Open Journal of Pathology
Vol.3 No.1(2013), Article ID:27274,11 pages DOI:10.4236/ojpathology.2013.31003
Telomere Maintenance Mechanisms: Prognostic and Therapeutic Implications for the Pathologist and Oncologist
![]()
Department of Pathology, Dunedin School of Medicine, University of Otago, Dunedin, New Zealand.
Email: *noelyn.hung@otago.ac.nz
Received August 30th, 2012; revised October 4th, 2012; accepted October 13th, 2012
Keywords: Telomere Maintenance Mechanism; Neoplasia; Sarcoma; Glioblastoma; Review
ABSTRACT
In neoplasia, telomere maintenance mechanisms (TMMs) can be prognostic and may direct therapy in the future. Two types of TMM, telomerase and recombination-based alternative lengthening of telomeres (ALT), result in four prognostic tumor groups when they occur individually, in combination, or in mutual absence. Correct designation of the TMM therefore requires an assessment of telomerase activity and for ALT telomere length distribution and ALT associated promyelocytic leukaemia protein (PML) bodies (APBs). The four groups are associated with differing prognoses that are dependent on the tumor type. As TMM inhibitors are developed, oncologists will require that pathologists determine the TMM, and the treatments will differ accordingly. Furthermore, any anti-TMM therapy administered has the potential to selectively change the TMM used by a tumor, necessitating reassessment of the therapeutic strategy. Herein, we review the telomere maintenance mechanisms, the current diagnostic measures and their utility as prognostic markers in the clinical setting.
1. Introduction
A predictable pattern of carcinogenesis is beginning to emerge based on a multitude of growth factors, cytoplasmic receptors, signal transducers, gene mutations and epigenetic changes that have been described in the cellular pathways of cancer [1]. One of these predominant cancer pathways, telomere maintenance, is best understood based on the actions of telomerase [2,3]. Telomeres protect the ends of linear chromosomes from replicative senescence, and thus structural telomere maintenance is an important aspect of cellular housekeeping activities. Consisting of tandem 5’TTAGGG3’ hexanucleotide repeats of up to 15 kilobases [4], the telomere is shortened at each round of cell division [5] because of the “end replication” problem [6]. This is not the case for cancer cells wherein fatal telomere shortening is circumvented by the activation of a telomere maintenance mechanism (TMM) [7-9]. The only two TMMs that have been described in mammalian cells are an enzymatic method that employs telomerase and DNA recombination methods (recently reviewed in detail) [10-13]. Telomerase is a reverse transcriptase consisting of a ribosomal RNA sequence (hTERC) and a protein catalytic component (hTERT) that synthesises telomeric DNA from its own template to replace telomeric DNA that is lost during cell division [14,15].
Evidence that telomere maintenance mechanisms exist, in addition to telomerase, came from investigations of telomerase-null yeast cells that carry out many divisions [16,17]. Two mechanisms for survival were identified, termed type I and type II. It is unknown whether a mechanism resembling that observed in the type I survivors exists in human tumors, but it is likely given that a similar mechanism has been identified in a human cell line that lacked both telomerase activity and the Werner syndrome protein [18]. Type II survivors have a mechanism that closely resembles the recombinant mechanism in human tumors [19]. The mammalian recombinant method was discovered in survivors of telomerase-null human cell lines that had sustained several hundreds of population doublings in culture [7,8]. This was called the alternative lengthening of telomeres (ALT) mechanism as it offers an alternative to telomerase [7].
Combinations of the two telomere maintenance mechanisms exist [20], and thus, human tumors can be divided into four groups, namely: telomerase (Tel)TMM or ALTTMM for tumors with one maintenance mechanism but not the other; TelTMM and ALTTMM (Tel/ALTTMM) for tumors with both mechanisms; or tumors with neither TelTMM nor ALTTMM, referred to as non-determined telomere maintenance mechanism (NDTMM). The TelTMM operates in most (and particularly aggressive) epithelial cancers [21-23]. The predominance of the TelTMM in human carcinoma was the impetus for developing telomerase-based therapies including telomerase inhibitors, anti-telomerase immunotherapies, and anti-telomerase viral therapies [24]. Telomerase inhibitors either block its catalytic activity or the ability of the complex to access telomeres. One welldescribed inhibitor, Imetelstat, is an oligonucleotide based compound designed to bind to the RNA component of telomerase preventing telomere access. Imetelstat inhibited telomerase activity in a wide range of human tumor cells and reduced tumor growth in mouse xenograft models. Imetelstat is currently in numerous clinical trials on a wide variety of tumors. Individuals with ALTTMM would not be expected to benefit from telomerase inhibitors, and they require the development of ALTTMM inhibitors. Those with NDTMM tumors would not be expected to benefit from TelTMM or ALTTMM inhibitor treatment. Anti-TMM treatment may alter the TMM used. Recent evidence using an animal model showed that telomerase extinction in established telomerase-positive tumors led to the development of ALTTMM positive tumors [25]. Therefore, it will be increasingly important to distinguish tumors according to their TMM when assigning the initial treatment and to continually monitor the TMM to assist in the selection of later treatment strategies.
2. Diversity of Telomere Maintenance Mechanisms
TelTMM is the principal mechanism in carcinomas of the breast (86%), colon (89%), prostate (88%), and pancreas (95%), as well as melanoma (91%) [26]. ALTTMM was first documented in osteosarcoma [8] and is most commonly found in neuroepithelial and mesenchymal tumors (reviewed in) [10,27,28]. Carcinomas with ALTTMM are rare and include cases of breast ductal carcinoma, hepatocellular carcinoma, clear cell carcinoma of the ovary, neuroendocrine carcinomas from varying locations, and squamous carcinoma of the cervix (reviewed in) [10]. ALTTMM has been inferred in a large cohort of different tumor types due to the detection of long telomeric lengths using fluorescent in situ hybridization (FISH) [29]. Using this method, no potential ALTTMM positive tumors were detected in tissue microarray samples of adenocarcinoma of the colon, small intestine, pancreas, prostate, or lungs, or in hematopoietic cancer. However, small tissue areas and tumor heterogeneity may have resulted in false negatives. The same group did report a small number of breast carcinomas with long telomeric lengths in association with human epidermal growth factor receptor 2 positivity, suggesting a link between ALTTMM and a particular aggressive carcinoma subtype [30].
In mesenchymal tumors, ALTTMM is found with frequency in osteosarcoma (35% - 47%), leiomyosarcoma (62%), liposarcoma (24% - 33% overall and 44% in grade 3), malignant fibrous histiocytoma (77%), diffuse malignant pleural mesothelioma (26%), and uterine sarcoma (46%), but it is rare in rhabdomyosarcoma (5%), and it is not found in Ewing’s sarcoma [31] (and reviewed in) [10,27,28,31]. It is suggested that sarcomas characterised by chromosomal translocations tend to maintain telomeres with telomerase, whereas sarcomas with complex karyotypes are associated with ALTTMM [32,33]. The most detailed analysis of TMM has been performed for astrocytoma. All four TMM groups are represented. Grade 1 astrocytoma, the predominant astrocytoma in children, is both TelTMM and ALTTMM negative with few exceptions [29,34,35]. Grade 2 - 3 astrocytomas are predominately ALTTMM positive [36]. However, in the aggressive grade 4 glioblastoma, ALTTMM is rare (15% - 25%) compared to TelTMM (> 40%) and the NDTMM (approximately 40%) [20,36,37]. Tel/ALTTMM asytrocytomas exist, but they are rarer (at most 5%) [20]. In glioblastoma, ALTTMM is a putative marker for secondary tumors because of the high incidence in lower grade astrocytomas, while telomerase activity marks de novo primary glioblastoma [36].
