Open Journal of Ecology
Vol.4 No.8(2014), Article ID:46467,12 pages DOI:10.4236/oje.2014.48039
Antarctic Cyanobacteria Biodiversity Based on ITS and TrnL Sequencing and Its Ecological Implication
Carla Micheli1*, Rossella Cianchi2, Raffaella Paperi3, Alessandro Belmonte1, Benjamin Pushparaj3
1ENEA Italian National Agency for New Technologies, Energy and Sustainable Economic Development, Research Center Casaccia, Rome, Italy
2Department of Genetics and Molecular Biology, University of Rome La Sapienza, Rome, Italy
3Institute of Ecosystem Studies CNR-ISE, Florence, Italy
Email: *carla.micheli@enea.it
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/


Received 20 March 2014; revised 20 April 2014; accepted 27 April 2014
ABSTRACT
Antarctic cyanobacteria biodiversity was investigated by simultaneous sequencing of the nuclear ribosomal internal transcribed spacer (ITS flanked by partial 16S and 23S), and Chloroplast tRNALeu UAA intron (TrnL), exploring whether such morphotypes constitute distinct species and explaining their current distribution. We identified Nostocales, Chrococcales and Oscillatoriales species, collected in different habitats (soil, algal mats, lake sediments, ice-water) after their growth in cultures. By comparative sequence analyses available in Genbank, our results proved to be mostly in agreement with both TrnL and ITS, in the identification of the strains, particularly for Nostocales. Although ITS demonstrated more usefully than TrnL did in identifying Oscillatoriales and Chroococcales, due to the frequent lack of the intron in these groups, our results lead us to support an independent phylogenetic dataset of ITS and TrnL (the latter based on conserved regions) producing not only concordant clusters but also a secondary structure. Specific assignments of the secondary structure evidenced by different cyanobacteria groups, especially the D1-D1’ region of ITS and the P6b region of TrnL. For the latter region, the sequences analyzed for Nostoc species could be divided into the two classes previously identified, on the basis of different heptanucleotide repeats in P6b which were not found in Oscillatoriales, which instead showed marked sequence length variation in region P5. A correlation between the two P6b intron classes and their ecological role is suggested for free-living Antarctic Nostoc, as it was previously found in symbiontic cyanobacteria from other regions.
Keywords:Antarctic Cyanobacteria, Biodiversity, ITS, tRNALeu UAA, Secondary Structure

1. Introduction
Biodiversity and evolutionary origins of Antarctic microbial ecosystems have recently stimulated interest since providing useful models of evolutionary ecology [1] [2] . In fact the Antarctic ice sheet provides unique, natural culture for study of microorganisms which have been isolated from the global gene pool over timescales of evolutionary significance. In this context cyanobacteria prove good bio-indicators since they adapt their growth to the dynamic interface between ice and water. Polar marine ecosystems are particularly sensitive to climatic change and small temperature differences can have large effects on the extent and thickness of sea ice [3] . Therefore Antarctic cyanobacteria diversity could be used as a possible biological indicator of global climate change. Cyanobacteria belong to an ancient group of oxygenic, photosynthetic organisms with a cosmopolitan distribution in many extreme environments [4] . They are characterized by physiological versatility and ample ecological tolerance, which allow them to compete successfully with other organisms in aquatic and terrestrial environments [5] . They are considered valid bio-indicators for their capacity to adapt to various stress environments, playing important roles in carbon and nitrogen cycles and modifying morphology, metabolism and lightharvesting systems.
Phylogenetic relationships among cyanobacteria are still not fully resolved [6] . At present three taxonomic schemes are used for cyanobacteria [7] -[11] which give varying emphasis to morphological, biochemical and genetic aspects of both wild and cultivated strains. Among these molecular markers are potentially most promising, especially in the identification of field samples without culturing [12] . So far, 16S rDNA has been widely used to infer phylogenetic relationships between species and strains as a single marker or combined with traditional techniques [13] [14] .
An increasing number of molecular markers have been proposed to provide a robust phylogeny such as the more variable ITS, the transcribed spacer between 16S and 23S [15] whilst another candidate is the chloroplast tRNALeu (UAA) intron (TrnL) by which one region is shown to be particularly useful for detecting species level variation [16] -[18] . The TrnL gene, including the intron, has been used as a phylogenetic marker for cyanobacteria and appears to be in agreement with phylogeny estimated using rRNA (e.g. 16S, [19] ). Moreover tRNALeu intron contains regions which have been highly conserved over a billion years of chloroplast evolution [20] [21] .
However, molecular markers are potentially useful in cyanobacteria, not only for genus and species identification, but also to better characterize different ecotypes. In fact recent studies revealed that cyanobacteria adapt physiologically to different environments, producing secondary active biological molecules such as fatty acids which are potentially useful for human health [22] -[25] . Some authors suggest correlations between TrnL variation, and the physiological role of various organisms such as nitrogen-fix, photosynthetic organisms and symbionts [26] -[30] .
The objective of this research was to analyze the genetic diversity of 13 strains of clonal filamentous cyanobacteria, coming from different locations of the Antarctic region of south polar lake (sediments, soil, algal mats and water), exploring whether such morphotypes constitute distinct species, comparing them to Mediterranean strains (fresh water and sea) and also to those available in Genbank. To this aim two regions were sequenced: internal transcribed spacers ITS (flanked by partial 16S and 23S rRNA), and part of the TrnL gene (including the intron). In a previous study, Pushparaj et al. [24] found that the fatty acid composition varied between these Antarctic strains, with relatively high quantities of polyunsaturated fatty acids, as well as the total lipid content (from 13% to 9%) and the C/N ratio (3.7 to 11.2). In the present study, the further aim was to identify the genetic features of these individual strains; therefore the utility of the two markers for phylogenetic inference and ecotype characterization, including secondary structure of TrnL, will be discussed.
