Advances in Biological Chemistry
Vol.4 No.5(2014), Article ID:48600,9 pages
DOI:10.4236/abc.2014.45037
Expression of SOX3 and SOX9 Genes in Gonads of Blue Gourami
Gad Degani1,2
1Faculty of Science and Technology, Tel-Hai Academic College, Kiryat Shmona, Israel
2MIGAL-Galilee Technology Center, Kiryat Shmona, Israel
Email: gad@migal.org.il
Copyright © 2014 by author and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 28 May 2014; revised 15 July 2014; accepted 1 August 2014
ABSTRACT
In vertebrates, SOX (SRY-related HMG box) genes are thought to be due to major gene duplication events, initially occurring during early stages of metazoan evolution and later during the transition between non-vertebrate chordates and vertebrates. The aim of this study is to examine SOX3 and SOX9 transcription in oogensis and spermatogenesis in a fish model, the blue gourami (Trichogaster trichopterus). In females during oogenesis, SOX9 mRNA levels were lower compared to SOX3 mRNA levels. In males, SOX9 mRNA levels were higher in testes compared to SOX3 mRNA levels, however no significant differences between SOX3 and SOX9 mRNA levels were observed in the gonads of males that were kept under non-reproductive conditions compared to males kept with females under reproductive conditions and that were nest-builders.
Keywords:SOX3, SOX9, mRNA and Blue Gourami

1. Introduction
Many transcription factors of the SOX (SRY-related HMG box) family are critical for a number of developmental processes, most notably sex determination, neural crest development and neurogenesis [1] [2] ). SOX genes are characterized by the presence of a single High Mobility Group box (HMG box), a 79 amino acid DNA-binding domain having affinity to the WWCAAW motif [3] . These factors are generally expressed in a dynamic, tissue-specific manner, and often interact with other transcription factors [4] .
Expansion in the number of vertebrate SOX genes is thought to be due to major gene duplication events, initially occurring during early stages of metazoan evolution and later during the transition between non-vertebrate chordates and vertebrates [5] . Gnathostomes (jawed vertebrates) have undergone two rounds of whole genome duplications [6] whereas estimates of the number of rounds of duplication in agnathans like lamprey range from one to two.
SOX genes were first classified by [7] ) using partial sequences from mouse SOX genes. This study defined the six paralogous groups (A - F) that were the basis of the current classification [7] . Four more groups were subsequently added to include recently identified paralogs. Nevertheless, members of the same groups did not always have similar roles or expression patterns, indicating that recent paralogs could adopt new functions with relative ease [8] [9] . A comparative amino acid analysis between lamprey and other vertebrate SOX genes provides insight into the evolutionary history of early vertebrates and duplication events occurring early in the vertebrate lineage.
When SRY was identified, it was initially assumed to be unique to the Y chromosome. A search for SRY in kangaroos, however, identified a homolog on the X chromosome, termed SOX3. The sequence of the HMG-box in SOX3 most closely resembled that of SRY, so it was suggested that SOX3 was the ancestor of SRY [10] . Most other genes on the Y (for example, RBMY, TSPY), were subsequently found to have homologs on the X from which they had obviously evolved [11] .
SOX3 was expressed strongly in gonads and the central nervous system, at least in mice and humans, but its deletion or duplication in human males affected fertility and intelligence rather than sex determination [12] . Its sequence similarity to SOX9 initially prompted the suggestion that SOX3 in its ancestor was originally a dosage-regulated inhibitor of SOX9 in females, and so a null mutant of SOX3 could have permitted activation of SOX9 and male development. Truncation of the null SOX3 allele was suggested to have subsequently turned it into an inhibitor of normal SOX3 and thus an activator of SOX9 [13] . However, it now seems more likely that SRY interacts with steroidogenic factor 1 (SF1) to activate SOX9 directly [13] , suggesting that acquisition of a testis-determining function is due to a changed SOX3 expression pattern or to its association with different binding partners. SOX3 is also found on the sex chromosomes of two distantly related non-mammalian vertebrates, the frog Rana rugosa [14] and the fish Oryzias dancena [15] . However, it has not yet been shown to be sex-determining in either species, and is not known to be sex-linked in any other vertebrate species. Mouse knockouts indicate that SOX3 is involved in spermatogenesis and is a developmental regulator like SOX1 and SOX2, specifying neuronal fate in fish and mammals.