Examples of other tumors in which all TMM groups exist include Wilms’ tumor [38], liposarcoma [39,40], osteosarcoma [41], neuroblastoma [42], and peripheral nerve sheath tumors [43]. In Tel/ALTTMM positive tumors, it is not clear whether both mechanisms operate in the same cell, or whether the tumor is represented by different TMM subpopulations. Not all tumors have a known telomere maintenance mechanism. This has been described in papillary carcinoma of the thyroid [36], grade 1 astrocytoma [34,35], subsets of liposarcoma (approximately 50% overall, but only 9% of grade 3 tumors) [39], osteosarcoma (18%) [41], Ewing’s sarcoma (30%) [40], peripheral nerve sheath tumors (40%) [43], and glioblastoma (approximately 40%) [20,44]. It is possible that early tumors do not experience sufficient telomeric shortening to activate a TMM. Low-grade astrocytomas have some, but not all markers of ALTTMM, suggesting that a predisposition for ALTTMM in NDTMM tumors may already exist in cell lineages prone to ALTTMM positive tumors [35]. A model for ALTTMM development is proposed in which normal cell function is deregulated through multiple key steps until a ALTTMM arises [45]. The NDTMM category may also contain tumors in which the TMM is yet to be discovered, such as a type I survivor-like mechanism [18].
3. Detection of Telomere Maintenance Mechanism
Methods to detect TelTMM and ALTTMM are routinely used in the research laboratory but are not always readily applicable to the diagnostic medical laboratory (reviewed in detail elsewhere) [10,28,46]. A combination of two methods is currently required, one that provides information on TelTMM and another that detects the ALTTMM status, to differentiate all four TMM groups. Each method has advantages and disadvantages, and their suitability to the clinical setting is summarised below. The key aspects of five assays (TRAP, TRF, APB, FISH, c-circle and TERRA) are given in Figure 1(a), and the application of these results to distinguish the four TMM tumor subtypes is outlined in Figure 1(b).
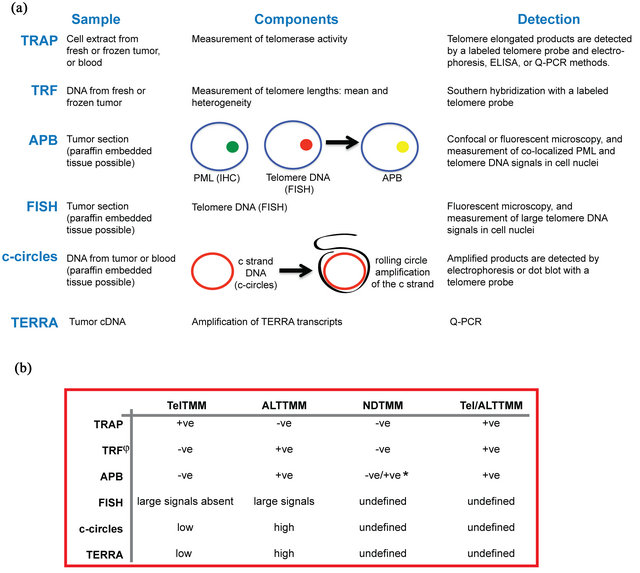
Figure 1. Assays for determining the telomere maintenance mechanism used in human tumors. (a) The TRAP, TRF, APB, FISH, c-circle, and TERRA TMM assays have different sample requirements, specific aspects, and methods of detection that ultimately provide information on whether a tumor has telomerase activity or uses the recombination method for telomere maintenance; (b) Assay results determine whether a tumor uses telomerase activity as a telomere maintenance mechanism (TelTMM), or the recombination-based method (ALTTMM). By using a combination of assays, tumors can be divided into all four telomere maintenance (TMM) subgroups: positive for one TMM (TelTMM (or ALTTMM), positive for both TMMs (Tel/ALTTMM), or positive for neither TMM (NDTMM). The use of at least two assays is preferred to ascertain certainty of the TMM. +ve, positive; -ve, negative; *, some low-grade tumors indicated as NDTMM based on the TRF length are positive for APBs. APB, ALT-associated PML bodies; ELISA, enzyme-linked immunosorbent assay; FISH, fluorescent in situ hybridization; PML, promyelocytic leukemia protein; Q-PCR, quantitative PCR; TRAP, telomere repeat amplification protocol; TRF, terminal restriction fragment; undetermined, the assay has yet to be tested on known Tel/ALTTMM or NDTMM tumors.
3.1. Detection of TelTMM Positive Tumors
TRAP assay: The determination of telomerase activity by the TRAP (telomere repeat amplification protocol) assay is a specific, well-established protocol (reviewed in) [46]. This technique is PCR based and quantifies enzyme processivity using tumor tissue [47], with high-throughput quantitative-PCR (Q-PCR) and ELISA-based kits (reviewed in) [46]. Although not as widely validated, blood samples may be used. Here, cancerous cells are first separated from leukocytes before the telomerase activity is assessed [48]. However, blood samples are only applicable if cancerous cells are in circulation.
Although, the TRAP assay is currently the gold standard for detecting TelTMM, it requires fresh or frozen tissue and has aspects that make it problematic for the diagnostic laboratory. Most adaptions of the TRAP assays allow only semi-quantitative measurements, falsenegative and false-positive results can occur, and the assays are technically demanding. Non-PCR based alternatives are available. An in situ-based adaption allows for the assessment of individual cells, and it can ensure that the telomerase activity is within cancerous cells and is not that from non-cancerous activated lymphocytes [49]. The TRE (telomere repeat elongation assay) assay allows sensitive and precise quantification of telomerase activity, but it does require specific equipment [50]. Continued improvement to the TRAP assay and wider validation of the alternative assays will promote the development of a routine assay for the diagnostic setting.
Immunohistochemistry: Immunohistochemistry-based methods, typically for hTERT, can be used on paraffinembedded tissues [51]. This method allows individual cells to be analyzed, but it is not as well validated as the TRAP assay. Successful hTERT detection depends on a number of factors, including the type of antibody used and the assay conditions [52]. The detection of hTERT does not necessarily equate to telomerase activity. The hTERT protein has been shown to participate in cell processes other than telomere maintenance [53,54]. Multiple isoforms of hTERT exist [55], and discrepancies between the level of hTERT transcript and the level of hTERT protein have been reported [56]. Other molecular characteristics of TelTMM, as have been found for ALTTMM, could be used for surrogate immunohistochemistry-based assays in the future.
TERRA assay: The telomeric repeat-containing RNA (TERRA) assay is a relatively new assay that may prove valuable for accessing whether a tumor is telomerase positive [57]. TERRAs are noncoding transcripts containing telomere repeats that occur as a result of transcription at subtelomeric regions [58,59]. TERRA expression is dependant on CpG island promoters found in many eukaryotic genes (reviewed in) [60]. Cytosines in CpG islands can be methylated by DNA methyltransferases (DNMT), and consequently transcription activity is negatively regulated (reviewed in) [61]. Methylation increases with advancing age, it is accelerated in the colon in response to chronic inflammation, and it might be an adaptive response to injury [62]. TERRA may inhibit telomerase activity [59,63]. A recent study demonstrated that TERRA is inversely proportional to telomerase activity [57]. TERRA transcript levels are quantified by PCR based methods, with low concentrations consistent with TelTMM.
Thus, TelTMM tumors can be identified using TRAP, immunohistochemistry and TERRA assays. The TRAP assay is the best characterised and the most widely validated, while developments in other telomerase activity assays, hTERT antibody design, and surrogate assays for the presence of telomerase, such as TERRA, provide additional options for adaption to the clinical setting.
3.2. Detection of ALTTMM Positive Tumors
The gold standard measurement for identifying ALTTMM is the presence of maintained telomere length by recombination in the absence of telomerase activity [7]. The absence of telomerase is an important distinction. As reviewed below, the prognosis for ALTTMM varies according to the presence or absence of telomerase. For human tumors, as opposed to cultured cells, the demonstration of recombination is not routinely possible, so the presence of heterogeneous telomeres suffices as a surrogate. Thus ALTTMM, assays rely on phenotypic ALTTMM features. Assays that are practicably applicable are the detection of 1) long and heterogeneous telomere lengths; 2) APBs and 3) c-circles.
