathogenicity assay has been performed in healthy fishes and found pathogenic bacteria such as A. hydrophila, A. bestiarum, A. salmonicida and A. veronii. All these Aeromonas isolates were isolated from diseased trout except one which from carp fry [11]. Therefore, it is urgent need to well characterize by molecular tools to discriminate pathogenic and non pathogenic isolates of A. hydrophila.
Since last decade, several molecular techniques have been developed for characterization of bacteria. The study of non-coding RNAs is important in searching the function or role in cells. In order to understand the function, we may find the secondary structure. The family of tRNAs is a type of RNA molecules which has particular function to translate amino acids into protein-building machinery. The 16S, 23S, ITS, gyrase, RNA polymerase and DNA ligase are highly conserved gene of bacteria and used for molecular characterization. The internal transcribed spacer (ITS) is also known as ISR present between 16S and 23S rDNA region of ribosomal genes. The arrangement of complete unit of ribosomal genes such as 16S-ITS-23S-ITS-5S are scattered in the genome of bacteria, which vary from 1 - 15 copy numbers. The recently developed DNA fingerprinting methods are based on enterobacterial repetitive intergenic consensus sequence (ERIC), the repetitive extragenic palindromic sequence (REP) and the BOX elements have been reported for the discrimination of bacterial strains [12].
Repetitive-element PCR (rep-PCR) with primers based on repetitive extragenic palindromic (REP) and enterobacterial repetitive intergenic consensus (ERIC) repeated DNA sequences has been reported for genomic fingerprinting of Bartonella species [13]. RFLP, ERIC and REP have been evaluated for typing of isolates of Aeromonas popoffii from different geographical origins [14]. However, molecular characterization of bacteria is performed using PCR analysis of length polymorphism of the intergenic spacers lying between tRNA genes (tDNAPCR) [15]. The tRNA genes are a highly conserved among species within a genus, so primers that allows a PCR product to be amplified from practically all members of a genus. The length of tRNA intergenic spacers were varied from 2 to 35 bp in Bacillus subtilis [16] and from 2 to 208 bp in Escherichia coli [17]. The order of tRNA genes within cistrons appears to be highly conserved within a genus. The tandem arrangements of tRNA gene clusters allow investigating efficiently using tDNA-PCR techniques [18]. The present study was to investigate the 16S-23S ITS PCR assay to discriminate and generate specific fingerprint at the strain level identification of diverse isolates of A. hydrophila recovered from diseased and apparently normal samples. We have also investigated the secondary structures of tRNA-Glu at different temperatures.
2. Materials and Methods
2.1. Collection, Bacterial Isolates Media and Chemicals
The complete genome sequence of A. hydrophila subsp. hydrophila ATCC 7966 was retrieved from the NCBIGenBank (Accession no. CP000462). We analyzed the copy number of ribosomal operon and their organizations in genome were verified. All the isolates of A. hydrophila used in this study have been earlier characterized on the basis of amplification of aerolysin gene [19]. All the isolates were preserved in 15% glycerol at −80˚C. Taq DNA polymerase, dNTPs, Proteinase K, DNA ladders, pTZ57- RT/A cloning vector (Fermentas), E. coli DH5α (Invitrogen) and other media, chemicals (Sigma, SRL, Himedia) were used.
2.2. Isolation of Genomic DNA
Genomic DNA of A. hydrophila was isolated by Hiney et al. [20] method with some modification. Briefly, a single colony of A. hydrophila was inoculated in 2 ml of Nutrient broth (NB) medium and incubated at 30˚C for over night. Well-grown culture was centrifuged at 10,000 rpm for 2 minutes at 4˚C. Total 500 µl lysis buffer (100 mM Tris pH 8.0, 10 mM EDTA pH 8.0, 1.25% NaCl and 0.25% Sucrose) was re-suspended the pellet properly and incubated at 65˚C for 2 hours. The 10 µl proteinase K (10 mg/ml) was added, inverted gently and again incubated at 37˚C for one hour. Equal amount of phenol: chloroform: isoamylalcohol (25:24:1) was added to cell lysate and mixed properly by the inversion of tube and the suspension was centrifuged at 10,000 rpm for 10 minutes at 4˚C. The supernatant was carefully transferred in a new tube and the step repeated twice. Added 1/10 volume sodium acetate (pH 5.2) and two volume of chilled ethanol subsequently incubated at −20˚C for 30 minutes to precipitate the DNA. Centrifuged at 10,000 rpm for 20 minutes at 20˚C, the DNA pellet was washed with 70% ethanol, air dried then dissolved in 50 μl of TE buffer (pH 8.0) and stored at −20˚C.
