Open Journal of Pediatrics
Vol. 2 No. 1 (2012) , Article ID: 17991 , 4 pages DOI:10.4236/ojped.2012.21003
High incidence of respiratory viruses in critically ill children*
![]()
1Department of Pediatrics, School of Medicine, Vanderbilt University, Nashville, USA
2Department of Pathology, Microbiology, and Immunology, School of Medicine, Vanderbilt University, Nashville, USA
Email: john.williams@vanderbilt.edu
Received 10 May 2011; revised 20 August 2011; accepted 20 October 2011
Keywords: Respiratory Virus; RT-PCR; Diagnostics
ABSTRACT
Background: Acute lower respiratory tract illness is a major cause of hospitalization, respiratory failure, and death in children worldwide. However, infectious agents are not identified in >50% of these clinical illnesses using traditional methods such as rapid antigen detection assays or viral culture. Objective: To conduct a pilot study using RT-PCR to determine the frequency of respiratory viruses in critically ill children with respiratory compromise. Methods: Critically ill children admitted to the Pediatric Critical Care Unit during 2 winter seasons with respiratory compromise or failure were prospectively enrolled. Respiratory tract specimens (tracheal lavage or nasal wash) were tested for 11 different viruses using realtime RT-PCR, including respiratory syncytial virus (RSV), human metapneumovirus (HMPV), influenza A (IAV) and B (IBV), parainfluenza viruses (PIV) 1-3, coronaviruses OC43, 229E, and Netherlands, and human rhinovirus (HRV). Results: Thirty-two children were enrolled, 28 (88%) of which required mechanical ventilation. Clinical diagnoses included bronchiolitis, croup, asthma, apnea, and pneumonia. Of these patients, 28/32 (88%) tested positive for one of eleven viruses by RT-PCR. Compared to antigen testing for RSV, IAV, and IBV, RT-PCR detected an additional nine infections for an increased yield of 53%. RT-PCR also detected coronavirus infections in five patients, including three patients with Netherlands coronavirus. Co-infection with more than one virus was detected in 8/32 (25%). None of the 32 patients tested positive for HMPV, PIV 1, or PIV 3. Conclusions: We detected a respiratory virus in 88% of children with respiratory failure. RT-PCR significantly increased the identification of viruses compared to antigen testing. RSV, influenza A, and coronaviruses were the most common viruses detected. Our data suggest that a viral etiology can be identified in the majority of childhood episodes of severe respiratory disease and that RT-PCR is a useful addition to the diagnostic approach.
1. INTRODUCTION
Lower respiratory illness (LRI) is a leading cause of morbidity and mortality in children worldwide [1]. Acute LRI including bronchiolitis and pneumonia are the most common reasons for hospitalization of infants in the US [2]. Previously identified major pathogens include respiratory syncytial virus (RSV), human metapneumovirus (HMPV), parainfluenza viruses (PIV) types 1 - 3, and influenza A (IAV) and B (IBV) viruses, all of which are associated with clinical syndromes of severe lower respiratory tract disease, such as bronchiolitis, pneumonia and laryngotracheobronchitis [3,4]. However, infectious agents are not identified in up to 50% of these clinical illnesses using traditional methods such as viral culture [5]. While rapid antigen assays for RSV and influenza and PCR-based methods increase detection, a number of LRI episodes in children have no virus identified [5,6]. It is unknown if these illnesses are due to identified pathogens that are simply not detected secondary to low virus titers in samples or if there are novel agents, which have yet to be identified.
Recent studies have identified previously known coronaviruses (HCoV) OC43 and 229E and the novel HCoV-NL63 as important causes of LRI [7,8]. Both classic and novel human rhinoviruses (HRV) have been associated with LRI in children and adults [9]. We hypothesized that a viral pathogen would be present in most children with respiratory failure and LRI. We conducted a prospective pilot study using highly sensitive real-time RT-PCR to test for respiratory viruses in critically ill children with respiratory compromise.