Notably zebrafish has duplicated SOX19 genes [16] whereas in most other teleosts for which SOX19 sequences are available, only a single copy appears to be present. Among vertebrates, orthologs in different species are highly similar to each other. Most of these groups are represented by a single gene in the invertebrate model organisms Drosophila melanogaster and Caenorhabditis elegans, suggesting the expansion of this single gene into multiple related genes during vertebrate evolution [8] . The blue gourami (T. trichopterus) is a tropical fish. It serves as a useful model for studying the role of endocrine regulation on reproduction since it is a multispawner with a synchronic ovary development [17] -[19] [20] .
Understanding the physiological perspectives of SOX3 and SOX9 gene systems on reproductive endocrinology will contribute to the general comprehension of basal mechanisms regulating reproductive activity and their relationships with other endocrine systems in non-mammalian vertebrate species.
2. Materials and Methods
2.1. Fish and Sampling
Trichogaster trichopterus (blue gourami) males and females purchased from local pet stores were separated into different containers and grown for about 2 - 3 months in aquaria at 27˚C under a light regime of 12 h light: 12 h darkness, fed twice a day with commercial tropical fish food (TetraBits, 47.5% protein, 6.5% fat) and once a day supplemented with frozen live food (Artemia salina). Males were divided into two groups: (1) grouped mature males (isolated from females); and (2) individual mature males under reproductive conditions, including plants adapted for nest-building. Females were divided into three groups: (1) non-mature females (BW = 1.12 ± 0.37) (n = 10), the ovaries of which contained mostly oocytes at their previtelogenic stage; (2) mature females that were kept in groups without males (6.80 ± 1.14) (n = 10); and (3) mature females that were kept with nestbuilding males for 24 hours (7.15 ± 1.10) (n = 6). The latter showed oocytes at their final maturation stage.
Fish were sampled and the gonads (whole testis or small pieces of the ovary) were dissected. The follicles were then isolated accurately using forceps under a microscope to form an enriched sample of the following follicular stages: previtellogenic follicles (from non-mature females), vitellogenic oocytes (from mature grouped females), and follicles at the final oocyte maturation stage (from mature paired females) [21] . The samples were kept in RNA save (Biological Industries, Israel) solution until RNA purification.
2.2. RNA Purification and cDNA Synthesis
Total RNA was extracted from female follicles (n = 26) using Tri-reagent (Invitrogen, CA), according to the manufacturer’s recommendations. First-strand cDNA was synthesized using the Verso-Reverse-IT 1st Strand Synthesis Kit (ABgene, UK) from 0.5 - 2 mg total RNA, with an incubation of 1 h at 57˚C, followed by 2 min at 94˚C.
2.3. SOX3 and SOX9 Partial Cloning
Gene-specific primers of SOX3 gene were designed for cloning of the partial cDNA sequence based on the fulllength sequence of Lates calcarifer (accession number DQ915951), Epinephelus coioides (accession number DQ219298), Acanthopagrus schlegelii (accession number EF605272) and Amphiprion melanopus (accession number EU908060) (Table 1).
Gene-specific primers of SOX9 gene were designed for cloning of the partial cDNA sequence based on the full-length sequence of Epinephelus coioides (accession number GQ232762), Epinephelus akaara (accession number AY676309) and Oreochromis aureus (accession number EU373500) (Table 1). The amplified PCR products were cloned into the pGEM-T vector (Promega, USA) that propagated in Escherichia coli cells. The recombinant plasmid was then extracted using the SV miniprep (Promega, USA), and the sequence of the amplified product was determined (HyLabs, Israel). Only one transcript of SOX3 and SOX9 was identified (Figure 1 and Figure 2, respectively). The polymerase chain reaction (PCR) was carried out in a total volume of 15 μL onsisting of 0.166 μM of each primer and 17.5 μL of Absolute Blue SYBR Green ROX Mix (ABgene). The PCR products were visualized on a UV-transilluminator following electrophoresis on a 1.5% agarose gel containing ethidium bromide. To confirm the specificity of the PCR reaction, the identity of each PCR product was verified by sequencing (Figure 2).