2.3. PCR Amplification of 16S-23S rDNA ITS
One set of PCR primer targeted the ITS region of A. hydrophila was synthesized from Integrated DNA Technology (IDT, USA) based on published sequences. The highly conserved sequences were adjacent to 3’ end region of the 16S rDNA (5’TGCGGCTGGATCACCTCCTT) [21] and 5’ end region of the 23S rDNA (5’GGTACTTAGATGTTTCAGTTC 3) [22]. The 50 μl reaction mixture was consisted of 10 ng of genomic DNA, 2.0 units of Taq DNA polymerase, 5 μl of 10X PCR buffer (100 mM Tris-HCl, 500 mM KCl pH 8.3), 200 μM dNTP, 10 pmoles of each primer (16S forward primer and 23S reverse primer) with 1.5 mM MgCl2 was used. Amplification included initial denaturation at 94˚C for 5 minutes, followed by 25 cycles of denaturation 94˚C for 30 seconds, annealing temperature of primers at 48˚C for 1 minute and extension at 72˚C for 2 minutes. A final extension at 72˚C for 10 minutes was used. Ten μl of PCR product was analyzed by agarose gel electrophoresis in 2.0% agarose with ethidium bromide, at 8 V/cm and the reaction product were visualized under Gel doc/UV Transilluminator.
2.4. Construction and Analysis of Dendrogram
It is intensive to relative concentration of bands between fingerprints, discontinuous noise and overall density of fingerprints. ITS-PCR patterns, a band-matching algorithm (Match-matching tolerance of 1.0%) were used to calculate pair wise similarity matrix with similarity coefficient. Cluster analysis of similarity matrices were performed by UPGMA (Unweighted Pair Group Method with Arithmetic Mean). The major DNA bands were considered for construction of phylogenetic tree with used of TFPGA (Tools for Population Genetic Analyses). Each isolates of A. hydrophila was considered as one population so there was total eight populations and four loci were considered for construction of dendogram.
2.5. Cloning and Sequencing of ITS Region
The size of 750 bp band was selected for cloning and cut the agarose gel for purification of PCR product (QIAquick Gel Extraction Kit, QIAGEN). The PCR product was ligated in pTZ57RT/A vector with T4 DNA ligase (Rapid ligation kit, Fermentas) and incubated at 16˚C for overnight subsequently inactivated for 15 minutes at 65˚C. Ligated plasmid was transformed into Escherichia coli DH5α chemically competent cells [23]. The clones were screened using 100 μg/ml concentration of Ampicillin antibiotic selection with X-gal (20 mg/ml) and IPTG (100 mM). The plasmid DNA was isolated from clones using QIAprep Spin Miniprep Kit (QIAGEN). Sequencing of ITS region was done by custom service from Chromous Biotech Pvt Ltd (Bangalore, India) using the forward and reverse primers of pTZ57RT/A cloning vector. The DNA sequence of ITS region was analyzed by BLASTN for the identification.
2.6. Identification of tRNA, Secondary Structure and Submission of Nucleotide Sequences
Different aminoacyl-tRNA synthetase is present for each amino acid. Accurate translation of the genetic code depends on attachment of each amino acid to an appropriate tRNA. The tRNAs are also undergoing substantial conformational changes within their ribosomal binding sites during protein synthesis. We used tRNAscan-SE tool for identification of tRNA sequences in ITS region, which identifies 99% - 100% of transfer RNA genes in DNA sequence while giving less than one false positive per 15 gigabases. The NUPACK tool was used for modeling of secondary structure at different temperatures (20˚C - 40˚C). The nucleotide sequence of ITS region from A. hydrophila has been submitted to NCBI-GenBank under accession number HM991866.