Our research is part of a multidisciplinary study of PNRA (Programma Nazionale Ricerche in Antartide) that included morphological identification, RAPD genetic diversity [31] and fatty acid composition [24] of Antarctic cyanobacteria.
2. Materials and Methods
2.1. Sampling and DNA Extraction
The cyanobacteria specimens were collected in the Antarctic region around Italian station, Mario Zucchelli, (ex-BTN, Baia Terranova), during various Italian expeditions in the years 2003 to 2005 (Austral summers) organized by the Programma Nazionale di Ricerche in Antartide (PNRA). Collecting data and geographical coordinates are given in Table 1. The strains were grown in laboratory conditions [24] and their morphological and biomass analyses (g/mL) were performed before RAPD genetic studies [31] .
DNA extraction was carried out, using the Magna Rack method (Kit of Invitrogen SpA, Italy) with some modifications involving reaction times. Briefly, fresh cells were harvested during exponential phase by centrifugation (10,000 × g for 10 min) then they were washed in buffer (10 mM Tris-HCl, 1 mM EDTA pH 8.5) and subsequently they were incubated for ten minutes at 37˚C in 500 μL Lysis Buffer containing 10 μL proteinase K and 5 μL lysozyme (a fresh solution of 50 μg/mL in water Sigma Aldrich L7651). Subsequently, the cells were incubated for 1 hour at 80˚C, before to start the followed re-suspending steps of the Charge Switch Magnetic Beads method (Invitrogen SpA, Italy). The supernatant containing the genomic DNA, was washed twice and eluited in a 200 μL of Tris-HCl buffer (pH 8.5). By spectrophotometric analyses (Perkin Elmer) the DNA content was determined (qualitatively and quantitatively).
Table 1. Cyanobacteria strains, geographical coordinate of sampling sites and substrate.
*(Codes in this paper, above, and Pushparaj et al., 2008, below); **Strains coming from Italian lakes and Mediterranean sea for comparison purpose; ***Strain used as outgroup.
2.2. TrnL and 16S-ITS-23S Amplification and Data Analyses
The amplification products were obtained by a thermal cycler (Perkin Elmer/Cetus 2400) and visualized after 1.4% agarose gel-electrophoresis (BIORAD, power pack 300, Italy).
Then sequencing was carried out by ABI automated dye-terminator system (Macrogen).
2.2.1. TrnL Amplification and Sequencing
For TrnL amplification, we used 5’-GGGGRTRTGGYGRAAT-3’ as forward primer, and 5’-GGGGRYRGRGGGACTT-3’ as reverse primer [12] . Amplification L volume containing 50 ng DNA in a mixer of reaction was performed in 50 Applied Biosystem reagents (1.25 U Taq polymerase, 0.2 mM dNTP, 2.5 mM MgCl2 and 1× buffer HCL). The cycling parameters for PCR were: initial denaturation at 95˚C for 5 min, then cycles repeated 35 times of 94˚C for 45', 53˚C for 45' annealing temperature, 72˚C for 1 min; finally one cycle at 72˚C for 10 min. The sequences were obtained with the primers: 5’-GGTAGACGCWRCGGACTT-3’ and 5’-TWTACARTCRACGGATTTT-3’.
2.2.2. 16S-ITS-23S Amplification and Sequencing
For both the amplification and sequencing of 16S-ITS-23S region, we used Wilmotte’s universal 322 and 340 primers [15] , respectively: 5’-TGTACACACCGCCCGTC-3’ and 5’-CTCTGTGTGCCTAGGTATCC-3’. L mixer (Applied Biosystem reagents) The reaction was obtained in a 50 containing 50 ng DNA (1.25 U Taq polymerase, 0.2 mM dNTP, 2.5 mM MgCl2 and 1× buffer HCL, 0.2 mM primers). PCR conditions were: initial denaturation at 95˚C for 5 min, then 35 cycles repeated at 94˚C for 45", 50˚C annealing temperature for 45", 72˚C for 1.20 min; finally one cycle at 72˚C for 10 min.
The obtained sequences were aligned using the program SEAVIEW version 2.4 [32] , which implements MUSCLE (multiple sequence comparison by log expectation) [33] , and by hand, also taking into account the secondary structures. Comparisons with sequences available in Genbank database were performed, using MEGABLAST version 3.1 available from http://www.ncbi.nlm.nih.gov/BLAST/. Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 4 [34] . Phylogenetic trees for the two regions were obtained using Maximum parsimony method, Neighbour Joining and Maximum Likelihood, with Bootstrap analysis. Predictions regarding secondary structures for the TrnL intron were made using the program RNA Structure version 4.6 [35] .
3. Results
3.1. Molecular Identification
Overall, the ITS types observed differed from those identified by [36] in a study of Antarctic cyanobacteria, thereby confirming the variability of this region. Figure 1 (left) and Figure 1 (right) show the best hits found in Gene-Bank database for the sequences obtained for ITS and TrnL respectively for the samples studied.
For Nostocaceae, identifications based on molecular markers were generally in close agreement with morphological ones: N0, N1, NM, corresponded to Nostoc sp. with both ITS and TrnL, and N5, N6 and N8 corresponded to Nostoc commune with both markers; N43 and N117 were successful sequenced only by TrnL and were related to N. sphaericum and N. commune, respectively.
In the case of Oscillatoriales, L, L5, LY and LM were identified by ITS as Leptolyngbya, and L4 as Phormidium, in agreement with morphological analysis.
The sequences obtained for TrnL region ranged from 272 bp (NM) to 389 bp (L, LY). In particular, in the TrnL region, a band of low molecular weight was consistently amplified, and a very short region sequenced with the forward primer (especially for L4, L5, G3, P2). This suggests a lack of the intron, as frequently observed in cyanobacteria [19] . Details of secondary structure are given below.