2. MATERIALS AND METHODS
2.1. Patients
Patients were eligible for enrollment if they were admitted to the Pediatric Intensive Care Unit (PICU) at Monroe Carell Jr. Children’s Hospital at Vanderbilt between September 1, 2004 and March 30, 2006 with a primary or secondary respiratory diagnosis. Specific diagnoses included respiratory distress, respiratory failure, pneumonia, bronchiolitis, reactive airway disease, asthma, status asthmaticus, and apnea. Patients were enrolled after parent/guardian informed consent. The Vanderbilt Institutional Review Board approved the study.
2.2. Clinical Characteristics
Clinical characteristics recorded included patient demographics, diagnosis, requirement for mechanical ventilation, and antigen testing results from the hospital clinical laboratory.
2.3. Specimen Collection and Processing
For mechanically ventilated patients, tracheal lavage specimens were collected during routine suctioning by the clinical staff using 3 ml of sterile saline collected in a suction trap connected to the suction catheter. If not mechanically ventilated, nasal wash specimens were obtained by instilling 3 ml of sterile saline into the patient’s nostril and collecting the nasal wash with a bulb syringe. All samples were immediately placed on ice and transported to the laboratory. Samples were aliquoted into lysis buffer (RLT, Qiagen). Original specimen and lysis buffer aliquots were stored at –70˚C for batch processing.
2.4. Nucleic Acid Extraction and Virus Genome Testing
Total RNA was extracted from 250 µl of nasal wash or tracheal lavage fluid using a commercial RNA extraction kit (RNeasy, Qiagen). Specimens were tested by real-time RT-PCR for 11 viruses: PIV1, PIV2, PIV3, HCoV OC43, HCoV 229E, HCoV NL63, IAV, IBV, RSV, HRV, and HMPV. Primers and dual-labeled probes were based on published assays for OC43 [10], 229E [10], NL63 [11], RSV [12], rhinovirus [13], and HMPV [14]. We developed primers and probes specific for PIV1, 2 and 3 (all targeting the N gene) using Primer Express 2.0 (ABI, Foster City, CA). PIV assays were tested in silico and against multiple other viruses to ensure specificity (not shown). GenBank reference sequences were M62850, D01070, and S38060 (PIV1); M55320.1, AF533010, AF533011, and AF533012 (PIV2); NC_001796, Z11575, X04612, and M14552 (PIV3). Influenza primer and probe sequences were provided by Steven Lindstrom of the Centers for Disease Control and Prevention [15]. All probes were labeled at the 5’ end with 6-carboxyfluorescein (FAM) and at the 3’ end with the non-fluorescent quencher Blackhole Quencher 1 (BHQ1) (Operon Biotechnologies). Real-time RT-PCR was performed with the Smart Cycler II (Cepheid). Reactions were performed as single assays using QuantiTect Probe RT-PCR kit (Qiagen) according to the manufacturer’s instructions. Extensive optimization was carried out to determine the optimal annealing temperatures, cycle times, and primer/ probe concentrations for each assay (data not shown). Cycling conditions were: reverse transcription for 30’ at 50˚C, HotStarTaq polymerase activation for 15’ at 95˚C, and 45 cycles of denaturation for 8 s at 94˚C and annealing/extension for 60 s at 60˚C. Fluorescent data was collected during the annealing/extension step. The sensitivity and specificity of each assay performed in our laboratory was determined using a panel of clinical isolates (data not shown). Samples were assayed in batches of 32. Each run included “no template” controls and positive viral RNA controls. The cycle threshold (Ct) was defined as the cycle at which the fluorescent signal crossed 10 times the background fluorescence. Specimens were considered positive if the Ct value was less than 40 cycles and all positive samples were confirmed by repeat assay.