3. Results and Discussion
The genome of A. hydrophila was analyzed for presence of ribosomal operon copy number. The 10 copy of ribosomal operon present in the genome of A. hydrophila was identified. In which six copies were localized from 5’-3’ direction and other four was reverse orientation. The ITS region was presented between the 16S and 23S rDNA sequences. The size of ITS was ranged between 500 - 1000 bp and it contains the number of tRNA encoding gene. Therefore, we amplified ITS region using the forward primers of 16S and reverse primers of 23S rDNA regions. Subsequently, we obtained the different bands pattern due to the copy number of ribosomal unit and tRNA encoding gene within ITS. The graphical representation of 16S-ITS-23S was shown in Figure 1(a). It was strategy to amplify the ITS for discrimination and typing of different isolates of A. hydrophila.
In the present investigation, there was one common band (750 bp) which was present in all these isolates of A. hydrophila except the AH21. Several other bands were also amplified and used for construction of the fingerprint that could be used for generation of phylogenetic relationship on the basis of results. The variation in intensity of the same size in different isolates results from point shift mutation, the template leading to reduce the stability or poor annealing of the primer to these templates. We have used different annealing temperature and Mg2+ concentration. The optimal result was obtained at 48˚C annealing temperature with 1.5 mM concentration of MgCl2 (Figure 1(b)). The size of 750 bp band could be considered as potential marker for A. hydrophila except one isolates AH21. All isolates showed good amplification and gave more prominent bands. In our previous study, A. hydrophila isolates AH14 has been potential pathogenic to fish based on molecular level amplification of aerolysin and hemolysin genes which was present in same isolates [24,25]. Total 160 strains belonging to four species such as A. hydrophila, A. bestiarum, A. salmonicida and A. popoffii were identified using 16S rDNA restriction fragment length polymorphism (RFLP). DNA sequencing of 16S rDNA and cluster analysis of the 16S-23S intergenic spacer region (ISR)-RFLP in selected strains of A. salmonicida and A. bestiarum indicated that the two species may share extremely conserved ribosomal operons and demonstrated that due to an extremely high degree of sequence conservation, 16S rDNA cannot be used to differentiate these two closely related species [26].
On the basis of this result we have constructed the phylogenetic tree using UPGMA method by considering presence, absence as well as highly intense bands pattern of A. hydrophila isolates. Total four loci were considered in 8 isolates and it showed good relationship between diseased and other isolates. The phylogeny of diverse isolates of A. hydrophila and two major clade A and B were obtained (Figure 2).
Isolate AH14 has been recovered from diseased fish
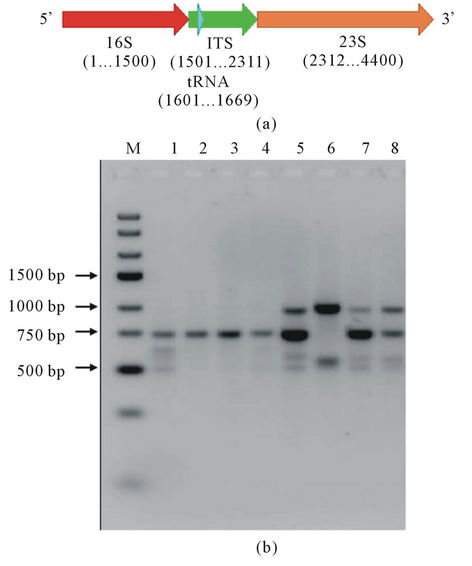
Figure 1. (a) Schematic representation of the position of ITS region in between 16S and 23S rDNA gene region; (b) PCR amplification of internal transcribed spacer (ITS) region from diverse isolates of A. hydrophila. Lane M: Generuler express DNA ladder 100 bp (Fermentas), Lane 1: Isolates AHPN 3-5, Lane 2: Isolates AH 13, Lane 3: Isolates AH 14, Lane 4: Isolates AH 16, Lane 5: Isolates AH 19, Lane 6: Isolates AH 21, Lane 7: Isolates AH 22, Lane 8: Isolates AH 24.