For the 16S-ITS-23S region, the obtained sequences ranged from 338 bp (N0) to 495 bp (LM). Amplification and sequencing of 3 strains (N43, N117, L11) were successful identified by only TrnL (Figure 1, right).
A single RNA operon, including both tRNA-Ile and tRNA-Ala, was found for L, LY, L4, L5, LM, P, P2, and G3, whereas a short one, not including these tRNAs, was amplified for N0, N1, N5, N6 and N8. Instead, two different operons were found for the Mediterranean strain NM: a short one, without the two tRNAs, and a longer one which included at least part of tRNA-Ile (Figure 1, right). TrnL provided less clear-cut identification for L, LY and LM, mainly corresponding to Oscillatoriales (Figure 1, right).
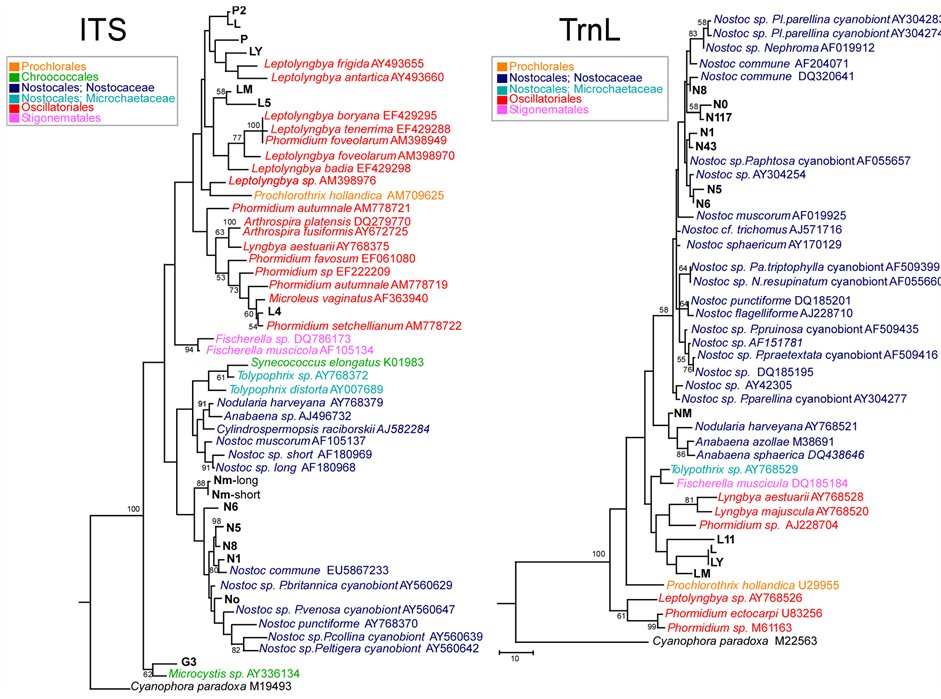
Figure 1. Left—ITS. Maximum Parsimony tree of ITS region for the samples studied, compared to taxa in Genbank. One of 12 most parsimonious trees is shown, with bootstrap values above 50% over 500 replicates indicated (final dataset of 576 bp, of which 244 parsimony informative). Tree drawn to scale, with branch lengths calculated using the average pathway method, as implemented in MEGA 4. Right—TrnL. Maximum Parsimony tree of TrnL region for the samples studied, compared to taxa in Genbank. Bootstrap consensus tree is shown, with values above 50% over 500 replicates (final dataset, of which 77 parsimony informative). Tree drawn to scale, with branch lengths calculated using the average pathway method, as implemented in MEGA 4. culated using the average pathway method, as implemented in MEGA 4. P. = Peltigera; N. = Nephroma; Pa. = Palmeliella; Pl. = Placopsis.
In the case of P, P2, the molecular identifications contrasted with those based on morphology that identified them as Pseudophormidium sp., whilst by ITS they proved related to Leptolyngbya. No TrnL intron was apparently recovered for these two samples according to the other Antarctic Oscillatoriales. Sample G3, identified morphologically as Gloeocapsa (Chroococcales), proved related to Microcystis (Chroococcales) by ITS and it was used as outgroup.
3.2. Phylogenetic Relationships
Evolutionary distances among the strains were estimated by Kimura-2-parameter.
In one case a same TrnL haplotype was shared by Antarctic samples geographically far apart (Leptolyngbya sp: L-Ly). A close relationship was detected at ITS between L and P2, also from different locations. The range of distance values (by pairwise deletion method) for the cyanobacteria studied at ITS were: 0.031 (N0-N8) - 0.100 (N5-NM) within Nostocales; 0 (L-P2) - 0.129 (L4-L, LY) within Oscillatoriales, and 0.102 (N6-LM) - 0.22602 (N1-L5) between the two groups.
The outgroup G3 (Chroococcales) showed values from 0.135 (with L) to 0.178 (with N1).
At TrnL the ranges of K-2 values were: 0.015 (N0-N117) - 0.112 (NM-N117) within Nostocales, 0 (L-LY) - 0.098 (L, LY-L11) within Oscillatoriales and 0.072 (L, LY-NM) - 0.152 (N5- LM) between the two groups.
Evolutionary relationships among the Antarctic and Mediterranean cyanobacterial strains studied are summarized in Figure 1 (left) and Figure 1 (right), which show trees for the ITS and TrnL regions respectively, obtained by the maximum parsimony method.
A number of cyanobacteria sequences available in Genbank were included for comparison: photosynthetic cyanelle of Cyanophora paradoxa, available for both regions, was used as outgroup.