3. RESULTS
We enrolled 32 patients in this pilot study. Of these 32 patients, 28 (88%) were intubated and mechanically ventilated for respiratory failure or apnea. The patient ages ranged from 12 days to 19 years old with a median age of 9 months. Over 50% (17/32) of the patients were less than 2 months of age and 78% (25/32) were less than 2 years of age. Male to female ratios were 19:13, (60% male). Patient diagnosis consisted of bronchiolitis (62.5%), pneumonia (21.9%), croup (6.3%), status asthmaticus (6.3%), and apnea (3.1%).
Of these patients, 27/32 (84.4%) tested positive for at least one of 11 viruses by RT-PCR testing (Table 1). Of the 11 viruses tested for in this cohort of critically ill children, eight viruses were identified. RSV was the most common virus identified at 21/32 (65.6%). IAV (4/32, 12.5%), IBV (1/32), PIV2 (1/32) and HRV (2/32) were identified in a small number of patients. Several HCoV strains were identified, including three patients infected with the recently discovered HCoV NL63. None of the patients tested positive for HMPV, PIV1, or PIV3.
RT-PCR increased the yield of viral detection signifycantly compared to viral antigen testing (Table 2). RSV viral antigen testing identified infection in 16 patients. RSV RT-PCR testing confirmed infection in these 16 patients and identified another five patients with negative RSV antigen tests. IAV was found by antigen testing in only 1 patient but RT-PCR confirmed this patient and identified an additional 3 patients infected with IAV. IBV was not found in any patient by antigen testing but was identified by RT-PCR in one patient.
Co-infection with more than one virus was found in 8/32 patients (25%) (Table 3). Five patients with RSV had a second virus identified and two patients with IAV infection had viral confection. One patient had three viruses identified: IAV, IBV and HCoV NL63. Of children with more than one virus detected, 8/8 (100%) were mechanically ventilated and 4/8 (50%) were premature infants.
4. DISCUSSION
In this pilot study of critically ill infants and children
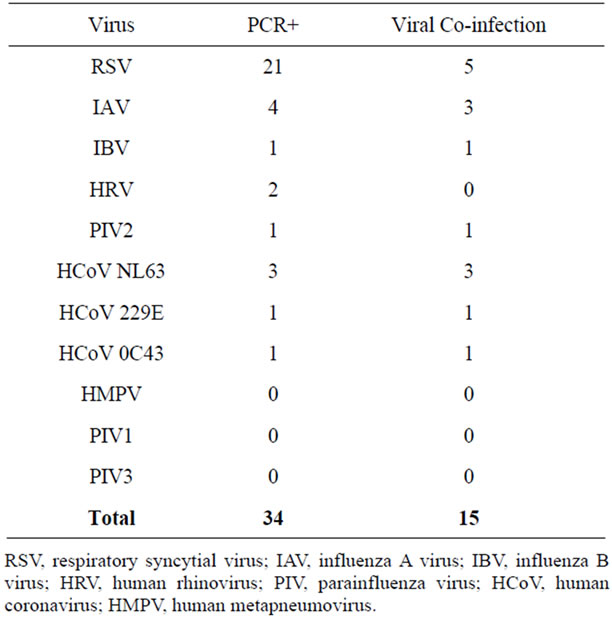
Table 1. Viruses detected and co-infections.
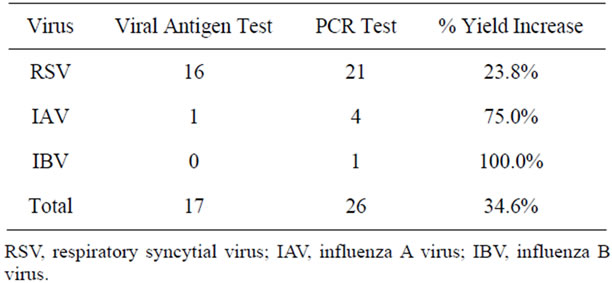
Table 2. Increase in viral detection by PCR testing compared to antigen testing.
