American Journal of Molecular Biology
Vol.1 No.3(2011), Article ID:7779,16 pages DOI:10.4236/ajmb.2011.13015
JnCML-like, an EF-hand motif-containing gene seasonally upregulated in the transition zone of black walnut (Juglans nigra L.)
![]()
1Department of Forestry and Natural Resources, Purdue University, West Lafayette, Indiana, USA;
2Department of Biological Sciences, Purdue University, West Lafayette, Indiana, USA;
3Department of Computer Science, Purdue University, West Lafayette, Indiana, USA;
4Markey Center of Structural Biology, Purdue University, West Lafayette, Indiana, USA;
5USDA Forest Service Northern Research Station Hardwood Tree Improvement and Regeneration Center, Purdue University, West Lafayette, Indiana, USA.
Email: *woeste@purdue.edu
Received 25 June 2011; revised 22 July 2011; accepted 3 August 2011.
Keywords: Transition Zone, M-Calpain, Juglans nigra, Grancalcin
ABSTRACT
The economic value of a black walnut (Juglans nigra L.) tree is strongly determined by the quality and quantity of darkly colored heartwood in its stem. To understand the regulation of heartwood formation, we analyzed the region of heartwood formation in walnut stems (i.e., the transition zone, TZ) for the expression of 80 ESTs. Semi-quantitative RT-PCR and real-time PCR was performed to detect expression changes of candidate genes in the TZ and sapwood of trees harvested in summer and fall. Results revealed that the transcript of a clone containing two presumed EF-hand motifs was expressed at higher levels in the TZ than in other xylem tissues. Analysis of the full-length coding sequence revealed that the black walnut gene JnCML-like is similar to grancalcin-like calcium-binding EF hand proteins in Arabidopsis thaliana (At3g10300) and Zea mays (NM 001153810). A model of the predicted structure of JnCML-like showed it is similar to grancalcin and m-calpain, penta-EF-hand family proteins associated with cell proliferation, differentiation and programmed cell death. JnCML-like transcript was detected in tissue from the region of the pith meristem, and in roots, embryogenic callus, vascular cambium, female flowers, male flowers, green leaves, and partially and fully senescent leaves of black walnut, although transcript abundance varied considerably among these tissues.
1. INTRODUCTION
Calcium is a critical second messenger in all eukaryotic cells responding to external stimuli. Calcium interacts with a large number of proteins, but among the most studied are proteins containing the EF-hand motif. Calcium-dependent EF-hand motif-containing proteins are common in animals, plants, and fungi [1,2]. The EF-hand was defined by Kretsinger and Nockolds (1973) [3] in carp (Cyprinus carpio L.) parvalbumin. The basic EF-hand motif is highly conserved and usually consists of two perpendicular 10 to 12 residue alpha helices with a 12-residue loop region. Together these three domains form a single helix-loop-helix calcium-binding site. In most cases, EF-hand motifs are found in pairs. Two hundred and fifty genes encoding EF-hand-containing proteins have been identified in Arabidopsis thaliana [4]. Penta-EF-hand (PEF) proteins are a family of proteins that, upon Ca2+ binding, are translocated to membranes. PEF proteins in mammals are divided into two subgroups: group I comprised of ALG-2 (apoptosis linked gene-2) and peflin, and group II (also called the Ca2+-dependent, Cys protease calpain subfamily), which includes sorcin and grancalcin [5]. Group I proteins are found in plants and fungi and have a clear role in regulating development, including apoptosis [6]. Group II PEF proteins are not well characterized in plants, although they have been identified in every angiosperm species with a completed genome. The best characterized phytocalpain, Defective Kernel 1 (DEK1), is described as calpainlike [7]; it appears to have a complex role in the regulation of plant growth and development.