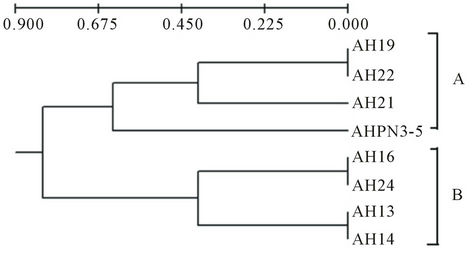
Figure 2. Dendogram constructed based on the DNA bands of the ITS region present in different isolates of A. hydrophila using UPGMA method.
sample and showed the homology with AH13, AH16 and AH24. It was indicated that these isolates were to be pathogenic and present in clades A. while the other isolates AHPN3-5 (it was water sample isolate), AH21, AH22 and AH19 present in clade B. This result could help to discriminate the diseased and apparently normal and water isolates. Similar study reported as characterization of ITS distribution using PCR and analyzed via a high-throughput capillary electrophoresis in 1191 Burkholderia pseudomallei strains. Three major type of ITS have been identified, two of which were commonly found in most B. pseudomallei strains from the endemic areas, whereas the third one was significantly correlated with worldwide sporadic strains [27]. RFLP of 16S-23S rDNA intergenic spacer region (ISR) of Aeromonas species have been used. Total of 69 isolates belonging to 18 DNA hybridization groups have been discriminated the isolates based on ISR amplification and RFLP [28].
The 120 strains isolated from stool specimens of gastroenteritis patient and environment of different geographical areas, these samples have been analyzed by PCR-RAPD, REP-PCR and ERIC-PCR on the species of A. hydrophila, A. bestiarum, A. salmonicida, A. caviae, A. media and A. veronii revealed clonal structure. No genetic similarities were observed between clinical and environmental strains of Aeromonas sp. isolated from distinct and same geographical area [29]. The BOX-PCR fingerprinting technique has been reported for discrimination of different isolates of A. hydrophila. All the studied isolates have shown major banding patterns ranged from 500 - 3000 bp and it could be advantageous to investigate the strain level specific fingerprints of A. hydrophila as potential genotypic markers [30]. Fifty isolates of Clostridium difficile have been analyzed by three PCR-based typing methods in order to determine genomic diversity within this strain that may form the basis of a sub typing method. The three methods were used like repetitive extragenic palindromic elements (REP), conserved repetitive DNA elements (BOX), and enterobacterial repetitive PCR intergenic consensus sequences (ERIC). These methods had satisfactory levels of type ability and reproducibility as determined by blind-coded repeats. The BOX-PCR generated between two and five major amplicons with four distinct BOX profiles [31].