2.4. Real-Time PCR
To compare mRNA levels of GnRH1, GnhRH3, SOX3, SOX9, βLH and βFSH in the gonads
of individual gourami, the relative abundance of their mRNA was normalized with
the endogenous reference gene, the 18S subunit of rRNA (18S rRNA), using the comparative
threshold cycle (CT) method according to Pfaffl et al. [22] . The relative amount of each gene was calculated by
the formula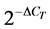 , where DCT corresponds to the difference
between the CT measured for each target hormone and that determined for
18S rRNA. To validate this method, serial dilutions were prepared from a gonad (testis
and ovary) cDNA sample and the efficiencies of each target gene and 18S rRNA amplifications
were compared by plotting DCT versus log (template) according to the
method of Muller et al. [23] . Linear regressions
of the plots showed the following R2 values and efficiency, respectively:
0.99 and 1 for GnRH1; 0.99 and 0.9 for GnRH3; 0.99 and 0.94 for 18S rRNA; 0.99 and
1 for βLH; 0.99 and 0.87 for βFSH; 0.99 and 1 for SOX3; and 0.99 and 0.94 for SOX9.
Gene-specific primers for real-time PCR were designed using Primer3 Software. Table 2 summarizes the primers used in the real-time
PCR. To each of the above PCR mixtures were added 7.5 ml of the Syber Green Master
Mix (ABgene) in a final volume
, where DCT corresponds to the difference
between the CT measured for each target hormone and that determined for
18S rRNA. To validate this method, serial dilutions were prepared from a gonad (testis
and ovary) cDNA sample and the efficiencies of each target gene and 18S rRNA amplifications
were compared by plotting DCT versus log (template) according to the
method of Muller et al. [23] . Linear regressions
of the plots showed the following R2 values and efficiency, respectively:
0.99 and 1 for GnRH1; 0.99 and 0.9 for GnRH3; 0.99 and 0.94 for 18S rRNA; 0.99 and
1 for βLH; 0.99 and 0.87 for βFSH; 0.99 and 1 for SOX3; and 0.99 and 0.94 for SOX9.
Gene-specific primers for real-time PCR were designed using Primer3 Software. Table 2 summarizes the primers used in the real-time
PCR. To each of the above PCR mixtures were added 7.5 ml of the Syber Green Master
Mix (ABgene) in a final volume
 (a)
(a)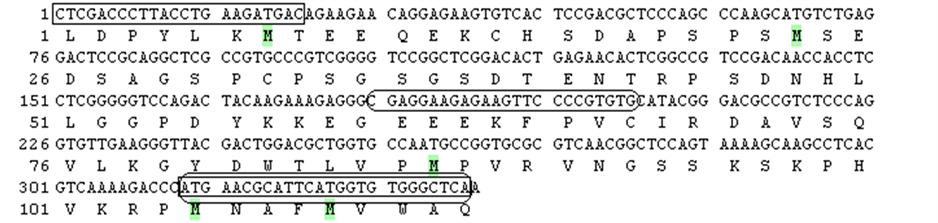 (b)
(b)
Figure 1. SOX3 (a) and
SOX9 (b) partial nucleotide and deduced amino acid sequences.
 —localization of primers for cDNA cloning.
—localization of primers for cDNA cloning.
 —localization of primers used for real-time PCR.
—localization of primers used for real-time PCR.
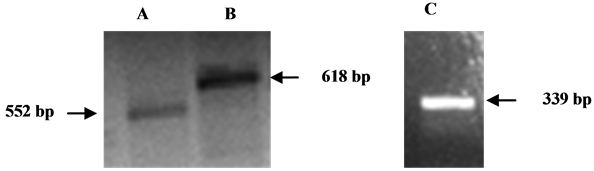
Figure 2. Detection of SOX3 and SOX9 transcription in blue gourami RTPCR. Total RNA from gourami testis or ovary was extracted, reverse transcribed and amplified by PCR. PCR products were separated on 1.5% agarose gel containing ethidium bromide. A: SOX3 partial fragment (primer sets: SOX3(fw)300 and SOX3(rev)854); B: SOX3 partial fragment (primer sets: SOX3(fw)145 and SOX3(rev)775); C: SOX9 partial fragment (primer sets: SOX9(fw)10 and SOX9(rev)359).