At ITS (Figure 1, left), Chroococcales (G3 and Microcystis sp.) form a basal well supported clade; in which two main clusters include respectively: 1) Nostocales, with Nostoc spp. (including all our Nostoc sp. samples) grouping together in a distinct subcluster; 2) Oscillatoriales, together with Stigonematales, and Prochlorales (Prochlorothrix hollandica). Within the latter cluster, three subclusters include: 1) Fischerella sp. (Stigonematales); 2) mainly Leptolyngbya spp., with L, LY, P, P2 related to other Antarctic Leptolyngbya strains, LM and L5 more related to Leptolyngbya faveolarum and L. badia; this subcluster also includes Prochlorothrix hollandica (Prochlorales); c) mainly Phormidium spp. and Microcoleus vaginatus, grouping with L4; this subcluster also includes Arthrospira spp. and Lyngbya estuarii.
TrnL phylogenetic analysis was carried out, excluding from analysis the most variable regions (loop sequences in P6b, P9, P5), as suggested by [6] and [19] (cf. Figure 2(b)). As shown in Figure 1 (right) (at TrnL) Nostocales form a distinct clade, whereas the relationships among Oscillatoriales, Stigonematales and Prochlorales, are less resolved, possibly due to the few sequences available: only one GenBank sequence of Leptolyngbya (well separated from the others with ITS) could be included.
 (a)
(a)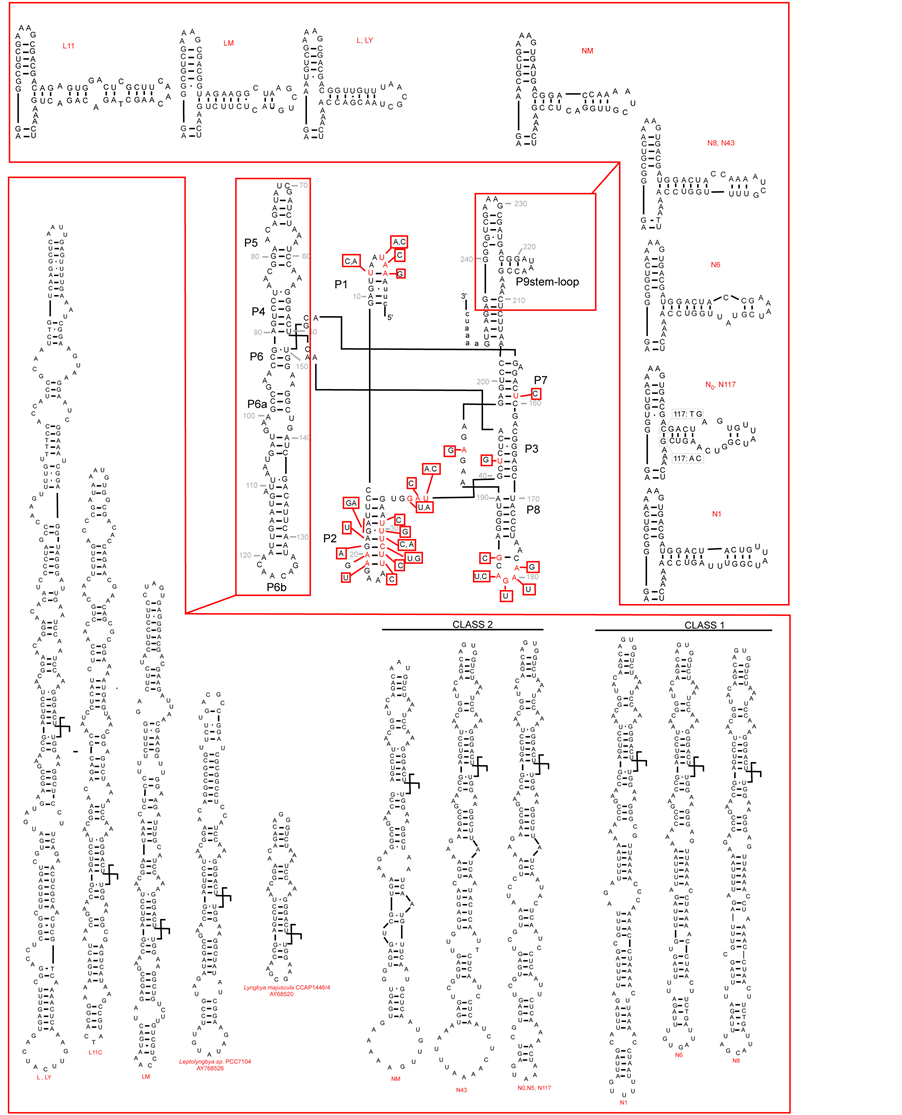 (b)
(b)
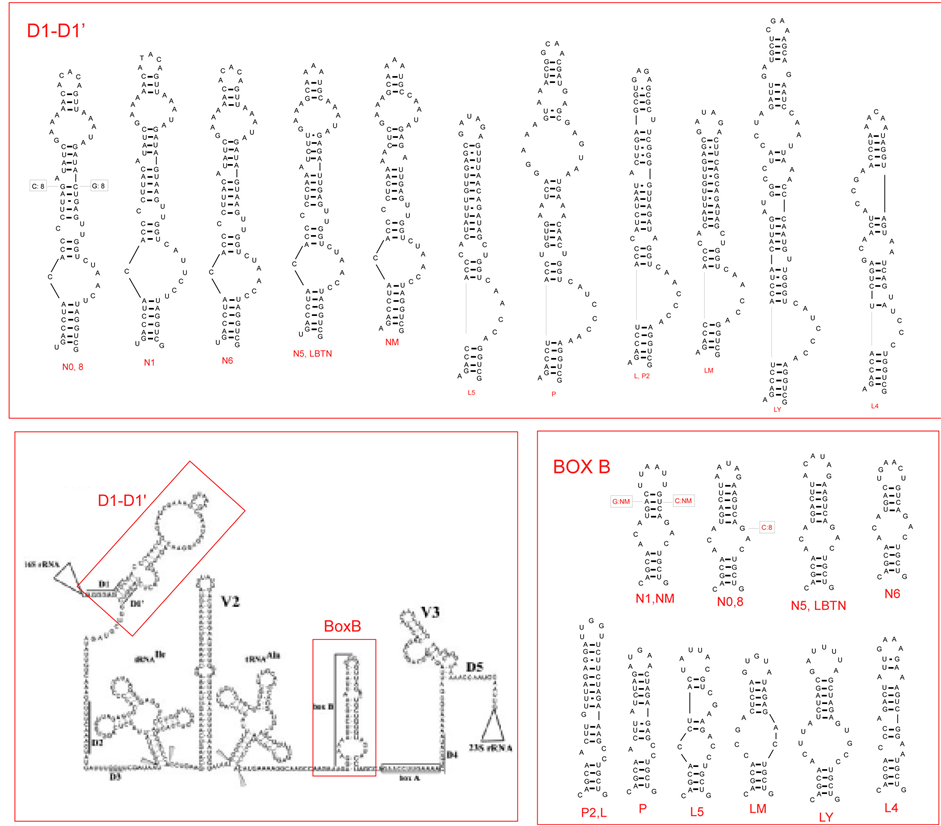
Figure 2. (a) Putative secondary structure of the 16S rRNA-23S rRNA ITS region in Anabaena PCC 7120 (central box, from Iteman et al., 2000) and details of the regions D1-D1’ and BoxB in the studied cyanobacteria (inserts). (b) Putative secondary structure of the TrnL region in the studied cyanobacteria. In the central part, variable sites are shown in red, compared to the model structure Anabaena sp.PCC7120 (modified from Paulsrud and Lindblad, 1998). Details of variable parts P6b and P9 stemloops, are shown for each specimen in the inserts. Secondary structures of Structures of Leptolyngbya sp. PCC7104 (AY768526) and Lyngbya majuscula CCAP1446/4 (AY68520) have been added for comparison. Class 1 and class 2 introns follow Paulsrud and Lindblad, 1998.
For TrnL, all Nostoc sp. samples form a cluster with Nostoc commune, whilst the Mediterranean strain NM clusters with Anabaena spp. and Tnodularia harveyana. The strains L, LY (with the same haplotype), L11 and the Mediterranean LM clustering together within Oscillatoriales.
3.3. Secondary Structure
3.3.1. ITS Region