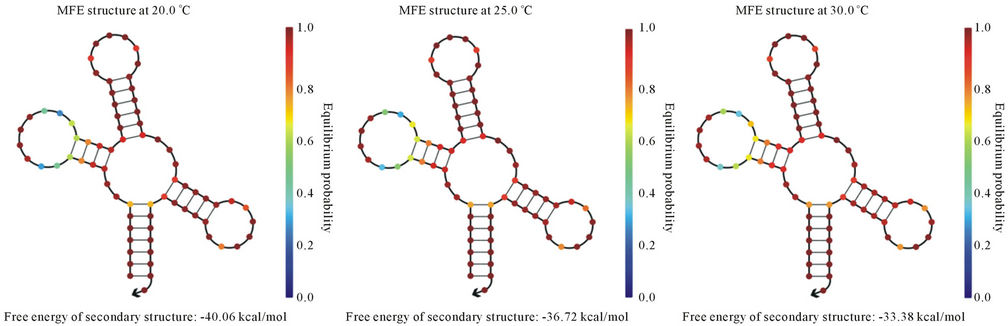
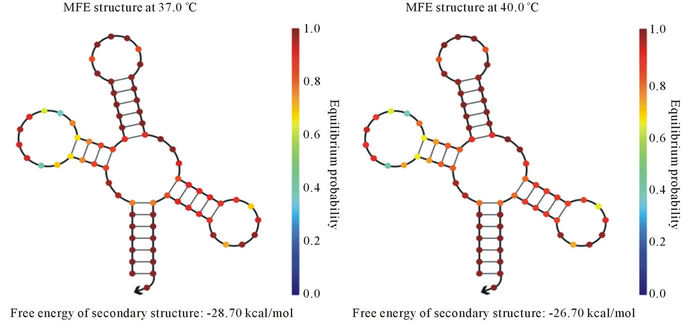
In the present study, we have cloned and sequenced the ITS region and was found homologous with A. hydrophila ITS gene sequences existing in NCBI-GenBank. Nucleotide sequence percent similarity was analyzed in between 90% - 100% (E value = 0) with other A. hydrophila strain 16S ribosomal DNA gene listed in NCBIGenBank. It indicates that ITS region may be potential molecular marker for A. hydrophila. Furthermore, we identified the tRNA-Glu in ITS region that could be conserved during evolution. The variations of different banding patterns were obtained of ITS regions due to presence of tRNA encoding genes. We also predicted the secondary structure of tRNA-Glu at temperature ranged from 20˚C - 40˚C (Figures 3(a)-(e)). The lowest free energy was −40.06 kcal/mol at 20˚C while −26.70 kcal/ mol at 40˚C.
We analyzed the total free energy difference −13.36 kcal/mol. We may assume that lower temperature may
(a) (b) (c) (d) (e)
Figure 3. Prediction of tRNA-Glu secondary structure at different temperatures (a) 20˚C; (b) 25˚C; (c) 30˚C; (d) 37˚C; and (e) 40˚C.
provide more support for RNA secondary structure stability in comparison to higher temperature. It may be potential risk for aquatic organisms because they are surviving at lower temperature and reservoir for A. hydrophila. Another report showed the secondary structures have been changed on different temperatures. For the structural gene the minimum free energy is problematic because it generates strong loops in secondary structure with lower energy [32]. However in case of ribosomal RNA we assume that they are less frequency of getting mutation during evolution. As ribosomes are the most ancient and important cell organelles contained the common structural features in all of organisms. It is composed of DNA especially ribosomal RNA coding genes and number of proteins. It plays major role in protein synthesis which is important in the cells viability, growth and development [33]. From last decades the ribosomal DNA has considered as a significant genetic marker because it is highly conserved throughout the evolution [34]. The lowest free energy of the 5S rRNA may reveal the most primitive bacteria and slow changes occurs throughout the evolution whereas higher free energy indicates less stability during the evolution [35]. In previous study, total 32 non-structural gene sequences of Influenza A virus H5N1 strain varied from 831 to 875 bp were used to construct the phylogeny and nine major clades were obtained. The computational tool has been used to model the RNA secondary structure of nine different strains of Influenza A virus and free energy ranges between −222.90 to −251.10 kcal/mol of the NS [36].
Thus, free energy of secondary structure is an important factor for stability of gene during evolution. PCRamplified 16S-23S rDNA spacer of Aeromonas species have been reported for total 69 isolates representing 18 DNA hybridization groups. The analysis of PCR products of 16S-23S rDNA spacers revealed patterns consisting of two to eight DNA fragments. The fragment sizes ranged from 730 to 1050 bp. DNA patterns revealed a considerable genetic diversity between species and within a species [28]. The 16S-23S intergenic spacer and 23S rRNA gene sequences have been determined for Aeromonas culicicola MTCC 3249T. Ten different ISR indicative of ten rRNA operons have been reported in the strain that were grouped in three major types. ISR I was non-coding while ISR II and III coded for tRNA-GluUUC [37].
4. Conclusion
We consider the techniques that would be useful for species and strains differentiation for a wide variety of bacteria and it should be applicable to studies of epidemiology, diagnosis, virulence and molecular taxonomy. It is rapid, easily performed and reproducible method that is appropriate for genotyping of A. hydrophila at the strain level.