of 15 ml. Amplification was carried out in a RotorGene 3000 Sequence Detection System (Corbett Research, Sydney, Australia) under the following conditions: for GnRH1—initial denaturation at 95˚C for 15 min, followed by 40 cycles of 95˚C for 10 s, 52˚C for 20 s, 72˚C for 20 s and 82˚C for 115 s; for GnRH3—initial denaturation at 95˚C for 15 min, followed by 40 cycles of 95˚C for 10 s, 53˚C for 20 s, 72˚C for 20 s and 82˚C for 10 s; for βLH—initial denaturation at 95˚C for 15 min, followed by 40 cycles of 95˚C for 10 s, 60˚C for 20 s, 72˚C for 25 s and 84˚C for 15 s; for βFSH—initial denaturation at 95˚C for 15 min, followed by 40 cycles of 95˚C for
10 s, 57˚C for 20 s and 72˚C 20 s; for SOX3—initial denaturation at 95˚C for 15 min, followed by 40 cycles of 95˚C for 15 s, 63˚C for 20 s, 72˚C for 20 s and 85˚C for 15 s; for SOX9—initial denaturation at 95˚C for 15 min, followed by 40 cycles of 95˚C for 10 s, 63˚C for 20 s, 72˚C for 25 s and 85˚C for 10 s; and for 18S rRNA—initial denaturation at 95˚C for 10 min, followed by 40 cycles of 95˚C for 20 s, 64˚C for 20 s and 71˚C for 20 s. Amplifications of each target gene and the reference gene 18S rRNA cDNAs were performed simultaneously in separate tubes in duplicate, and the results were analyzed using Q-Gene software (BioTechniques Software Library at: www.BioTechniques.com). Dissociation-curve analysis was run after each real-time experiment to ensure that there was only one product. To control for false positives, a non template negative control was run for each primer pair.
2.5. Statistical Analysis
Data are presented as mean ± SEM. The significance of the differences between group means of hormone mRNA levels was determined either by a one-way analysis of variance (ANOVA) followed by an a posteriori Bonferroni post-hoc test or a student T-test in males using SPSS 17.0 software. Differences were considered statistically significant at P < 0.05.
3. Results
3.1. SOX3 and SOX9 Gene Expression in the Gonads of Blue Gourami
In order to determine if SOX3 and SOX9 are expressed in the gonads of blue gourami, total RNA from the organs of gourami fish was reverse-transcribed. Reactions using cDNA derived from ovary RNA samples using two sets of primers specific to partial segments of the gourami SOX3 revealed two bands: 552 bp and 618 bp. One primer set of primers specific to a partial SOX9 segment revealed a 338 bp band (Figure 2). Similar results were obtained from samples derived from testis (data not shown).
3.2. Variations in SOX3 and SOX9 mRNA Levels in Oocytes during Different Stages of Oogenesis
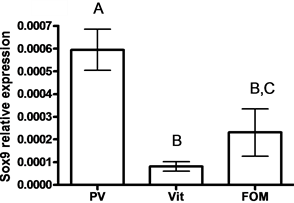
SOX9 mRNA levels were lower in oocytes compared to SOX3 mRNA levels (33.24 fold) (P < 0.05, by student t-test) (Figure 3(a)). Lower levels of SOX3 were measured in vitellogenic and FOM oocytes compared to previtelogenic oocytes (11- and 52-fold, respectively) (Figure 3(b)), whereas lower levels of SOX9 were measured in vitellogenic oocytes compared to previtelogenic oocytes (3.3-fold) (Figure 3(c)) (P < 0.05, by ANOVA, Bonferroni post-hoc test). However, the level of SOX9 transcription in ovary found in FOM was significantly higher than during vitellogenesis.
3.3. SOX3 and SOX9 mRNA Levels in the Testis of Blue Gourami
In males, SOX9 mRNA levels were higher in testes compared to SOX3 mRNA levels (9.21-fold) (P < 0.05, by student t-test) (Figure 4(a)). However, no significant differences between SOX3 and SOX9 mRNA levels were observed in the gonads of males that were kept in a group compared to males kept with females and that were nest-builders (P < 0.05, student T-test)
4. Discussion
In the current study, the transcription of both SOX3 and SOX9 was found in both testis and ovary, but at different levels. The SOX gene signature domain HMG was highly conserved across different vertebrates. Recent studies in teleosts such as zebrafish, medaka, rice field eel and carp also reported two forms of SOX9 [24] -[27] .