TRF measurement: The terminal restriction fragment (TRF) assay is most commonly used as an indication of the telomere length distribution. Based on Southern hybridization, telomere DNA is separated using electrophoresis, and a probe recognising the telomere DNA allows for visualisation of the telomere length distribution [7]. ALTTMM positive tumors possess long (increased mean length) and heterogeneous (ranging from critically short to extremely long in length) lengths compared to TelTMM or NDTMM tumors. The TRF assay is labor intensive and can produce equivocal results when used as a single TMM assay [36]. For telomere length, higher throughput versions using Q-PCR-based methods are available with a good correlation with telomere length measurement using the traditional TRF method [64,65]. False-positive interpretations occur, as some telomerasepositive cells do produce long telomere lengths, consistent with that observed for ALTTMM (reviewed in) [66].
FISH: More recently, the increased length of the telomeres is detected by telomere DNA fluorescence in situ hybridisation (FISH) [29,30]. In this assay, ALTTMM tumors produce large fluorescent signals compared to noncancerous cells. FISH is within the capabilities of many diagnostic laboratories, and therefore this method could be applicable to the clinical setting. Again, as outlined earlier, false-positive interpretation can occur if the telomere length alone is used to assay for TMM.
APB assay: In addition to telomere DNA FISH detection combined with co-localized detection of the promyelocytic leukaemia protein (PML), is the APB assay. “ALT-associated” nuclear PML Bodies (APBs) in large form are unique to ALTTMM with few exceptions [19, 35,36]. The development of the APB detection assay by the Reddel group marked a substantial advance in identification of ALTTMM tumors because paraffin embedded sections, or even cytopathological samples, could be used [36]. Immunofluorescence detects the PML protein (or an associated telomere protein such as telomere repeat factor 1, telomere repeat factor 2, or a PML body associated protein such as sp100) (reviewed in) [12,45], and FISH detects telomere DNA, typically using a protein nucleic acid (PNA) probe consisting of three telomeric DNA repeats.
An APB is defined as a large (≥1.4 µm2) co-localized PML protein and telomere DNA signal in the cell nucleus [35,36]. A tumor is typically considered positive if the percentage of cells containing APBs is >0.5%. Large co-localizations of PML and telomere DNA are tumor specific, whereas smaller co-localizations have been found in both neoplastic and non-neoplastic cells [67, 68]. The function of both large and small co-localizations is unknown, but upon malignant transformation to ALTTMM, large APBs have been shown to appear and disappear precisely with the gross deregulated telomere length phenotype [19] (and reviewed in) [28]. Tumors with only large co-localizations and no heterogeneous telomeres have also been described [35]. Consistent with the ALTTMM-negative status, grade 1 astrocytomas do not show the gross aberrant telomere length phenoltype. However, these tumors do have large co-localizations found in ALTTMM tumors, suggesting that the APB assay may be able to detect early changes toward ALTTMM as it provides an assessment of individual cells [35]. Standard fluorescent or confocal microscopy is required, which is labor intensive, and some tumors show recombination activity without APB formation [29,69,70].
C-circle assay: C-circles are ALTTMM specific and are comprised of circular telomeric C-strand (i.e., (CCCTAA)n circles) [71]. C-circles are self-priming, an attribute that is utilized in the c-circle assay. The c-circle assay is quantitative and involves amplification of circular DNA using rolling circle amplification with the C-telomere strand as the template and an isothermic polymerase. The G-strand products can be visualised by electrophoresis or using a dot blot format using a telomere DNA probe. The c-circle assay has been correlated with ALTTMM activity in solid tumors [31]. Although this assay has not been widely used, no false negative results have been reported. The c-circle assay can be used with tumor tissue, including paraffin-embedded tissue [31], or peripheral blood genomic DNA samples [71]. Detection of cancer-derived products in the blood is a distinct advantage of this assay for the clinical setting.
TERRA assay: The TERRA assay outlined earlier for TelTMM detection may also provide insight into the ALTTMM status. Cells deficient for DNMT (expected to express high levels of TERRA) were shown to possess increased heterogeneity of telomere length and increased APB frequency (hallmarks of the ALTTMM positive tumor) [72]. Thus, a loss of methylation and increased TERRA transcripts promote recombination. High concentrations of TERRA expression were found in tumors with long telomere lengths that lacked telomerase activeity [57]. These results are consistent with increased TERRA levels (determined by a Q-PCR based method described earlier) being a marker of ALTTMM [57].
Mutation assay: Large-scale genomic analyses have discovered new markers associated with ALTTMM [73,74]. One of these is mutation in the isocitrate dehydrogenase 1 (1DH1) gene [74-76]. Mutant IDH1 is much more frequent in grade 2 - 3 astrocytomas and secondary glioblastoma compared to primary gliboblastomas [75, 76]. In ALTTMM-positive tumors, IDH1 mutations are found at a frequency of 19% - 59% and were rare in TelTMM and NDTMM glioblastomas (<5%) [37,77]. In brain tumors, the R132H variant is the most prominent IDH1 mutation and can be detected by immunohistochemistry or sequencing of tumor DNA [75,78].
Mutations in the ATRX, DAXX, and histone H3.3 (H3F3A) genes are also associated with ALTTMM [73, 79-81]. Mutations can be identified by sequencing, or in the case of ATRX and DAXX mutations, by a lack of protein expression using immunohistochemistry [79,80]. Screening for ATRX/DAXX mutations using immunohistochemistry in pancreatic neuroendocrine tumors revealed that 100% of tumors with ATRX/DAXX mutations were ALTTMM positive [79]. In pediatric glioblastomas, recurrent H3F3A mutations were found in approximately 36% of tumors, but only in 3.4% of adult glioblastomas. In glioblastoma, H3F3A mutations were concurrent with ATRX/DAXX mutations and were mutually exclusive with IDH1 mutations [80]. For detection of all ALTTMMs by associated mutation analysis, a panel of antibodies to the various mutations would be required.
ALTTMM tumors are identified using the TRF, FISH, APB, c-circle and TERRA assays. The TRF and APB assays are the best characterized and the most widely validated, but the other methods are more readily adapted for the diagnostic laboratory. In the near future, ALTTMM tumors may be identified, or the TMM may be supported using a panel of antibodies to the various mutations that are characteristic of ALTTMM.
4. Prognosis and Telomere Maintenance Mechanism
The prognostic significance of the TMM is dependent on the tumor type. Here we focus our attention on tumors with all four TMM subtypes. With the exception of glioblastoma, tumors with NDTMM are consistently associated with the best survival as might be expected, whilst the presence of the ALTTMM is often associated with the worst prognosis [20,39-42,82].
4.1. Prognosis in Glioblastoma
In glioblastoma, the TRAP, TRF and the APB assays can be used to separate tumors into TelTMM, ALTTMM, Tel/ALTTMM and NDTMM [20,36,37,83]. Patients with ALTTMM tumors have an improved prognosis compared to those with TelTMM and NDTMM [20]. Patients with Tel/ALTTMM tumors had the worst mean survival, but they are too rare for a statistical comparison with the other TMM tumor types [20]. Further subtyping of the TelTMM, ALTTMM and NDTMM subgroups based on other molecular parameters showed that patients with ALTTMM-positive tumors with p53 mutations had an improved survival compared to ALTTMM positive tumors with wild-type p53 [83]. In contrast, a p53 mutation conferred a poorer survival for those with TelTMM, and no difference in survival was detected for those with NDTMM [83]. In ALTTMM-positive glioblastomas, IDH1 mutations increased the overall survivability by approximately 3 months [76]. The ALTTMM H3F3A, ATRX, DAXX, and IDH1 mutation phenotypes are thought to contribute to tumor development by inhibiting demethylation, therefore leading to increased methylation, altered gene transcription, and a more progenitor cell-like tumor signature [80,84]. Thus the possibility that these mutations also alter the TERRA promoter methylation pattern is raised. In NDTMM glioblastoma, the frequency of a sequence variant (G500) in the 3’ untranslated region of the cyclin-dependent kinase inhibitor 2A (CDKN2A) gene was increased and associated with a poorer survival, an increased age, and loss of CDKN2A gene dosage in carriers [37].