5. Acknowledgements
The authors are thankful to G. Rathore, A. K. Singh, Pritee Singh and Reena for providing the suggestions, fruitful discussion and encouragement during preparation of the manuscript.
REFERENCES
- R. P. Kokka, J. M. Janda, L. S. Oshiro, M. Altwegg, T. Shimada, R. Sakazaki and D. J. Brenner, “Biochemical and Genetic Characterization of Autoagglutinating Phenotypes of Aeromonas Species Associated with Invasive and Noninvasive Disease,” Journal of Infectious Disease, Vol. 163, No. 4, 1991, pp. 890-894. doi:10.1093/infdis/163.4.890
- M. Dorsch, N. J. Ashbolt, P. T. Cox and A. E. Goodman, “Rapid Identification of Aeromonas Species Using 16S rDNA Targeted Oligonucleotide Primers: A Molecular Approach Based on Screening of Environmental Isolates,” Journal of Applied Bacteriology, Vol. 77, No. 6, 1994, pp. 722-726. doi:10.1111/j.1365-2672.1994.tb02825.x
- D. Qian. Y. Chen, J. Shen and Z. Shen, “Serogroups, Virulence and Hemolytic Activity of Aeromonas hydrophila Which Caused Fish Bacterial Septicaemia,” Wei Sheng Wu Xue Bao, Vol. 35, No. 6, 1995, pp. 460-464.
- A. S. Yadav and A. Kumar, “Prevalence of Enterotoxigenic Motile Aeromonads in Children, Fish, Milk and Ice-Cream and Their Public Health Significance,” Southeast Asian Journal of Tropical Medical and Public Health, Vol. 31, Suppl. 1, 2000, pp. 153-156.
- H. Saeki, N. Matsuda, T. Tamura, N. Masuda and A. Yonei, “A Case of Severe Septicemia Due to Aeromonas hydrophila,” Masui, Vol. 51, No. 2, 2002, pp. 193-195.
- V. Minnaganti, P. Patet, D. Iancu, P. Schoch and B. Cunha, “Necrotizing Fasciitis Caused by Aeromonas hydrophila,” Heart and Lung, Vol. 29, No. 4, 2000, pp. 306-308. doi:10.1067/mhl.2000.106723
- B. Austin and D. A. Austin, “Bacterial Fish Pathogens: Diseases in Farmed and Wild Fish,” Praxis Publishing, Chichester, 1999.
- J. M. Janda, S. L. Abbott, W. K. Cheung and D. F. Hanson, “Biochemical Identification of Citrobacteria in the Clinical Laboratory,” Journal of Clinical Microbiology, Vol. 32, No. 8, 1994, pp. 1850-1854.
- T. R. Karl, R. W. Knight and N. Plummer, “Trends in High-Frequency Climate Variability in the Twentieth Century,” Nature, Vol. 377, No. 6546, 1995, pp. 217-220. doi:10.1038/377217a0
- R. M. Carr, U. J. Blumenthal and D. D. Mara, “Guidelines for the Safe Use of Wastewater in Agriculture: Revisiting WHO Guidelines,” Water Science and Technology, Vol. 50, No. 2, 2004, pp. 31-38.
- A. Kozinska, “Dominant Pathogenic Species of Mesophilic Aeromonads Isolated from Diseased and Healthy Fish Cultured in Poland,” Journal of Fish Disease, Vol. 30, No. 5, 2007, pp. 293-301. doi:10.1111/j.1365-2761.2007.00813.x
- M. Tacao, A. Alves, M. J. Saavedra and A. Correia, “BOXPCR Is an Adequate Tool for Typing Aeromonas spp.,” Antonie van Leeuwenhoek, Vol. 88, No. 2, 2005, pp. 173- 179. doi:10.1007/s10482-005-3450-9
- M. C. Rodriguez-Barradas, R. J. Hamill, E. D. Houston, P. R. Georghiou, J. E. Clarridge, R. L. Regneryn and J. E. Koehler, “Genomic Fingerprinting of Bartonella Species by Repetitive Element PCR for Distinguishing Species and Isolates,” Journal of Clinical Microbiology, Vol. 33, No. 5, 1995, pp. 1089-1093.