In the current study, the transcription of SOX9 was high in males and that of SOX3 in females. Moreover, the level of both transcription factors SOX3 and SOX9 in testis changed during oogenesis and the sexual behavior of females but not of males. In catfish, semi-quantitative RT-PCR analysis in adult gonads revealed sexual
 (a)
(a)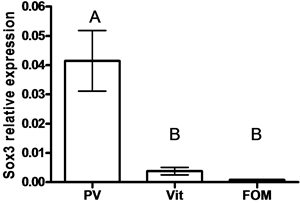 (b)
(b)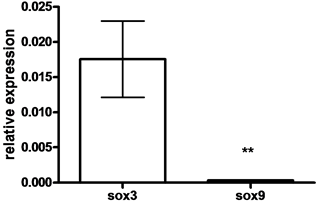 (c)
(c)
Figure 3. The expression of SOX3 and SOX9 genes
in follicles of females (a) and in oocytes at each of the following stages of oogenesis:
previtellogenesis (PV), vitellogenesis (Vit) and final oocyte maturation (FOM)
(b, c). Females were kept in groups or with males for one day. Total RNA was
extracted and reverse-transcribed to cDNA, which was used as a template for real-time
PCR. SOX3 and SOX9 mRNA levels were normalized to that of 18S rRNA by the CT
cycle method, where
![]() reflects the relative amount of specific gene precursor transcripts. Each histogram
represents the average of independent measurements (mean ± SEM; n = 6 - 26). A different
letter above the histogram denotes its significant difference from the mRNA levels
of the other histograms [P < 0.05 student t-test (a) or ANOVA and Bonfferoni post-hoc
test (b, c)].
reflects the relative amount of specific gene precursor transcripts. Each histogram
represents the average of independent measurements (mean ± SEM; n = 6 - 26). A different
letter above the histogram denotes its significant difference from the mRNA levels
of the other histograms [P < 0.05 student t-test (a) or ANOVA and Bonfferoni post-hoc
test (b, c)].
 (a)
(a)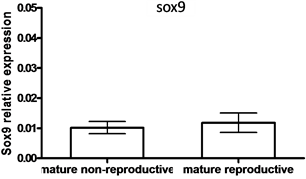 (b)
(b)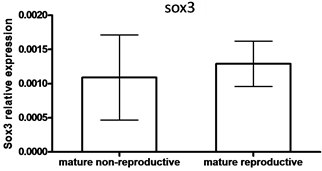 (c)
(c)
Figure 4. The expression of SOX3 and SOX9 genes
in testis (a) and mature males under non-reproductive and reproductive conditions
(paired with females) (b, c). Total RNA was extracted and reverse-transcribed to
cDNA, which was used as a template for real-time PCR. SOX3 and SOX9 mRNA levels
were normalized to that of 18S rRNA by the CT cycle method, where
![]() reflects the relative amount of specific gene precursor transcripts. Each histogram
represents the average of independent measurements (mean ± SEM; n = 6 - 15). A different
letter above the histogram denotes its significant difference from the mRNA levels
of the other histograms [P < 0.05 student t-test (a) or ANOVA and Bonfferoni post-hoc
test (b, c)].
reflects the relative amount of specific gene precursor transcripts. Each histogram
represents the average of independent measurements (mean ± SEM; n = 6 - 15). A different
letter above the histogram denotes its significant difference from the mRNA levels
of the other histograms [P < 0.05 student t-test (a) or ANOVA and Bonfferoni post-hoc
test (b, c)].
dimorphic expression of SOX9a and SOX9b in adult and developing gonads, whereas SOX9a was expressed preferably in testis, while SOX9b was expressed abundantly in ovary [24] . This type of differential expression pattern of duplicated genes might attribute to more flexible gene regulation and function during catfish development. A similar kind of expression pattern was also noticed in zebrafish using PCR Southern and in situ hybridization [28] .
Most studies of those genes examined the mRNA levels of SOX3 and SOX9 during sex determination and differentiation, but not during spermatogenesis and oogenesis, as was done in the current study [29] . The results of this study are in agreement with a previous finding that SOX9 is present in both sexes, but in one of the first molecular events in the male pathway is upregulated in Sertoli cell precursors (which are essential for testis differentiation) by the primary sex-determining trigger interacting with an evolutionarily conserved sequence within SOX9 [29] . In mammals, the SOX8 and SOX9 transcription factors are involved, among other things, in sex differentiation, male gonad development and adult maintenance of spermatogenesis [30] .