The TRAP and TRF assays require frozen tissue, however this is not always possible when large cohorts are sought. In a cohort of 573 glioblastomas, the APB assay alone confirmed a 2-year increase in the survival rate for those with APB-positive tumors (consistent with ALTTMM) at 22% compared to 11% for those with APB negative tumors [77]. The use of a large cohort resulted in the discovery of important implications for treatment. For ALTTMM tumors, surgery and radiotherapy offered no statistically significant difference in survival compared to surgery alone, whilst for ALTTMM-negative tumors, surgery and radiotherapy were associated with a significant improvement in survival compared to surgery alone [77]. That the ALTTMM is associated with resistance to radiotherapy is further evident in studies with glioma stem cells, which are resistant to radiotherapy, compared to telomerase-positive tumor stem cells [85]. Resistance to radiotherapy is suggested to result from an impaired DNA damage response [85]. The disadvantage of using the APB assay alone is that the NDTMM and TelTMM tumors are not distinguished.
As suggested by the findings of Sampl et al. (2012), in an analysis of a smaller glioblastoma cohort (28 cases), the survival of individuals based on a single TMM assays did not result in a correlation with survival [57]. However, analysis of survival based on a combination of assays (TERRA levels, telomere length by Q-PCR and telomerase activity by TRAP) was associated with a trend for survival. This confirms that at least two assays are required to maintain certainty of the mechanism and resolve the TMM category.
4.2. Prognosis in Other Tumor Types
In contrast to gliomagenesis, TMM in other non-epithetlial tumors offers a quite different prognosis. Three sarcomas, (uterine sarcoma) [31], liposarcoma [39,40,70,82], and osteosarcoma [36,41]) as well as neuroblastoma [41] have been studied with enough clinical follow-up and in sufficient detail to highlight the major prognostic differences, at least in uni-variant analyzes. In liposarcoma, the use of the APB and/or TRF, and TRAP assays revealed that ALTTMM and Tel/ALTTMM are associated with poor survival, if not the poorest, TelTMM is associated with poor or intermediate survival, and NDTMM is associated with the best survival for patients [39,40,70,82]. In osterosarcoma, NDTMM was associated with good progression-free survival compared to the other TMM subgroups combined [41]. In uterine sarcoma, patients with ALTTMM were associated with a poorer disease-free survival and overall survival compared to those with non-ALTTMMs [31]. For neuroblastoma, NDTMM was associated with 100% progression-free survival compared to 46% for Tel/ALTTMM, 67% for ALTTMM, and 78% for TelTMM [42]. Whether ALTTMM-positive sarcomas are also associated with IDH1 mutations, as was recently reported for glioblastoma, is unknown. IDH1 mutations have been detected in chondrosarcomas that have a high prevalence of ALTTMM but were not detected in other ALTTMM-positive types of osteosarcoma or soft tissue mesenchymal malignancies [86]. This warrants further investigation, as IDH1 mutations are relatively easy to identify both in the laboratory and with radiological imaging and could offer improved prognostic detail.
In sarcoma, it is not merely the presence of a TMM, but the presence of ALTTMM that is prognostically important. The poorer prognosis for ALTTMM-positive tumors could be due to a more aggressive tumor phenoltype. The ALTTMM is more frequent in high-grade compared to low-grade tumors, and it has an increased mitotic index and tumor size compared to non-ALTTMMs [31,39,87]. The picture is less clear with disease progresssion, and metastases are frequently telomerase positive [88,89].
5. Conclusions
Although telomerase inhibition is an attractive option for treating cancer, patients with ALTTMM and NDTMM tumors would not benefit. Stratifying patients based on the TMM will be required for assigning the correct treatment, either a ALTTMM or TelTMM inhibitor, or neither. Furthermore, the prognostic data presented in this review emphasise the need to diagnostically consider and determine all four groups of tumor TMM. The methods used to detect these groups are also important, and at least two methods are currently required. The ALTTMM can be inferred from heterogeneous and long telomere lengths, high TERRA, the presence of APBs, or the c-circle assay. Telomerase activity is determined by TRAP and inferred based on low TERRA or the hTERT immunohistochemistry assay. The ability to monitor the TMM using blood samples is an attractive option in the clinical setting, and it may prove important for early tumor detection.
As yet, all combinations of TMM detection methods have not been formally validated against known ALTTMM tissues, and this evidence is urgently required to progress to clinical trials. From the above studies, a combination of telomere length and TRAP, or the APB assay and TRAP appear adequate. Newer tests such as TERRA, immunohistochemistry for multiple markers, and the c-circle analysis are available, and combinations includeing these may be better adapted to the diagnostic laboratory. Hopefully a combination can be found that will prove clinically predictive, with good sensitivity and specificity. Targeted and effective telomere maintenance pathway treatment will then become a useful addition to the oncologist’s armamentarium.
REFERENCES
- D. Hanahan and R. A. Weinberg, “Hallmarks of Cancer: The Next Generation,” Cell, Vol. 144, No. 5, 2011, pp. 646-674. doi:10.1016/j.cell.2011.02.013
- E. H. Blackburn, C. W. Greider and J. W. Szostak, “Telomeres and Telomerase: The Path from Maize, Tetrahymena and Yeast to Human Cancer and Aging,” Nature Medicine, Vol. 12, No. 10, 2006, pp. 1133-1138. doi:10.1038/nm1006-1133
- S. E. Artandi and R. A. DePinho, “Telomeres and Telomerase in Cancer,” Carcinogenesis, Vol. 31, No. 1, 2010, pp. 9-18. doi:10.1093/carcin/bgp268
- R. K. Moyzis, J. M. Buckingham, L. S. Cram, M. Dani, L. L. Deaven, M. D. Jones, J. Meyne, R. L. Ratliff and J. R. Wu, “A Highly Conserved Repetitive DNA Sequence, (TTAGGG)n, Present at the Telomeres of Human Chromosomes,” Proceedings of the National Academy of Sciences of the United States, Vol. 85, No. 18, 1988, pp. 6622-6626. doi:10.1073/pnas.85.18.6622
- C. B. Harley, A. B. Futcher and C. W. Greider, “Telomeres Shorten during Ageing of Human Fibroblasts,” Nature, Vol. 345, No. 6274, 1990, pp. 458-460. doi:10.1038/345458a0
- A. M. Olovnikov, “A Theory of Marginotomy. The Incomplete Copying of Template Margin in Enzymic Synthesis of Polynucleotides and Biological Significance of the Phenomenon,” Journal of Theoretical Biology, Vol. 41, No. 1, 1973, pp. 181-190. doi:10.1016/0022-5193(73)90198-7
- T. M. Bryan, A. Englezou, J. Gupta, S. Bacchetti and R. R. Reddel, “Telomere Elongation in Immortal Human Cells without Detectable Telomerase Activity,” The EMBO Journal, Vol. 14, No. 17, 1995, pp. 4240-4248.
- T. M. Bryan, A. Englezou, L. Dalla-Pozza, M. A. Dunham and R. R. Reddel, “Evidence for an Alternative Mechanism for Maintaining Telomere Length in Human Tumors and Tumor-Derived Cell Lines,” Nature Medicine, Vol. 3, No. 11, 1997, pp. 1271-1274. doi:10.1038/nm1197-1271
- J. W. Shay and S. Bacchetti, “A Survey of Telomerase Activity in Human Cancer,” European Journal of Cancer, Vol. 33, No. 5, 1997, pp. 787-791. doi:10.1016/S0959-8049(97)00062-2
- S. T. Durant, “Telomerase-Independent Paths to Immortality in Predictable Cancer Subtypes,” Journal of Cancer, Vol. 3, 2012, pp. 67-82. doi:10.7150/jca.3965
- A. J. Cesare and R. R. Reddel, “Alternative Lengthening of Telomeres: Models, Mechanisms and Implications,” Nature Reviews. Genetics, Vol. 11, No. 5, 2010, pp. 319- 330. doi:10.1038/nrg2763
- A. Nabetani and F. Ishikawa, “Alternative Lengthening of Telomeres Pathway: Recombination-Mediated Telomere Maintenance Mechanism in Human Cells,” Journal of Biochemistry, Vol. 149, No. 1, 2011, pp. 5-14. doi:10.1093/jb/mvq119
- J. W. Shay and W. E. Wright, “Role of Telomeres and Telomerase in Cancer,” Seminars in Cancer Biology, Vol. 21, No. 6, 2011, pp. 349-353. doi:10.1016/j.semcancer.2011.10.001
- C. W. Greider and E. H. Blackburn, “Identification of a Specific Telomere Terminal Transferase Activity in Tetrahymena Extracts,” Cell, Vol. 43, No. 2, 1985, pp. 405- 413. doi:10.1016/0092-8674(85)90170-9
- C. W. Greider and E. H. Blackburn, “A Telomeric Sequence in the RNA of Tetrahymena Telomerase Required for Telomere Repeat Synthesis,” Nature, Vol. 337, No. 6205, 1989, pp. 331-337. doi:10.1038/337331a0
- S. C. Teng and V. A. Zakian, “Telomere-Telomere Recombination Is an Efficient Bypass Pathway for Telomere Maintenance in Saccharomyces cerevisiae,” Molecular and Cellular Biology, Vol. 19, No. 12, 1999, pp. 8083- 8093.