Two semi-conserved regions of the internal transcribed spaces of the rRNA operon were analyzed: 1) the stem included between the conserved domains D1 (immediately following the 16S RNA) and D1’; and 2) the Box B stem, preceding the conserved anti-terminator Box A sequences, involved in preventing a premature termination of transcription (cf. [15] -[37] ). The putative secondary structures of the samples studied are shown in Figure 2(a), together with that of the 16S-23S rRNA region for Nostoc PCC 7120 (data from [15] ). The D1-D1’ region shows marked differences between Nostocales and Oscillatoriales, with characteristic one-sided loops in the basal part of the stem and allows a ready distinction of the two groups. Box B appears to be more variable among both Nostocales and Oscillatoriales, but also for this region the two groups can be distinguished. The patterns observed are consistent with previous findings in Nostocales [38] and Leptolyngbia [39] .
3.3.2. TrnL Intron Region
The predicted secondary structures of the TrnL intron for the Antarctic and Mediterranean cyanobacteria studied, are given in Figure 2(b), and compared to the model structure proposed by [40] for Nostoc (Anabaena) PCC 7120. Our data confirm that the TrnL intron core structure is highly conserved, with variability restricted to some regions, especially P6b, P9, P5 loops, characterized by striking sequence length variation [19] . In the Nostoc sequences analyzed we could recognize the two classes, identified by [26] , characterized by different heptanucleotide repeats in P6b: class 1, shared by N1, N6 and N8, and class 2, shared by N0, N5, N43, N177 and NM. This was not shown by Oscillatoriales, which instead showed marked sequence length variation in region P5, from a very short sequence e.g. in Lyngbya majuscula CCAP1446/4 (Gene-bank accession AY68520T), to a very long one in L1 and LY (so far not observed elsewhere). Such differences in the regions P6b and P5 might be useful in distinguishing between Nostocaceae and Oscillatoriales, if confirmed in a higher number of strains.
4. Discussion
Comparative sequence analyses of Antarctic cyanobacteria proved very useful for identifying species of Nostocales, Chrococcales and Oscillatoriales, and for analyzing their genetic diversity, especially when these strains were sequenced at ITS and TrnL regions. Molecular identifications were generally in agreement with both TrnL and ITS, particularly for Nostocales. ITS proved more useful than TrnL for the assignments of Oscillatoriales and Chroococcales, due to the frequent lack of the intron in these groups (e.g. [19] ). On the other hand, by microscope similar morphologies we found for genetically different strains, e.g. P and P2 that at first showed the highest similarity to Pseudophormidium sp. in Oscillatoriales, but subsequently they were recognized as Leptolyngbia by ITS.