The expression level of the SOX9 gene varied among several organs of adult Cyprinus carpio, being highest in the brain and testis [27] . Similar structure and identity of SOX9 and SOX17 genes in mammals, chickens and fish suggest that these genes have evolutionarily conserved roles, potentially including sex determination and differentiation [31] . Based on the current study, we suggested that SOX9 genes were involved in spermatogenesis. In this study, it was found that the level of SOX9 transcription was high in mature males before sexual behavior and reproduction. A high level of sperm was found in the testis in both these stages [32] , and spermatogenesis was controlled by gonadotropins (FSH and LH) and androgens hormones [20] [33] . Raghuveer and Senthilkumaran [34] observed that in breathing catfish Clarias gariepinus, which underwent an annual reproductive cycle, a dimorphic expression pattern of SOX9a and SOX9b was found in both adult and developing gonads using RT-PCR, indicating that SOX9a retained its function in testis while SOX9b might play a new role in ovary, as was revealed in the current study in blue gourami using real-time PCR.
In the current study, it was found that the levels of both SOX9 and SOX3 were high in the ovary where the oocytes were found in PV compared to VT and FOM. In rainbow trout ovary, on the basis of the HMG box amino acid sequence, SOX24 can be categorized into the same subgroup of SOX proteins as SOX4, SOX11 and SOX22. These results suggest that SOX24 plays a role as a transcriptional regulator during oogenesis [35] . Our results regarding the change of mRNA in PV compared to VT and FOM as was found in blue gourami are in agreement with the result for SOX9 transcription in oogensis in catfish [34] .
In mature female catfish, SOX9 expression was restricted to the perinucleolar stage of developing oocytes and pre-vitellogenic oocytes, but not in mature oocytes. Raghuveer and Senthikumaran (2010) suggested that the decrease in SOX9 transcript in mature oocytes might be due to the dilution of transcripts with increasing mass/size of mature oocytes that accumulated large amounts of vitellogenin and maternal RNAs. These results combined indicated that SOX9 might play an important role in early gonadal development and recrudescence.
5. Conclusion
In conclusion, it is suggested that SOX genes that are involved in sex differentiation where cell structure and function change are found during oogenesis and spermatogensis might be involved similarly as during sex differentiation in fish.
References
- Hong, C.S. and Saint-Jeannet, J.P. (2005) Sox Proteins and Neural Crest Development. Seminars in Cell & Developmental Biology, 16, 694-703. http://dx.doi.org/10.1016/j.semcdb.2005.06.005
- Betancur, P., Bronner-Fraser, M. and Sauka-Spengler, T. (2010) Genomic Code for Sox10 Activation Reveals a Key Regulatory Enhancer for Cranial Neural Crest, Proceedings of the National Academy of Sciences of the United States, 107, 3570-3575. http://dx.doi.org/10.1073/pnas.0906596107
- Laudet, V., Stehelin, D. and Clevers, H. (1993) Ancestry and Diversity of the HMG Box Superfamily. Nucleic Acids Research, 21, 2493-2501. http://dx.doi.org/10.1093/nar/21.10.2493
- Prior, H.M. and Walter, M.A. (1996) SOX Genes: Architects of Development. Molecular Medicine, 2, 405-412.
- Dehal, P. and Boore, J.L. (2005) Two Rounds of Whole Genome Duplication in the Ancestral Vertebrate, PLoS Biology, 3, e314. http://dx.doi.org/10.1371/journal.pbio.0030314
- Escriva, H., Manzon, L., Youson, J. and Laudet, V. (2002) Analysis of Lamprey and Hagfish Genes Reveals a Complex History of Gene Duplications during Early Vertebrate Evolution. Molecular Biology and Evolution, 19, 1440-1450. http://dx.doi.org/10.1093/oxfordjournals.molbev.a004207
- Wright, E.M., Snopek, B. and Koopman, P. (1993) Seven New Members of the Sox Gene Family Expressed during Mouse Development. Nucleic Acids Research, 21, 744. http://dx.doi.org/10.1093/nar/21.3.744
- Bowles, J., Schepers, G. and Koopman, P. (2000) Phylogeny of the SOX Family of Developmental Transcription Factors Based on Sequence and Structural Indicators. Developmental Biology, 227, 239-255. http://dx.doi.org/10.1006/dbio.2000.9883
- Uy, B.R., Simoes-Costa, M., Sauka-Spengler, T. and Bronner, M.E. (2012) Expression of Sox Family Genes in Early Lamprey Development. The International Journal of Developmental Biology, 56, 377-383. http://dx.doi.org/10.1387/ijdb.113416bu
- Collignon, J., Sockanathan, S., Hacker, A., Cohen-Tannoudji, M., Norris, D., Rastan, S., Stevanovic, M., Goodfellow, P.N. and Lovell-Badge, R. (1996) A Comparison of the properties of Sox-3 with Sry and Two Related Genes, Sox-1 and Sox-2. Development, 122, 509-520.