- V. Lundblad and J. W. Szostak, “A Mutant with a Defect in Telomere Elongation Leads to Senescence in Yeast,” Cell, Vol. 57, No. 4, 1989, pp. 633-643. doi:10.1016/0092-8674(89)90132-3
- A. Siddiqa, D. Cavazos, J. Chavez, L. Long and R. A. Marciniak, “Modulation of Telomeres in Alternative Lengthening of Telomeres Type I Like Human Cells by the Expression of Werner Protein and Telomerase,” Journal of Oncology, Vol. 2012, 2012, Article ID: 806382. doi:10.1155/2012/806382
- T. R. Yeager, A. A. Neumann, A. Englezou, L. I. Huschtscha, J. R. Noble and R. R. Reddel, “TelomeraseNegative Immortalized Human Cells Contain a Novel Type of Promyelocytic Leukemia (PML) Body,” Cancer Research, Vol. 59, No. 17, 1999, pp. 4175-4179.
- V. Hakin-Smith, D. A. Jellinek, D. Levy, T. Carroll, M. Teo, W. R. Timperley, M. J. McKay, R. R. Reddel and J. A. Royds, “Alternative Lengthening of Telomeres and Survival in Patients with Glioblastoma Multiforme,” Lancet, Vol. 361, No. 9360, 2003, pp. 836-838. doi:10.1016/S0140-6736(03)12681-5
- K. Hiyama, E. Hiyama, S. Ishioka, M. Yamakido, K. Inai, A. F. Gazdar, M. A. Piatyszek and J. W. Shay, “Telomerase Activity in Small-Cell and Non-Small-Cell Lung Cancers,” Journal of the National Cancer Institute, Vol. 87, No. 12, 1995, pp. 895-902. doi:10.1093/jnci/87.12.895
- E. Hiyama, L. Gollahon, T. Kataoka, K. Kuroi, T. Yokoyama, A. F. Gazdar, K. Hiyama, M. A. Piatyszek and J. W. Shay, “Telomerase Activity in Human Breast Tumors,” Journal of the National Cancer Institute, Vol. 88, No. 2, pp. 1996, pp. 116-122. doi:10.1093/jnci/88.2.116
- N. Tatsumoto, E. Hiyama, Y. Murakami, Y. Imamura, J. W. Shay, Y. Matsuura and T. Yokoyama, “High Telomerase Activity Is an Independent Prognostic Indicator of Poor Outcome in Colorectal Cancer,” Clinical Cancer Research: An Official Journal of the American Association for Cancer Research, Vol. 6, No. 7, 2000, pp. 2696- 2701.
- C. M. Buseman, W. E. Wright and J. W. Shay, “Is Telomerase a Viable Target in Cancer?” Mutation Research, Vol. 730, No. 1-2, 2012, pp. 90-97. doi:10.1016/j.mrfmmm.2011.07.006.
- J. Hu, S. S. Hwang, M. Liesa, B. Gan, E. Sahin, M. Jaskelioff, Z. Ding, H. Ying, A. T. Boutin, H. Zhang, S. Johnson, E. Ivanova, M. Kost-Alimova, A. Protopopov, Y. A. Wang, O. S. Shirihai, L. Chin and R. A. DePinho, “Antitelomerase Therapy Provokes ALT and Mitochondrial Adaptive Mechanisms in Cancer,” Cell, Vol. 148, No. 4, 2012, pp. 651-663. doi:10.1016/j.cell.2011.12.028
- A. K. Meeker and D. S. Coffey, “Telomerase: A Promising Marker of Biological Immortality of Germ, Stem, and Cancer Cells. A Review,” Biochemistry (Moscow), Vol. 62, No. 11, 1997, pp. 1323-1331.
- J. E. Johnson and D. Broccoli, “Telomere Maintenance in Sarcomas,” Current Opinion in Oncology, Vol. 19, No. 4, 2007, pp. 377-382. doi:10.1097/CCO.0b013e3281214423
- J. D. Henson and R. R. Reddel, “Assaying and Investigating Alternative Lengthening of Telomeres Activity in Human Cells and Cancers,” FEBS Letters, Vol. 584, No. 17, 2010, pp. 3800-3811. doi:10.1016/j.febslet.2010.06.009
- C. M. Heaphy, A. P. Subhawong, S. M. Hong, M. G. Goggins, E. A. Montgomery, E. Gabrielson, G. J. Netto, J. I. Epstein, T. L. Lotan, W. H. Westra, M. S. Ie, C. A. Iacobuzio-Donahue, A. Maitra, Q. K. Li, C. G. Eberhart, J. M. Taube, D. Rakheja, R. J. Kurman, T. C. Wu, R. B. Roden, P. Argani, A. M. De Marzo, L. Terracciano, M. Torbenson and A. K. Meeker, “Prevalence of the Alternative Lengthening of Telomeres Telomere Maintenance Mechanism in Human Cancer Subtypes,” American Journal of Pathology, Vol. 179, No. 4, 2011, pp. 1608-1615. doi:10.1016/j.ajpath.2011.06.018
- A. P. Subhawong, C. M. Heaphy, P. Argani, Y. Konishi, N. Kouprina, H. Nassar, R. Vang and A. K. Meeker, “The Alternative Lengthening of Telomeres Phenotype in Breast Carcinoma Is Associated with HER-2 Overexpression,” Modern Pathology: An Official Journal of the United States and Canadian Academy of Pathology, Inc., Vol. 22, No. 11, 2009, pp. 1423-1431.
- Y. K. Lee, N. H. Park and H. Lee, “Prognostic Value of Alternative Lengthening of Telomeres-Associated Biomarkers in Uterine Sarcoma and Uterine Carcinosarcoma,” International Journal of Gynecological Cancer, Vol. 22, No. 3, 2012, pp. 434-441. doi:10.1097/IGC.0b013e31823ca017
- G. A. Ulaner, A. R. Hoffman, J. Otero, H. Y. Huang, Z. Zhao, M. Mazumdar, R. Gorlick, P. Meyers, J. H. Healey and M. Ladanyi, “Divergent Patterns of Telomere Maintenance Mechanisms among Human Sarcomas: Sharply Contrasting Prevalence of the Alternative Lengthening of Telomeres Mechanism in Ewing’s Sarcomas and Osteosarcomas,” Genes Chromosomes Cancer, Vol. 41, No. 2, 2004, pp. 155-162. doi:10.1002/gcc.20074
- E. Montgomery, P. Argani, J. L. Hicks, A. M. DeMarzo and A. K. Meeker, “Telomere Lengths of TranslocationAssociated and Nontranslocation-Associated Sarcomas Differ Dramatically,” American Journal of Pathology, Vol. 164, No. 5, 2004, pp. 1523-1529. doi:10.1016/S0002-9440(10)63710-8
- U. Tabori, B. Vukovic, M. Zielenska, C. Hawkins, I. Braude, J. Rutka, E. Bouffet, J. Squire and D. Malkin, “The Role of Telomere Maintenance in the Spontaneous Growth Arrest of Pediatric Low-Grade Gliomas,” Neoplasia, Vol. 8, No. 2, 2006, pp. 136-142. doi:10.1593/neo.05715
- T. Slatter, J. Gifford-Garner, A. Wiles, X. Tan, Y. J. Chen, M. MacFarlane, M. Sullivan, J. Royds and N. Hung, “Pilocytic Astrocytomas Have Telomere-Associated Promyelocytic Leukemia Bodies without Alternatively Lengthened Telomeres,” American Journal of Pathology, Vol. 177, No. 6, 2010, pp. 2694-2700. doi:10.2353/ajpath.2010.100468
- J. D. Henson, J. A. Hannay, S. W. McCarthy, J. A. Royds, T. R. Yeager, R. A. Robinson, S. B. Wharton, D. A. Jellinek, S. M. Arbuckle, J. Yoo, B. G. Robinson, D. L. Learoyd, P. D. Stalley, S. F. Bonar, D. Yu, R. E. Pollock and R. R. Reddel, “A Robust Assay for Alternative Lengthening of Telomeres in Tumors Shows the Significance of Alternative Lengthening of Telomeres in Sarcomas and Astrocytomas,” Clinical Cancer Research, Vol. 11, No. 1, 2005, pp. 217-225.