- L. Soler, M. J. Figueras, M. R. Chacón, J. Guarro and A. J. Martinez-Murcia, “Comparison of Three Molecular Methods for Typing Aeromonas popoffii Isolates,” Antonie Van Leeuwenhoek, Vol. 83, No. 4, 2003, pp. 341-349. doi:10.1023/A:1023312415276
- B. Vold, “Structure and Organization of Genes for Transfer Ribonucleic Acid in Bacillus subtilis,” Microbiology Review, Vol. 49, No. 1, 1985, pp. 71-80.
- J. Welsh and M. McClelland, “Genomic Fingerprints Produced by PCR with Consensus tRNA Gene Primers,” Nucleic Acids Research, Vol. 19, No. 4, 1991, pp. 861-866. doi:10.1093/nar/19.4.861
- S. Jinks-Robertson and M. Nomura, “Ribosomes and tRNA,” In: F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter and H. E. Umbarger, Eds., Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology, American Society for Microbiology, Washington DC, 1987, pp. 1358-1385.
- M. McClelland, C. Petersen and J. Welsh, “Length Polymorphism in tRNA Intergenic Spacers Detected by Using the Polymerase Chain Reaction Can Distinguish Streptococcal Strains and Species,” Journal of Clinical Microbiology, Vol. 30, No. 6, 1992, pp. 1499-1504.
- V. Singh, G. Rathore, D. Kapoor, B. N. Mishra and W. S. Lakra, “Detection of Aerolysin Gene in Aeromonas hydrophila Isolated from Fish and Pond Water,” Indian Journal of Microbiology, Vol. 48, No. 4, 2008, pp. 453- 458. doi:10.1007/s12088-008-0056-8
- M. Hiney, M. T. Dawson, D. M. Heery, R. P. Smith, F. Gannon and R. Powell, “DNA Probe for Aeromonas salmonicida,” Applied and Environmental Microbiology, Vol. 58, No. 3, 1992, pp. 1039-1042.
- M. A. Parker, “Bradyrhizobia from Wild Phaseolus, Desmodium and Macroptilium Species in Northern Mexico,” Applied and Environmental Microbiology, Vol. 68, No. 4, 2002, pp. 2044-2048. doi:10.1128/AEM.68.4.2044-2048.2002
- C. Rachman, P. Kabadjova, R. Valcheva, H. Prévost and X. Dousset, “Identification of Carnobacterium Species by Restriction Fragment Length Polymorphism of the 16S- 23S rRNA Gene Intergenic Spacer Region and SpeciesSpecific PCR,” Applied and Environmental Microbiology, Vol. 70, No. 8, 2004, pp. 4468-4477. doi:10.1128/AEM.70.8.4468-4477.2004
- J. Sambrook, E. F. Fritsch and T. Maniatis, “Molecular Cloning: A Laboratory Manual,” 2nd Edition, Cold Spring Harbor Laboratory Press, New York, 1989.
- V. Singh and P. Somvanshi, “Inhibition of Oligomerization of Aerolysin from Aeromonas hydrophila: Homology Modeling and Docking Approach for Exploration of Hemorrhagic Septicemia,” Letters in Drug Design and Discovery, Vol. 6, No. 3, 2009, pp. 215-223. doi:10.2174/157018009787847864
- V. Singh, P. Somvanshi, G. Rathore, D. Kapoor and B. N. Mishra, “Gene Cloning, Expression and Homology Modeling of Hemolysin Gene from Aeromonas hydrophila,” Protein Expression and Purification, Vol. 65, No. 1, 2009, pp. 1-7. doi:10.1016/j.pep.2008.11.015
- A. J. Martínez-Murcia, L. Soler, M. J. Saavedra, M. R. Chacón, J. Guarro, E. Stackebrandt and M. J. Figueras, “Phenotypic, Genotypic, and Phylogenetic Discrepancies to Differentiate Aeromonas salmonicida from Aeromonas bestiarum,” International Microbiology, Vol. 8, No. 4, 2005, pp. 259-269.