- Graves, J.A. (2006) Sex Chromosome Specialization and Degeneration in Mammals. Cell, 124, 901-914.http://dx.doi.org/10.1016/j.cell.2006.02.024
- Weiss, J., Meeks, J.J., Hurley, L., Raverot, G., Frassetto, A. and Jameson, J.L. (2003) Sox3 Is Required for Gonadal Function, but Not Sex Determination, in Males and Females. Molecular and Cellular Biology, 23, 8084-8091.http://dx.doi.org/10.1128/MCB.23.22.8084-8091.2003
- Sekido, R. and Lovell-Badge, R. (2008) Sex Determination Involves Synergistic Action of SRY and SF1 on a Specific Sox9 Enhancer. Nature, 453, 930-934. http://dx.doi.org/10.1038/nature06944
- Uno, Y., Nishida, C., Oshima, Y., Yokoyama, S., Miura, I., Matsuda, Y. and Nakamura, M. (2008) Comparative Chromosome Mapping of Sex-Linked Genes and Identification of Sex Chromosomal Rearrangements in the Japanese Wrinkled Frog (Rana rugosa, Ranidae) with ZW and XY Sex Chromosome Systems. Chromosome Research, 16, 637-647. http://dx.doi.org/10.1007/s10577-008-1217-7
- Takehana, Y., Demiyah, D., Naruse, K., Hamaguchi, S. and Sakaizumi, M. (2007) Evolution of Different Y Chromosomes in Two Medaka Species, Oryzias dancena and O. latipes. Genetics, 175, 1335-1340.http://dx.doi.org/10.1534/genetics.106.068247
- Okuda, Y., Yoda, H., Uchikawa, M., Furutani-Seiki, M., Takeda, H., Kondoh, H. and Kamachi, Y. (2006) Comparative Genomic and Expression Analysis of Group B1 Sox Genes in Zebrafish Indicates Their Diversification during Vertebrate Evolution. Developmental Dynamics, 235, 811-825. http://dx.doi.org/10.1002/dvdy.20678
- Jackson, K., Goldberg, D., Ofir, M., Abraham, M. and Degani, G. (1999) Blue Gourami (Trichogaster trichopterus) Gonadotropic Beta Subunits (I and II) cDNA Sequences and Expression during Oogenesis. Journal of Molecular Endocrinology, 23, 177-187. http://dx.doi.org/10.1677/jme.0.0230177
- Levy, G., Gothilf, Y. and Degani, G. (2009) Brain Gonadotropin Releasing Hormone3 Expression Variation during Oogenesis and Sexual Behavior and Its Effect on Pituitary Hormonal Expression in the Blue Gourami. Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology, 154, 241-248. http://dx.doi.org/10.1016/j.cbpa.2009.06.010
- Levy, G., Jackson, K. and Degani, G. (2010) Association between Pituitary Adenylate Cyclase-Activating Polypeptide and Reproduction in the Blue Gourami. General and Comparative Endocrinology, 166, 83-93. http://dx.doi.org/10.1016/j.ygcen.2009.09.015
- Degani, G. (2001) Blue Gourami (Trichogaster trichopterus) Model for Labyrinth Fish. Laser Pages Publishing, Jerusalem.
- Jackson, K., Goldberg, D., Ofir, M., Abraham, M. and Degani, G. (1999) Blue Gourami (Trichogaster trichopterus) Gonadotropic Beta Subunits (I and II) cDNA Sequences and Expression during Oogenesis. Journal of Molecular Endocrinology, 23, 177-187. http://dx.doi.org/10.1677/jme.0.0230177
- Pfaffl, M.W. (2001) A New Mathematical Model for Relative Quantification in Real-Time RT-PCR. Nucleic Acids Research, 29, e45. http://dx.doi.org/10.1093/nar/29.9.e45
- Muller, P.Y., Janovjak, H., Miserez, A.R. and Dobbie, Z. (2002) Processing of Gene Expression Data Generated by Quantitative Real-Time RT-PCR. Biotechniques, 32, 1372-1379.