- J. A. Royds, S. Al Nadaf, A. K. Wiles, Y. J. Chen, A. Ahn, A. Shaw, S. Bowie, F. Lam, B. C. Baguley, A. W. Braithwaite, M. R. MacFarlane, N. A. Hung and T. L. Slatter, “The CDKN2A G500 Allele Is More Frequent in GBM Patients with No Defined Telomere Maintenance Mechanism Tumors and Is Associated with Poorer Survival,” PLoS One, Vol. 6, No. 10, 2011, Article ID: e26737. doi:10.1371/journal.pone.0026737
- L. Venturini, M. G. Daidone, R. Motta, P. Collini, F. Spreafico, M. Terenziani, L. Piva, P. Radice, D. Perotti and N. Zaffaroni, “Telomere Maintenance in Wilms Tumors: First Evidence for the Presence of Alternative Lengthening of Telomeres Mechanism,” Genes, Chromosomes & Cancer, Vol. 50, No. 10, 2011, pp. 823-829. doi:10.1002/gcc.20903
- A. Costa, M. G. Daidone, L. Daprai, R. Villa, S. Cantu, S. Pilotti, L. Mariani, A. Gronchi, J. D. Henson, R. R. Reddel and N. Zaffaroni, “Telomere Maintenance Mechanisms in Liposarcomas: Association with Histologic Subtypes and Disease Progression,” Cancer Research, Vol. 66, No. 17, 2006, pp. 8918-8924. doi:10.1158/0008-5472.CAN-06-0273
- L. Venturini, R. Motta, A. Gronchi, M. Daidone and N. Zaffaroni, “Prognostic Relevance of ALT-Associated Markers in Liposarcoma: A Comparative Analysis,” BMC Cancer, Vol. 10, 2010, p. 254. doi:10.1186/1471-2407-10-254
- G. A. Ulaner, H. Y. Huang, J. Otero, Z. Zhao, L. Ben-Porat, J. M. Satagopan, R. Gorlick, P. Meyers, J. H. Healey, A. G. Huvos, A. R. Hoffman and M. Ladanyi, “Absence of a Telomere Maintenance Mechanism as a Favorable Prognostic Factor in Patients with Osteosarcoma,” Cancer Research, Vol. 63, No. 8, 2003, pp. 1759- 1763.
- A. Ohali, S. Avigad, S. Ash, Y. Goshen, D. Luria, M. Feinmesser, R. Zaizov and I. Yaniv, “Telomere Length Is A Prognostic Factor in Neuroblastoma,” Cancer, Vol. 107, No. 6, 2006, pp. 1391-1399. doi:10.1002/cncr.22132
- L. Venturini, M. G. Daidone, R. Motta, G. Cimino-Reale, S. F. Hoare, A. Gronchi, M. Folini, W. N. Keith and N. Zaffaroni, “Telomere Maintenance Mechanisms in Malignant Peripheral Nerve Sheath Tumors: Expression and Prognostic Relevance,” Neuro-Oncology, Vol. 14, No. 6, 2012, pp. 736-744. doi:10.1093/neuonc/nos083
- R. Villa, M. G. Daidone, R. Motta, L. Venturini, C. De Marco, A. Vannelli, S. Kusamura, D. Baratti, M. Deraco, A. Costa, R. R. Reddel and N. Zaffaroni, “Multiple Mechanisms of Telomere Maintenance Exist and Differentially Affect Clinical Outcome in Diffuse Malignant Peritoneal Mesothelioma,” Clinical Cancer Research, Vol. 14, No. 13, 2008, pp. 4134-4140. doi:10.1158/1078-0432.CCR-08-0099
- I. Chung, S. Osterwald, K. I. Deeg and K. Rippe, “PML Body Meets Telomere: The Beginning of an ALTernate Ending?” Nucleus, Vol. 3, No. 3, 2012, pp. 263-275. doi:10.4161/nucl.20326
- J. Fajkus, “Detection of Telomerase Activity by the TRAP Assay and Its Variants and Alternatives,” Clinica chimica Acta; International Journal of Clinical Chemistry, Vol. 371, No. 1-2, 2006, pp. 25-31.
- N. W. Kim, M. A. Piatyszek, K. R. Prowse, C. B. Harley, M. D. West, P. L. Ho, G. M. Coviello, W. E. Wright, S. L. Weinrich and J. W. Shay, “Specific Association of Human Telomerase Activity with Immortal Cells and Cancer,” Science, Vol. 266, No. 5193, 1994, pp. 2011-2015. doi:10.1126/science.7605428
- L. R. Gauthier, C. Granotier, J. C. Soria, S. Faivre, V. Boige, E. Raymond and F. D. Boussin, “Detection of Circulating Carcinoma Cells by Telomerase Activity,” British Journal of Cancer, Vol. 84, No. 5, 2001, pp. 631-635. doi:10.1054/bjoc.2000.1662
- K. Ohyashiki, J. H. Ohyashiki, J. Nishimaki, K. Toyama, Y. Ebihara, H. Kato, W. E. Wright and J. W. Shay, “Cytological Detection of Telomerase Activity Using an in Situ Telomeric Repeat Amplification Protocol Assay,” Cancer Research, Vol. 57, No. 11, 1997, pp. 2100-2103.
- C. Maesawa, T. Inaba, H. Sato, S. Iijima, K. Ishida, M. Terashima, R. Sato, M. Suzuki, A. Yashima, S. Ogasawara, H. Oikawa, N. Sato, K. Saito and T. Masuda, “A Rapid Biosensor Chip Assay for Measuring of Telomerase Activity Using Surface Plasmon Resonance,” Nucleic Acids Research, Vol. 31, No. 2, 2003, p. E4. doi:10.1093/nar/gng004
- E. Hiyama, K. Hiyama, T. Yokoyama and J. W. Shay, “Immunohistochemical Detection of Telomerase (hTERT) Protein in Human Cancer Tissues and a Subset of Cells in Normal Tissues,” Neoplasia, Vol. 3, No. 1, 2001, pp. 17- 26. doi:10.1038/sj.neo.7900134
- P. Yan, J. Benhattar, W. Seelentag, J. C. Stehle and F. T. Bosman, “Immunohistochemical Localization of hTERT Protein in Human Tissues,” Histochemistry and Cell Biology, Vol. 121, No. 5, 2004, pp. 391-397. doi:10.1007/s00418-004-0645-5
- C. Dudognon, F. Pendino, J. Hillion, A. Saumet, M. Lanotte and E. Segal-Bendirdjian, “Death Receptor Signaling Regulatory Function for Telomerase: hTERT Abolishes TRAIL-Induced Apoptosis, Independently of Telomere Maintenance,” Oncogene, Vol. 23, No. 45, 2004, pp. 7469-7474. doi:10.1038/sj.onc.1208029
- W. Fu, J. G. Begley, M. W. Killen and M. P. Mattson, “Anti-Apoptotic Role of Telomerase in Pheochromocytoma Cells,” The Journal of Biological Chemistry, Vol. 274, No. 11, 1999, pp. 7264-7271. doi:10.1074/jbc.274.11.7264
- A. Kilian, D. D. Bowtell, H. E. Abud, G. R. Hime, D. J. Venter, P. K. Keese, E. L. Duncan, R. R. Reddel and R. A. Jefferson, “Isolation of a Candidate Human Telomerase Catalytic Subunit Gene, Which Reveals Complex Splicing Patterns in Different Cell Types,” Human Molecular Genetics, Vol. 6, No. 12, 1997, pp. 2011-2019. doi:10.1093/hmg/6.12.2011
- J. S. Shin, T. Foo, A. Hong, M. Zhang, T. Lum, M. J. Solomon and C. S. Lee, “Telomerase Expression as a Predictive Marker of Radiotherapy Response in Rectal Cancer,” Pathology, Vol. 44, No. 3, 2012, pp. 209-215. doi:10.1097/PAT.0b013e3283511cd5
- S. Sampl, S. Pramhas, C. Stern, M. Preusser, C. Marosi and K. Holzmann, “Expression of Telomeres in Astrocytoma WHO Grade 2 to 4: TERRA Level Correlates with Telomere Length, Telomerase Activity and Advanced Clinical Grade,” Translational Oncology, Vol. 5, No. 1, 2012, pp. 56-65.