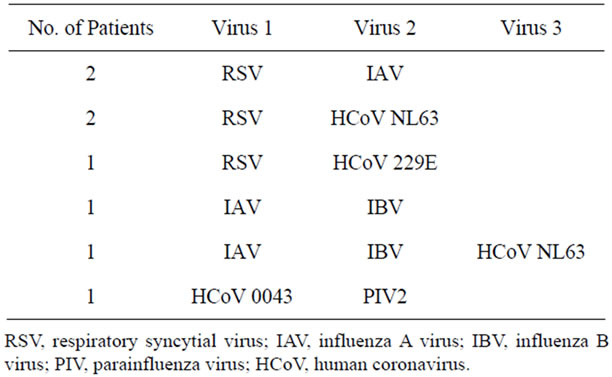
Table 3. Viruses detected by RT-PCR in co-infected patients.
with respiratory failure, we identified a viral respiratory pathogen by RT-PCR in almost all patients in this cohort. This finding supports our hypothesis that respiratory viruses at least contribute to, if not cause, most episodes of LRI in children. Reliable identification of bacterial respiratory pathogens is problematic and some of these children may have had complicating bacterial pneumonias. Nonetheless, large field studies of conjugated pneumococcal vaccine showed reduction in viral LRI as well as pneumococcal pneumonia [16]. Hence, the identification of viruses in these patients likely indicates a role in pathogenesis.
The use of RT-PCR significantly improved the detection of respiratory viruses for which antigen tests are available including RSV and influenza viruses. Identification of these viruses is important because influenza is treatable with oseltamivir, and many high-risk infants receive immunoprophylaxis with palivizumab against RSV. Furthermore, both of these cause nosocomial outbreaks. While universal precautions should be used on every patient, especially during the winter season, knowledge of a patient’s virus-infected status could increase compliance and encourage placement of virusinfected patients in separate areas from critically ill cardiac or postsurgical patients.
RT-PCR testing also allowed the detection of other viral infections in critically ill children including HRV, HCoV, and PIV2. An unexpected finding in our population was that over 15% (5/32) of the critically ill children had a coronavirus detected by RT-PCR. In three of these five children, we identified HCoV NL63, a recently discovered virus. All three HCoV NL63-infected patients were co-infected with either RSV or IAV, both well-established primary pathogens, thus raising questions about the contribution of HCoV NL63 to these illnesses. One of our patients infected with HCoV NL63 had underlying malignancy and treatment-induced immunosuppression and developed ARDS. The original reports of HCoV NL63 included patients with underlying medical conditions and severe LRI [8]. In those studies, no other respiratory pathogen was identified in the HCoV NL63- infected patients, suggesting a primary role for the virus. This raises the question whether underlying disease increases the risk of severe disease with HCoV NL63.
In conclusion, the use of real-time RT-PCR enabled us to identify a respiratory virus in the majority of children with acute respiratory failure. Highly sensitive molecular assays would enhance detection of viral infections in the PCCU population, including the identification of viruses that are difficult to grow and for which there are no rapid antigen tests. This information could be useful to guide therapy, including the choice of antimicrobial therapy. Greater knowledge of the burden of severe disease associated with these viruses also could inform intervention strategies such as vaccine development. Larger studies are warranted to define the epidemiology of these viruses in more detail and with greater statistical rigor. Our data show that expanded RT-PCR detection of respiratory viruses could be beneficial in the diagnosis and treatment of critically ill children with respiratory distress and respiratory failure.