- A. P. Liguori, S. D. Warrington, J. L. Ginther, T. Pearson, J. Bowers, M. B. Glass, M. Mayo, V. Wuthiekanun, D. Engelthaler, S. J. Peacock, B. J. Currie, D. M. Wagner, P. Keim and A. Tuanyok, “Diversity of 16S-23S rDNA Internal Transcribed Spacer (ITS) Reveals Phylogenetic Relationships in Burkholderia pseudomallei and Its NearNeighbors,” Plos One, Vol. 6, No. 12, 2011, p. e29323. doi:10.1371/journal.pone.0029323
- M. Laganowska and A. Kaznowski, “Restriction Fragment Length Polymorphism of 16S-23S rDNA Intergenic Spacer of Aeromonas spp.,” Systematic and Applied Microbiology, Vol. 27, No. 5, 2004, pp. 549-557. doi:10.1078/0723202041748226
- E. Szczuka and A. Kaznowski, “Typing of Clinical and Environmental Aeromonas sp. Strains by Random Amplified Polymorphic DNA PCR, Repetitive Extragenic Palindromic PCR, and Enterobacterial Repetitive Intergenic Consensus Sequence PCR,” Journal of Clinical Microbiology, Vol. 42, No. 1, 2004, pp. 220-228. doi:10.1128/JCM.42.1.220-228.2004
- V. Singh, D. K. Chaudhary, I. Mani, P. Somvanshi, G. Rathore and N. Sood, “Genotyping of Aeromonas hydrophila by Box Elements,” Microbiology, Vol. 79, No. 3, 2010, pp. 370-373. doi:10.1134/S0026261710030136
- A. Rahmati, M. Gal, G. Northey and J. S. Brazier, “Subtyping of Clostridium difficile Polymerase Chain Reaction (PCR) Ribotype 001 by Repetitive Extragenic Palindromic PCR Genomic Fingerprinting,” Journal of Hospital Infection, Vol. 60, No. 1, 2005, pp. 56-60. doi:10.1016/j.jhin.2004.09.034
- M. Huynen, R. Gutell and D. Konings, “Assessing the Reliability of RNA Folding Using Statistical Mechanics,” Journal of Molecular Biology, Vol. 267, No. 5, 1997, pp. 1104-1112. doi:10.1006/jmbi.1997.0889
- N. S. Kupriyanova, “Conservation and Variation of Ribosomal DNA in Eukaryotes,” Molecular Biology, Vol. 34, No. 5, 2000, pp. 637-647. doi:10.1007/BF02759600
- D. M. Hillis and M. T. Dixon, “Ribosomal DNA: Molecular Evolution and Phylogenetic Inference,” The Quarterly Review of Biology, Vol. 66, No. 4, 1991, pp. 411-453. doi:10.1086/417338
- V. Singh and P. Somvanshi, “Computational Modeling Analyses of RNA Secondary Structures and Phylogenetic Inference of Evolutionary Conserved 5S rRNA in the Prokaryotes,” Journal of Molecular Graphics and Modeling, Vol. 27, No. 7, 2009, pp. 770-776. doi:10.1016/j.jmgm.2008.11.012
- P. Somvanshi, V. Singh and M. Arshad, “Modeling of RNA Secondary Structure of Non Structural Gene and Phylogenetic Analysis of Influenza A Virus through the in Silico Methods,” Journal of Proteomics Bioinformatics, Vol. 1, 2008, pp. 219-226. doi:10.4172/jpb.1000026
- V. J. Pidiyar, K. Jangid, M. S. Patole and Y. S. Shouche, “Analysis of 16S-23S Intergenic Spacer Regions and rrn Operon Copy Number of Aeromonas culicicola MTCC 3249T,” DNA Sequences, Vol. 14, No. 3, 2003, pp. 183- 194. doi:10.1080/1042517031000101257
NOTES
*Corresponding author.