Our data confirm the need for multiple taxonomic criteria, including molecular markers, for a successful classification of cyanobacteria. In particular our findings support the validity of the TrnL intron as a phylogenetic marker for cyanobacteria and especially for Nostocales. Independent analyses of ITS and TrnL datasets (the latter based on conserved regions) produced concordant clusters, especially when the most variable regions of the intron (parts of the helix P6, P9, P5) were excluded, as suggested by [19] and [6] . For both markers, the Antarctic strains studied in the present paper proved related, not only to Antarctic species, but also to those from other parts of the world, including the Mediterranean region [36] -[41] . The analysis of TrnL secondary structure in the cyanobacteria studied confirmed the presence of two classes in Nostoc sp., characterized by different heptanucleotide repeats in the region P6b, shared by Antarctic and Mediterranean species. Interestingly, class 2 was observed in strains from three Antarctic samples isolated from soil, moss, and a Mediterranean strain from seawater; whereas class 1 was detected in Antarctic strains isolated from algal mats, moss, and fresh water. These findings support the correlation found by [26] between the two intron classes and physiological roles in symbiotic cyanobacteria, particularly nitrogen fixation (class 2), or both photosynthesis and nitrogen fixation (class 1). Previous analysis also suggests differences in haplotypes and other characteristics, such as C/N ratios and fatty acid composition; these were found to vary among Antarctic strains, including those studied in the present paper [24] . Interestingly, strains L and Ly, which shared the same TrnL haplotype, both showed high values of palmitoleic oil (16:01) and only traces of 17.04 (IUPAC), in contrast to P and P2 (possibly intronless), which showed values more similar to Nostocales. A correlation between fatty acid composition and RAPD genetic diversity at genera level was detected by [31] .
Moreover in further ad hoc studies we found a correlation between cyanobacteria haplotypes and their ecophysiological characteristics on the basis of sequences available in GenBank, as shown for Nostoc species in the Figure 3.
We can conclude that the combined use of ITS and TrnL markers has proved useful to identify Antarctic cyanobacterial samples. The secondary structure of the D1-D1’ region appears particularly promising for a rapid routine distinction between Nostocales and Oscillatoriales, whereas the secondary structure of the P6b region can provide insight into the functional role in Nostocales. Such tools can contribute to the build-up of a database
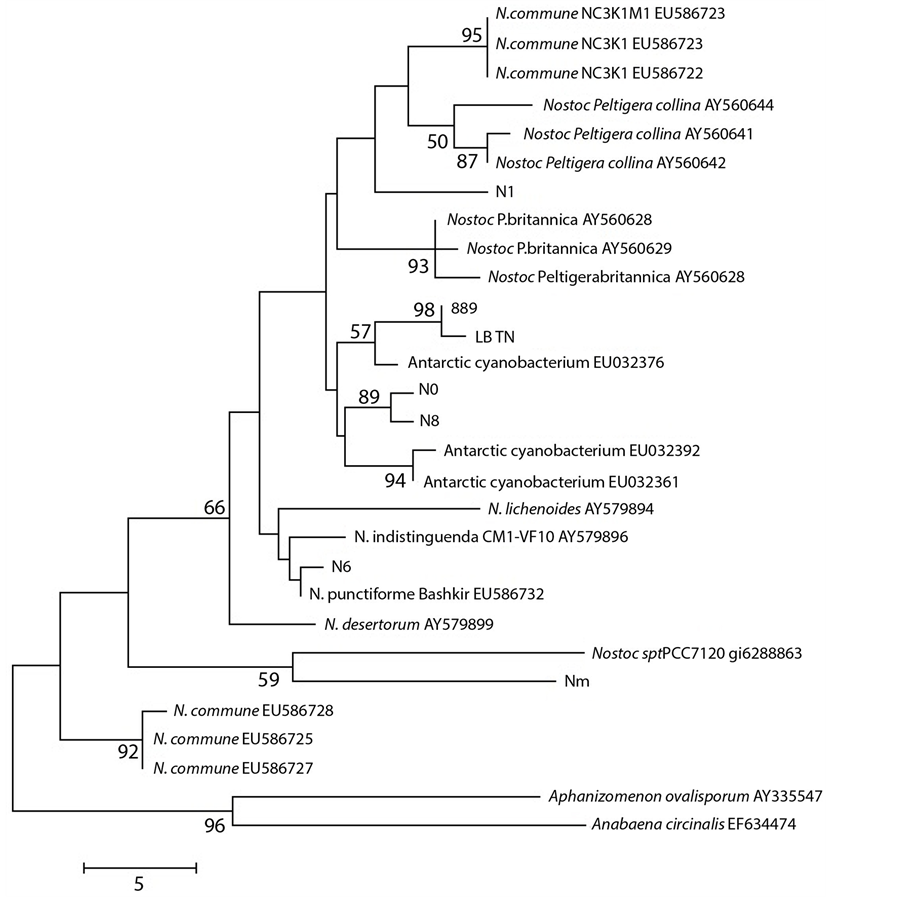
Figure 3. Maximum Parsimony tree of ITS region for the samples studied, compared to taxa in GenBank. Consensus tree (from 28 most parsimonious trees) is shown, with bootstrap values above 50% over 100 replicates. Tree drawn to scale (branch lengths calculated using the average pathway method, as implemented in MEGA 4; complete deletion; 248 bp of which 48 were parsimony informative).
of genetic diversity of cyanobacteria in the still relatively undisturbed Antarctic ecosystem. This will make it possible to monitor and track changes in genetic diversity, and relate them to local, regional and global climate changes habitat disturbance. With increasing concern regarding global climate change, evaluation of biodiversity (genetic, species, ecosystem), is necessary in order to track changes within the climate system: it affects interpretations of future global, and especially polar climate change. In this context cyanobacteria diversity monitoring could be important as a possible biological indicator, in particular regarding diversity and connectivity between diverse habitats across a range of geographic scales.
Acknowledgements
This research was funded by Programma Nazionale di Ricerche in Antartide (PNRA). Project LUP-PUS 1.4/03: Monitoring and screening of bioactive compounds and toxins produced by Antarctic cyanobacteria strains. The authors thanks Dr. Claudio Sili, CNR-ISE of Firenze, for examining the cultures microscopically and providing his opinion on their taxonomy.