REFERENCES
- Mulholland, K. (2007) Childhood pneumonia mortality— A permanent global emergency. Lancet, 370, 285-289. doi:10.1016/S0140-6736(07)61130-1
- Shay, D.K., Holman, R.C., Newman, R.D., et al. (1999) Bronchiolitis-associated hospitalizations among US children, 1980-1996. Journal of the American Medical Association, 282, 1440-1446. doi:10.1001/jama.282.15.1440
- Williams, J.V., Harris, P.A., Tollefson, S.J., et al. (2004) Human metapneumovirus and lower respiratory tract disease in otherwise healthy infants and children. The New England Journal of Medicine, 350, 443-450. doi:10.1056/NEJMoa025472
- Williams, J.V., Edwards, K.M., Weinberg, G.A., et al. (2010) Population-based incidence of human metapneumovirus infection among hospitalized children. Journal of Infectious Diseases, 201, 1890-1898. doi:10.1086/652782
- Weinberg, G.A., Erdman, D.D., Edwards, K.M., et al. (2004) Superiority of reverse-transcription polymerase chain reaction to conventional viral culture in the diagnosis of acute respiratory tract infections in children. Journal of Infectious Diseases, 189, 706-710. doi:10.1086/381456
- Griffin, M.R., Walker, F.J., Iwane, M.K., et al. (2004) Epidemiology of respiratory infections in young children: Insights from the new vaccine surveillance network. Pediatric Infectious Disease Journal, 23, S188-S192. doi:10.1097/01.inf.0000144660.53024.64
- Sizun, J., Gagneur, A., Legrand, M.C. and Baron R. (2001) Respiratory coronavirus infections in children. Pediatric Infectious Disease Journal, 20, 555-556. doi:10.1097/00006454-200105000-00026
- Van der Hoek, L., Pyrc, K., Jebbink, M.F., et al. (2004) Identification of a new human coronavirus. Nature Medicine, 10, 368-373. doi:10.1038/nm1024
- Arden, K.E., McErlean, P., Nissen, M.D., et al. (2006) Frequent detection of human rhinoviruses, paramyxoviruses, coronaviruses, and bocavirus during acute respiratory tract infections. Journal of Medical Virology, 78, 1232-1240. doi:10.1002/jmv.20689
- Van Elden, L.J., Van Loon, A.M., Van Alphen, F., et al. (2004) Frequent detection of human coronaviruses in clinical specimens from patients with respiratory tract infection by use of a novel real-time reverse-transcriptase polymerase chain reaction. Journal of Infectious Diseases, 189, 652-657. doi:10.1086/381207
- Fouchier, R.A., Hartwig, N.G., Bestebroer, T.M., et al. (2004) A previously undescribed coronavirus associated with respiratory disease in humans. Proceedings of the National Academy of Sciences USA, 101, 6212-6216. doi:10.1073/pnas.0400762101
- Mentel, R., Wegner, U., Bruns, R. and Gurtler, L. (2003) Real-time PCR to improve the diagnosis of respiratory syncytial virus infection. Journal of Medical Microbiology, 52, 893-896. doi:10.1099/jmm.0.05290-0
- Deffernez, C., Wunderli, W., Thomas, Y., et al. (2004) Amplicon sequencing and improved detection of human rhinovirus in respiratory samples. Journal of Clinical Microbiology, 42, 3212-3218. doi:10.1128/JCM.42.7.3212-3218.2004
- Maertzdorf, J., Wang, C.K., Brown, J.B., et al. (2004) Real-time reverse transcriptase PCR assay for detection of human metapneumoviruses from all known genetic lineages. Journal of Clinical Microbiology, 42, 981-986. doi:10.1128/JCM.42.3.981-986.2004
- Kandun, I.N., Wibisono, H., Sedyaningsih, E.R., et al. (2006) Three Indonesian clusters of H5N1 virus infection in 2005. The New England Journal of Medicine, 355, 2186-2194. doi:10.1056/NEJMoa060930
- Madhi, S.A. and Klugman, K.P. (2004) A role for streptococcus pneumoniae in virus-associated pneumonia. Nature Medicine, 10, 811-813. doi:10.1038/nm1077
NOTES
*Supported by a MedImmune Fellowship Grant 2004-2006 to MVJ.