- Chiang, E.F., Pai, C.I., Wyatt, M., Yan, Y.L., Postlethwait, J. and Chung, B. (2001) Two Sox9 Genes on Duplicated Zebrafish Chromosomes: Expression of Similar Transcription Activators in Distinct Sites. Developmental Biology, 231, 149-163. http://dx.doi.org/10.1006/dbio.2000.0129
- Zhou, R., Liu, L., Guo, Y., Yu, H., Cheng, H., Huang, X., Tiersch, T.R. and Berta, P. (2003) Similar Gene Structure of Two Sox9a Genes and Their Expression Patterns during Gonadal Differentiation in a Teleost Fish, Rice Field Eel (Monopterus albus). Molecular Reproduction and Development, 66, 211-217. http://dx.doi.org/10.1002/mrd.10271
- Kluver, N., Kondo, M., Herpin, A., Mitani, H. and Schartl, M. (2005) Divergent Expression Patterns of Sox9 Duplicates in Teleosts Indicate a Lineage Specific Subfunctionalization. Development Genes and Evolution, 215, 297-305.http://dx.doi.org/10.1007/s00427-005-0477-x
- Du, Q.Y., Wang, F.Y., Hua, H.Y. and Chang, Z.J. (2007) Cloning and Study of Adult-Tissue-Specific Expression of Sox9 in Cyprinus carpio. Journal of Genetics, 86, 85-91. http://dx.doi.org/10.1007/s12041-007-0013-z
- Chiang, E.F., Yan, Y.L., Guiguen, Y., Postlethwait, J. and Chung, B. (2001) Two Cyp19 (P450 Aromatase) Genes on Duplicated Zebrafish Chromosomes Are Expressed in Ovary or Brain. Molecular Biology and Evolution, 18, 542-550.http://dx.doi.org/10.1093/oxfordjournals.molbev.a003833
- Bagheri-Fam, S., Sinclair, A.H., Koopman, P. and Harley, V.R. (2010) Conserved Regulatory Modules in the Sox9 Testis-Specific Enhancer Predict Roles for SOX, TCF/LEF, Forkhead, DMRT, and GATA Proteins in Vertebrate Sex Determination. The International Journal of Biochemistry & Cell Biology, 42, 472-477. http://dx.doi.org/10.1016/j.biocel.2009.07.001
- Chalmel, F., Lardenois, A., Georg, I., Barrionuevo, F., Demougin, P., Jegou, B., Scherer, G. and Primig, M. (2013) Genome-Wide Identification of Sox8-, and Sox9-Dependent Genes during Early Post-Natal Testis Development in the Mouse. Andrology, 1, 281-292. http://dx.doi.org/10.1111/j.2047-2927.2012.00049.x
- Zhou, R., Cheng, H., Zhang, Q., Guo, Y., Cooper, R.K. and Tiersch, T.R. (2002) SRY-Related Genes in the Genome of the Rice Field Eel (Monopterus albus). Genetics Selection Evolution, 34, 129-137. http://dx.doi.org/10.1186/1297-9686-34-1-129
- Degani, G., Jackson, K., Goldberg, D., Sarfati, R. and Avtalion, R.R. (2003) βFSH, βLH and Growth Hormone Gene Expression in Blue Gourami (Trichogaster trichopterus, Pallas 1770) during Spermatogenesis and Male Sexual Behavior. Zoological Science, 20, 737-743. http://dx.doi.org/10.2108/zsj.20.737
- Degani, G. (1993) Reproduction Control in Multi-Spawning Asynchronic Trichogaster trichopterus (Pallas) as a Model for the Anabantidae Family. Trends in Comparative Biochemistry & Physiology, 1, 1269-1275.
- Raghuveer, K. and Senthilkumaran, B. (2010) Isolation of Sox9 Duplicates in Catfish: Localization, Differential Expression Pattern during Gonadal Development and Recrudescence, and hCG-Induced Up-Regulation of Sox9 in Testicular Slices. Reproduction, 140, 477-487. http://dx.doi.org/10.1530/REP-10-0200
- Kanda, H., Kojima, M., Miyamoto, N., Ito, M., Takamatsu, N., Yamashita, S. and Shiba, T. (1998) Rainbow Trout Sox24, a Novel Member of the Sox Family, Is a Transcriptional Regulator during Oogenesis. Gene, 211, 251-257.http://dx.doi.org/10.1016/S0378-1119(98)00100-0