- C. M. Azzalin, P. Reichenbach, L. Khoriauli, E. Giulotto and J. Lingner, “Telomeric Repeat Containing RNA and RNA Surveillance Factors at Mammalian Chromosome Ends,” Science, Vol. 318, No. 5851, 2007, pp. 798-801. doi:10.1126/science.1147182
- S. Schoeftner and M. A. Blasco, “Developmentally Regulated Transcription of Mammalian Telomeres by DNADependent RNA Polymerase II,” Nature Cell Biology, Vol. 10, No. 2, 2008, pp. 228-236. doi:10.1038/ncb1685
- [61] J. P. Issa, “CpG Island Methylator Phenotype in Cancer,” Nature Reviews Cancer, Vol. 4, No. 12, 2004, pp. 988- 993. doi:10.1038/nrc1507
- [62] M. Esteller, “Cancer Epigenomics: DNA Methylomes and Histone-Modification Maps,” Nature Reviews Genetics, Vol. 8, No. 4, 2007, pp. 286-298. doi:10.1038/nrg2005
- [63] J. P. Issa, N. Ahuja, M. Toyota, M. P. Bronner and T. A. Brentnall, “Accelerated Age-Related CpG Island Methylation in Ulcerative Colitis,” Cancer Research, Vol. 61, No. 9, 2001, pp. 3573-3577.
- [64] B. Luke, A. Panza, S. Redon, N. Iglesias, Z. Li and J. Lingner, “The Rat1p 5’to 3’ Exonuclease Degrades Telomeric Repeat-Containing RNA and Promotes Telomere Elongation in Saccharomyces cerevisiae,” Molecular Cell, Vol. 32, No. 4, 2008, pp. 465-477. doi:10.1016/j.molcel.2008.10.019
- [65] R. M. Cawthon, “Telomere Measurement by Quantitative PCR,” Nucleic Acids Research, Vol. 30, No. 10, 2002, Article ID: e47. doi:10.1093/nar/30.10.e47
- [66] N. O’Callaghan, V. Dhillon, P. Thomas and M. Fenech, “A Quantitative Real-Time PCR Method for Absolute Telomere Length,” BioTechniques, Vol. 44, No. 6, 2008, pp. 807-809. doi:10.2144/000112761
- [67] J. D. Henson and R. R. Reddel, “Assaying and Investigating Alternative Lengthening of Telomeres Activity in Human Cells and Cancers,” FEBS Letters, Vol. 584, No. 17, 2010, pp. 3800-3811. doi:10.1016/j.febslet.2010.06.009
- [68] T. L. Slatter, X. Tan, Y. C. Yuen, S. Gunningham, S. S. Ma, E. Daly, S. Packer, C. Devenish, J. A. Royds and N. A. Hung, “The Alternative Lengthening of Telomeres Pathway May Operate in Non-Neoplastic Human Cells,” The Journal of Pathology, Vol. 226, No. 3, 2012, pp. 509- 518. doi:10.1002/path.2981
- [69] C. L. Fasching, A. A. Neumann, A. Muntoni, T. R. Yeager and R. R. Reddel, “DNA Damage Induces Alternative Lengthening of Telomeres (ALT) associated Promyelocytic Leukemia Bodies That Preferentially Associate with Linear Telomeric DNA,” Cancer Research, Vol. 67, No. 15, 2007, pp. 7072-7077. doi:10.1158/0008-5472.CAN-07-1556
- [70] C. L. Fasching, K. Bower and R. R. Reddel, “Telomerase-Independent Telomere Length Maintenance in the Absence of Alternative Lengthening of Telomeres-Associated Promyelocytic Leukemia Bodies,” Cancer Research, Vol. 65, No. 7, 2005, pp. 2722-2729. doi:10.1158/0008-5472.CAN-04-2881
- [71] J. N. Jeyapalan, A. Mendez-Bermudez, N. Zaffaroni, Y. E. Dubrova and N. J. Royle, “Evidence for alternative Lengthening of Telomeres in Liposarcomas in the Absence of ALT-Associated PML Bodies,” International Journal of Cancer, Vol. 122, No. 11, 2008, pp. 2414- 2421. doi:10.1002/ijc.23412
- [72] J. D. Henson, Y. Cao, L. I. Huschtscha, A. C. Chang, A. Y. Au, H. A. Pickett and R. R. Reddel, “DNA C-Circles Are Specific and Quantifiable Markers of AlternativeLengthening-of-Telomeres Activity,” Nature Biotechnology, Vol. 27, No. 12, 2009, pp. 1181-1185. doi:10.1038/nbt.1587
- [73] S. Gonzalo, I. Jaco, M. F. Fraga, T. Chen, E. Li, M. Esteller and M. A. Blasco, “DNA Methyltransferases Control Telomere Length and Telomere Recombination in Mammalian Cells,” Nature Cell Biology, Vol. 8, No. 4, 2006, pp. 416-424. doi:10.1038/ncb1386
- [74] R. G. Verhaak, K. A. Hoadley, E. Purdom, V. Wang, Y. Qi, M. D. Wilkerson, C. R. Miller, L. Ding, T. Golub, J. P. Mesirov, G. Alexe, M. Lawrence, M. O’Kelly, P. Tamayo, B. A. Weir, S. Gabriel, W. Winckler, S. Gupta, L. Jakkula, H. S. Feiler, J. G. Hodgson, C. D. James, J. N. Sarkaria, C. Brennan, A. Kahn, P. T. Spellman, R. K. Wilson, T. P. Speed, J. W. Gray, M. Meyerson, G. Getz, C. M. Perou and D. N. Hayes, “Integrated Genomic Analysis Identifies Clinically Relevant Subtypes of Glioblastoma Characterized by Abnormalities in PDGFRA, IDH1, EGFR and NF1,” Cancer Cell, Vol. 17, No. 1, 2010, pp. 98-110. doi:10.1016/j.ccr.2009.12.020
- [75] D. W. Parsons, S. Jones, X. Zhang, J. C. Lin, R. J. Leary, P. Angenendt, P. Mankoo, H. Carter, I. M. Siu, G. L. Gallia, A. Olivi, R. McLendon, B. A. Rasheed, S. Keir, T. Nikolskaya, Y. Nikolsky, D. A. Busam, H. Tekleab, L. A. Diaz, Jr., J. Hartigan, D. R. Smith, R. L. Strausberg, S. K. Marie, S. M. Shinjo, H. Yan, G. J. Riggins, D. D. Bigner, R. Karchin, N. Papadopoulos, G. Parmigiani, B. Vogelstein, V. E. Velculescu and K. W. Kinzler, “An Integrated Genomic Analysis of Human Glioblastoma Multiforme,” Science, Vol. 321, No. 5897, 2008, pp. 1807-1812. doi:10.1126/science.1164382
- [76] H. Yan, D. W. Parsons, G. Jin, R. McLendon, B. A. Rasheed, W. Yuan, I. Kos, I. Batinic-Haberle, S. Jones, G. J. Riggins, H. Friedman, A. Friedman, D. Reardon, J. Herndon, K. W. Kinzler, V. E. Velculescu, B. Vogelstein and D. D. Bigner, “IDH1 and IDH2 Mutations in Gliomas,” The New England Journal of Medicine, Vol. 360, 2009, pp. 765-773. doi:10.1056/NEJMoa0808710
- [77] J. Balss, J. Meyer, W. Mueller, A. Korshunov, C. Hartmann and A. von Deimling, “Analysis of the IDH1 Codon 132 Mutation in Brain Tumors,” Acta Neuropathologica, Vol. 116, No. 6, 2008, pp. 597-602. doi:10.1007/s00401-008-0455-2