References
- Warwick, F.V. (2000) Evolutionary Origins of Antarctic Microbiota: Invasion, Selection and Endemism. Antarctic Science, 12, 374-385. http://dx.doi.org/10.1016/j.cosust.2009.06.001
- Trenberth, K.E. (2009) An Imperative for Climate Change Planning: Tracking Earth’s Global Energy. Current Opinion in Environmental Sustainability, 1, 19-27.
- Smetacek, V. and Nicol, S. (2005) Polar Ocean Ecosystems in a Changing World. Nature, 437, 362-368. http://dx.doi.org/10.1038/nature04161
- Graham, L.E. and Wilcox, L.W. (2000) Algae. Prentice Hall, Upper Saddle River, 640.
- Honda, D., Yokota, A. and Sugiyava, J. (1998) Detection of Seven Major Evolutionary Lineages in Cyanobacteria Based on the 16S tRNA Gene Sequence Analysis with New Sequences of Five Marine Synecococcus Strains. Journal of Molecular Evolution, 48, 723-739. http://dx.doi.org/10.1007/PL00006517
- Oksanen, I., Lohtander, K., Sivonen, K. and Rikkinen, J. (2004) Repeat-Type Distribution in TrnL Intron Does Not Correspond with Species Phylogeny; Comparison of the Genetic Markers 16S rRNA and TrnL Intron in Heterocystous Cyanobacteria. International Journal of Systematic and Evolutionary Microbiology, 54, 765-772. http://dx.doi.org/10.1099/ijs.0.02928-0
- Komárek, J. and Anagnostidis, K. (2005) Cyanoprokaryota 2.Teil: Oscillatoriales. Elsevier GmbH, Munchen, 759 p.
- Komarek, J. and Anagnostic, K. (1989) Modern Approach to the Classification System of Cyanophites 4-Nostocales. Algological Studies/Archiv für Hydrobiologie, 56, 247-345.
- Hoffmann, L., Komárek, J. and Kaštovský, J. (2005) System of Cyanoprokaryotes (Cyanobacteria)—State 2004. Algological Studies, 117, 95-115. http://dx.doi.org/10.1127/1864-1318/2005/0117-0095
- Anagnostidis, K. and Komárek, J. (1985) Modern Approach to the Classification System of Cyanophytes 1—Introduction. Archiv für Hydrobiologie, Supplement, 71, 291-302.
- Rippka, R, Deruelles, J., Waterbury, J.B., Herdmann, M. and Stanier, R.Y. (1979) Generic Assignments, Strain Histories and Properties of Pure Coltures of Cyanobacteria. Journal of General Microbiology, 3, 1-61. http://dx.doi.org/10.1099/00221287-111-1-1
- Wirtz, N., Lumbsch, H.T., Gree, T.G.A., Turk, R., Pindado, A., Sancho, L. and Schroeter, B. (2003) Lichen Fungi Have Low Cyanobiont Selectivity in Maritime Antarctica. New Phytologist, 160, 177-183. http://dx.doi.org/10.1046/j.1469-8137.2003.00859.x
- Wilmotte, A. (1994) Molecular Evolution and Taxonomy of Cyanobacteria. In: Bryant, D.A., Ed., The Molecular Biology of Cyanobacteria, Kluwer, Dordrecht, 1-25. http://dx.doi.org/10.1007/978-94-011-0227-8_1
- Caroppo, C., Albertano, P., Bruno, L., Montinari, M., Rizzi, M., Vigliotta, G. and Pagliara, P. (2012) Identification and Characterization of a New Halomicronema Species (Cyanobacteria) Isolated from the Mediterranean Marine Sponge Petrosia ficiformis (Porifera) Fottea. Olomouc, 12, 315-326.
- Iteman, I., Rippka, R., de Marsac, N.T. and Herdman, M. (2001) Comparison of Conserved Structural and Regulatory Domains within Divergent 16S rRNA-23S rRNA Spacer Sequences of Cyanobacteria. Microbiology, 146, 1275-1286.
- Taberlet, P., Gielly, L., Pautou, G. and Bouvet, J. (1991) University Primers for Amplification of Three Non-Coding Regions of Chloroplast DNA. Plant Molecular Biology, 17, 1105-1109. http://dx.doi.org/10.1007/BF00037152
- Ferris, C., Oliver, R.P, Davy, A.J. and Hewitt, G.M. (1993) Native Oak Chloroplast Reveals an Ancient Divide across Europe. Molecular Ecology, 2, 337-344. http://dx.doi.org/10.1111/j.1365-294X.1993.tb00026.x
- Gielly, L. and Taberlet, P. (1994) The Use of Chloroplast DNA to Resolve Plant Phylogenies: Noncoding versus rbcL Sequences. Molecular Biology and Evolution, 11, 769-777.
- Paquin, B., Kathe, S.D., Nierzwick-Bauer, S.A. and Shub, D.A. (1997) Origin and Evolution of Group I Introns in Cyanobacterial tRNA Genes. Journal of Bacteriology, 179, 6798-6806.