- [78] K. L. McDonald, J. McDonnell, A. Muntoni, J. D. Henson, M. E. Hegi, A. von Deimling, H. R. Wheeler, R. J. Cook, M. T. Biggs, N. S. Little, B. G. Robinson, R. R. Reddel and J. A. Royds, “Presence of Alternative Lengthening of Telomeres Mechanism in Patients with Glioblastoma Identifies a Less Aggressive Tumor Type with Longer Survival,” Journal of Neuropathology & Experimental Neurology, Vol. 69, No. 7, 2010, pp. 729-736. doi:10.1097/NEN.0b013e3181e576cf
- [79] D. Capper, H. Zentgraf, J. Balss, C. Hartmann and A. von Deimling, “Monoclonal Antibody Specific for IDH1 R132H Mutation,” Acta Neuropathologica, Vol. 118, No. 5, 2009, pp. 599-601. doi:10.1007/s00401-009-0595-z
- [80] C. M. Heaphy, R. F. de Wilde, Y. Jiao, A. P. Klein, B. H. Edil, C. Shi, C. Bettegowda, F. J. Rodriguez, C. G. Eberhart, S. Hebbar, G. J. Offerhaus, R. McLendon, B. A. Rasheed, Y. He, H. Yan, D. D. Bigner, S. M. Oba-Shinjo, S. K. Marie, G. J. Riggins, K. W. Kinzler, B. Vogelstein, R. H. Hruban, A. Maitra, N. Papadopoulos and A. K. Meeker, “Altered Telomeres in Tumors with ATRX and DAXX Mutations,” Science, Vol. 333, No. 6041, 2011, p. 425. doi:10.1126/science.1207313
- [81] J. Schwartzentruber, A. Korshunov, X. Y. Liu, D. T. Jones, E. Pfaff, K. Jacob, D. Sturm, A. M. Fontebasso, D. A. Quang, M. Tonjes, V. Hovestadt, S. Albrecht, M. Kool, A. Nantel, C. Konermann, A. Lindroth, N. Jager, T. Rausch, M. Ryzhova, J. O. Korbel, T. Hielscher, P. Hauser, M. Garami, A. Klekner, L. Bognar, M. Ebinger, M. U. Schuhmann, W. Scheurlen, A. Pekrun, M. C. Fruhwald, W. Roggendorf, C. Kramm, M. Durken, J. Atkinson, P. Lepage, A. Montpetit, M. Zakrzewska, K. Zakrzewski, P. P. Liberski, Z. Dong, P. Siegel, A. E. Kulozik, M. Zapatka, A. Guha, D. Malkin, J. Felsberg, G. Reifenberger, A. von Deimling, K. Ichimura, V. P. Collins, H. Witt, T. Milde, O. Witt, C. Zhang, P. Castelo-Branco, P. Lichter, D. Faury, U. Tabori, C. Plass, J. Majewski, S. M. Pfister and N. Jabado, “Driver Mutations in Histone H3.3 and Chromatin Remodelling Genes in Paediatric Glioblastoma,” Nature, Vol. 482, No. 7384, 2012, pp. 226-231. doi:10.1038/nature10833
- [82] G. Wu, A. Broniscer, T. A. McEachron, C. Lu, B. S. Paugh, J. Becksfort, C. Qu, L. Ding, R. Huether, M. Parker, J. Zhang, A. Gajjar, M. A. Dyer, C. G. Mullighan, R. J. Gilbertson, E. R. Mardis, R. K. Wilson, J. R. Downing, D. W. Ellison and S. J. Baker, “Somatic Histone H3 Alterations in Pediatric Diffuse Intrinsic Pontine Gliomas and Non-Brainstem Glioblastomas,” Nature Genetics, Vol. 44, No. 3, 2012, pp. 251-253. doi:10.1038/ng.1102
- [83] C. J. Cairney, S. F. Hoare, M. G. Daidone, N. Zaffaroni and W. N. Keith, “High Level of Telomerase RNA Gene Expression Is Associated with Chromatin Modification, the ALT Phenotype and Poor Prognosis in Liposarcoma,” British Journal of Cancer, Vol. 98, No. 8, 2008, pp. 1467- 1474. doi:10.1038/sj.bjc.6604328
- [84] Y. J. Chen, V. Hakin-Smith, M. Teo, G. E. Xinarianos, D. A. Jellinek, T. Carroll, D. McDowell, M. R. MacFarlane, R. Boet, B. C. Baguley, A. W. Braithwaite, R. R. Reddel and J. A. Royds, “Association of Mutant TP53 with Alternative Lengthening of Telomeres and Favorable Prognosis in Glioma,” Cancer Research, Vol. 66, No. 13, 2006, pp. 6473-6476. doi:10.1158/0008-5472.CAN-06-0910
- [85] C. Lu, P. S. Ward, G. S. Kapoor, D. Rohle, S. Turcan, O. Abdel-Wahab, C. R. Edwards, R. Khanin, M. E. Figueroa, A. Melnick, K. E. Wellen, D. M. O’Rourke, S. L. Berger, T. A. Chan, R. L. Levine, I. K. Mellinghoff and C. B. Thompson, “IDH Mutation Impairs Histone Demethylation and Results in a Block to Cell Differentiation,” Nature, Vol. 483, No. 7390, 2012, pp. 474-478. doi:10.1038/nature10860
- [86] D. C. Silvestre, J. R. Pineda, F. Hoffschir, J. M. Studler, M. A. Mouthon, F. Pflumio, M. P. Junier, H. Chneiweiss and F. D. Boussin, “Alternative Lengthening of Telomeres in Human Glioma Stem Cells,” Stem Cells, Vol. 29, No. 3, 2011, pp. 440-451. doi:10.1002/stem.600
- [87] M. F. Amary, K. Bacsi, F. Maggiani, S. Damato, D. Halai, F. Berisha, R. Pollock, P. O’Donnell, A. Grigoriadis, T. Diss, M. Eskandarpour, N. Presneau, P. C. Hogendoorn, A. Futreal, R. Tirabosco and A. M. Flanagan, “IDH1 and IDH2 Mutations Are Frequent Events in Central Chondrosarcoma and Central and Periosteal Chondromas But Not in Other Mesenchymal Tumors,” The Journal of Pathology, Vol. 224, No. 3, 2011, pp. 334-343. doi:10.1002/path.2913
- [88] R. L. Giuntoli 2nd, D. S. Metzinger, C. S. DiMarco, S. S. Cha, J. A. Sloan, G. L. Keeney and B. S. Gostout, “Retrospective Review of 208 Patients with Leiomyosarcoma of the Uterus: Prognostic Indicators, Surgical Management, and Adjuvant Therapy,” Gynecologic Oncology, Vol. 89, No. 3, 2003, pp. 460-469. doi:10.1016/S0090-8258(03)00137-9
- [89] R. Tomoda, M. Seto, H. Tsumuki, K. Iida, T. Yamazaki, J. Sonoda, A. Matsumine and A. Uchida, “Telomerase Activity and Human Telomerase Reverse Transcriptase mRNA Expression Are Correlated with Clinical Aggressiveness in Soft Tissue Tumors,” Cancer, Vol. 95, No. 5, 2002, pp. 1127-1133. doi:10.1002/cncr.10793
- [90] K. Aogi, A. Woodman, V. Urquidi, D. C. Mangham, D. Tarin and S. Goodison, “Telomerase Activity in SoftTissue and Bone Sarcomas,” Clinical Cancer Research: An Official Journal of the American Association for Cancer Research, Vol. 6, No. 12, 2000, pp. 4776-4781.
NOTES
*Corresponding author.