- Kuhsel, M.G., Strickland, R. and Palmer, J.D. (1990) An Ancient Group I Intron Shared by Eubacteria and Chloroplasts. Science, 250, 1570-1573. http://dx.doi.org/10.1007/BF00037152
- Xu, E.M., Kathe, S.D., Goodrich-Blair, H., Nierzwicki-Bauer, S.A. and Shub, D.A. (1990) Bacterial Origin of a Chloroplast Intron: Conserved Self-Splicing Group I Introns in Cyanobacteria. Science, 250, 1570-1573. http://dx.doi.org/10.1126/science.2125747
- Amoutzias, G.D., Van de Peer, Y. and Mossialos, D. (2008) Evolution and Taxonomic Distribution of Noribosomal Peptide and Polyketide Synthases. Future Microbiology, 3, 361-370. http://dx.doi.org/10.2217/17460913.3.3.361
- Zhao, J., Yang, N. and Zeng, R. (2008) Phylogenetic Analysis of Type I Polyketide Synthase and Non-Ribosomal Peptide Synthetase Genes in Antarctic Sediment. Extremophiles, 12, 97-105. http://dx.doi.org/10.1007/s00792-007-0107-9
- Pushparaj, B., Buccioni, A., Paperi, R., Piccardi, R., Ena, A., Carrozzi, P. and Sili, C. (2008) Fatty Acid Composition of Antarctic Cyanobacteria. Phycologia, 47, 430-434. http://dx.doi.org/10.1007/s00792-007-0107-9
- Dembitsky, V.M. and Řezanka, T. (2005) Metabolites Produced by Nitrogen Fixing Nostoc Species. Folia Microbiologica, 50, 363-391.
- Paulsrud, P. and Lindblad, P. (1997) Sequence Variation of the tRNA(Leu) Intron as a Marker for Genetic Diversity and Specificity of Symbiotic Cyanobacteria in Some Lichens. Applied and Environmental Microbiology, 64, 310-315.
- Paulsrud, P., Rikkinen, J. and Lindblad, P. (1998) Cyanobiont Specificity in Some Nostoc-Containing Lichens and in a Peltigera aphthosa Photosymbiodeme. New Phytologist, 139, 517-524. http://dx.doi.org/10.1046/j.1469-8137.1998.00220.x
- Costa, J.L., Paulsrud, P., Rikkinen, J. and Lindblad, P. (2001) Genetic Diversity of Nostoc Symbionts Endophytically Associated with Two Bryophyte Species. Applied and Environmental Microbiology, 67, 4393-4396. http://dx.doi.org/10.1128/AEM.67.9.4393-4396.2001
- Costa, J.L., Paulsrud, P. and Lindblad, P. (2002) The Cyanobacterial tRNALeu (UAA) Intron: Evolutionary Patterns in a Genetic Marker. Molecular Biology and Evolution, 19, 850-857. http://dx.doi.org/10.1093/oxfordjournals.molbev.a004142
- Hoshina, R. and Imamura, N. (2008) Eu-Chlorella Large Subunit rDNA Sequences and Group I Intron in Ribosomal DNA of the Paramecian Symbiotic Alga NC64A. Phycological Research, 56, 21-32. http://dx.doi.org/10.1111/j.1440-1835.2008.00481.x
- Micheli, C., Spinosa, F., Paperi, R., Buccioni, A. and Pushparaj, B. (2007) Biodiversity and Fatty Acid Production in Cyanobacteria. Rapports et Proces-Verbaux des Rèunions Commission Internationale pour l’Exploration Scientifique de la Mer Méditerranée, 38, 380.
- Galtier, N., Gouy, M. and Gautier, C. (1996) SEAVIEW and PHYLO_WIN: Two Graphic Tools for Sequence Alignment and Molecular Phylogeny. Computer Applications in the Biosciences, 12, 543-548.
- Edgar, R.C. (2004) MUSCLE: Multiple Sequence Alignment with High Accuracy and High throughput. Nucleic Acids Research, 32, 1792-1797. http://dx.doi.org/10.1093/nar/gkh340
- Tamura, K., Dudley, J., Nei, M. and Kumar, S. (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Molecular Biology and Evolution, 24, 1596-1599. http://dx.doi.org/10.1093/molbev/msm092
- Mathews, D.H., Disney, M.D., Childs, J.L., Schroeder, S.J., Zuker, M. and Turner, D.H. (2004) Incorporating Chemical Modification Constraints into a Dynamic Programming Algorithm for Prediction of RNA Secondary Structure. Proceedings of the National Academy of Sciences of the United States of America, 101, 7287-7292. http://dx.doi.org/10.1073/pnas.0401799101
- Taton, A., Grubisic, S., Balthasart, P., Hodgson, D.A., Laybourn-Parry, Wilmotte, A. (2006) Biogeographycal Distribution and Ecological Ranges of Benthic Cyanobacteria in East Antarctica Lakes. FEMS Microbiology Ecology, 57, 272-289. http://dx.doi.org/10.1111/j.1574-6941.2006.00110.x
- Condon, C., Squires, C. and Squires, C.L. (1995) Control of rRNA Transcription in Escherichia coli. Microbiological Reviews, 59, 623-645.
- Řeháková, K., Johansen, J.R., Casamatta, D.A., Li, X.S. and Vincent, J. (2007) Morphological and Molecular Characterization of Selected Desert Soil Cyanobacteria: Three Species New to Science Including Mojavia pulchra Gen. et sp. Nov. Phycologia, 46, 481-502. http://dx.doi.org/10.2216/06-92.1
- Johansen, J.R., Olsen, C.E., Lowe, R.L., Fučíková, K. and Casamatta, D.A. (2008) Leptolyngbya Species from Selected Seep Walls in the Great Smoky Mountains National Park. Algological Studies, 126, 21-36. http://dx.doi.org/10.1127/1864-1318/2008/0126-0021
- Cech, T.R., Damberger, S.H. and Gutell, R.R. (1994) Representation of the Secondary and Tertiary Structures of Group I Introns. Nature Structural Biology, 1, 273-280. http://dx.doi.org/10.1038/nsb0594-273
- Taton, A., Grubisic, S., Brambilla, B., De Vit, R. and Wilmotte, A. (2003) Cyanobacterial Diversity in Natural and Artificial Microbial Mats of Lake Fryxell (McMurdo Dry Valley, Antarctica): A Morphological and Molecular Approach. Applied and Environmental Microbiology, 69, 5157-5169. http://dx.doi.org/10.1128/AEM.69.9.5157-5169.2003
NOTES

*Corresponding author.