In many fine hardwoods, the quantity and quality of heartwood is an important factor determining wood value. Black walnut heartwood is a zone of darkly colored xylem at the center of a mature stem that is surrounded by pale-colored sapwood. In fully differentiated sapwood, only ray and axial parenchyma cells are alive [8]. Ray parenchyma cells span the stem cross-section from phloem to the heartwood, and play an important role in heartwood formation, nutrient cycling, and other functions [8-9]. Most research on heartwood formation has focused on a narrow transition zone (TZ) located between the sapwood and heartwood [10-12] because the abrupt death of all parenchyma cells at the TZ of angiosperms results in the formation of heartwood [8, 13-14]. Heartwood formation in black walnut probably occurs just prior to cambial dormancy in the fall [11, 15-16]. Yang et al. [12] used a microarray to show that 569 genes out of a total of 1873 examined from the TZ of mature black locust (Robinia pseudoacacia L.) trees were differentially expressed in summer and fall, including 276 genes that were up-regulated in fall and 293 that were up-regulated in summer. They also identified two calcium-related clones in the TZ, a calcium-dependent protein kinase (BI677735) and a calmodulinrelated protein (BI678222). It is not clear how cell death in the TZ is related to the formation and deposition of extractives that produce the familiar colors of hardwood lumber, since not all tree species have darkly colored heartwood, but the formation of heartwood resembles programmed cell death (PCD) in that both processes involve loss of cell integrity, nuclear fragmentation, mitochondrial degeneration, vacuolar collapse, and release of extractives [8,17-19]. Extractives are primarily phenolic compounds polymerized to varying degrees; most colored heartwood is associated with the accumulation of flavonoids and quinones. In black walnut, the TZ can be identified macroscopically because it fluoresces blue under UV light [11], perhaps because of the accumulation of sinapic acid esters [20].
Juglans nigra L., commonly known as black walnut, is one of the most valuable hardwood tree species in the U.S. because of the excellent qualities of its heartwood. Despite its economic value, the molecular mechanisms regulating heartwood formation in black walnut are poorly understood. In this study, we present the isolation and basic characterization of an EF-hand motif-containing gene from black walnut, JnCML-like, which is upregulated in the transition zone of black walnut in a seasonally dependent manner.
2. MATERIALS AND METHODS
2.1. Plant Materials and Growth Conditions
On July 1, 2004, and October 14, 2004, two 39-year-old black walnut trees growing at the Martell Research Forest, Tippecanoe County, IN, were cut down. The trees were labeled “summer tree” and “fall tree” respectively.
Another four black walnut trees were felled on the same dates in 2006. Immediately after felling, stem cross-sections (“cookies”) about 2.5 cm thick and 20 cm in diameter were cut with a chainsaw. The cookies were immediately submerged in liquid nitrogen. After returning to the lab, the cookies were transferred to an ultra-low freezer (–80˚C) for storage. Transition zone xylem, the region of sapwood immediately outside the heartwood zone, was identified under UV light and carefully chiseled out of the cookies from each tree. These cells were in the immediate periphery of the darkly colored heartwood and they emitted a bright, blue fluorescence under UV light, showing they were undergoing the physiological conversion to heartwood. In order to investigate the expression of JnCML-like in the TZ versus sapwood in more detail, the sapwood portion of black walnut logs harvested in summer and fall were divided into three zones: an interior sapwood zone, which was next to the transition zone; a middle sapwood zone; and an exterior sapwood zone, close to the cambium (Figure 1). About 100 g of xylem was sampled from each xylem region (TZ, IS, ES, cambium). Multiple cookies from a single tree were used to produce RNA, multiple extractions were required to obtain sufficient RNA for use as a single replicate. Cambium zone tissue (which probably included small amounts of newly differentiated phloem and xylem) was removed from beneath the bark of young trees in the spring when the bark was slipping. Roots were collected from about ten young walnut trees growing in a greenhouse, RNA was extracted from each separately. Embryogenic calli were from in vitro cultures of a single source genotype, RNA was extracted from multiple plates of calli and pooled. Pith meristem tissues were collected from multiple branches of three genotypes and the RNA extracted from the pooled tissue sample. RNA from female flowers, male flowers, green leaves, and partially and fully senescent leaves were obtained by pooling multiple samples taken from many branches of a 15-year-old black walnut tree growing on the Purdue University campus. The region of the pith meristem was identified as the tissue subtending the apical meristem superior to the pith and inside the vascular cylinder (Figure 1(b)).
2.2. RNA Isolation
RNA was isolated as described previously [21]. Xylem tissue was ground to a fine powder in a 6750 SPEX CertiPrep freezer mill (SPEX CertiPrep, Inc., Metuchen, NJ). Extraction buffer consisted of the following: 200 mM Tris, pH 8.5, 1.5% lithium dodecyl sulfate, 300 mM lithium chloride, 10 mM disodium salt EDTA, 1% sodium deoxycholate, 1% tergito powder P-40, 5 mM thiourea, 1 mM aurintricarboxylic acid, 10 mM dithiothreitol,
 (a)
(a) (b)
(b)
Figure 1. (a) Cross-section of a black walnut stem under white light. The transition zone fluoresces blue under UV light; (b) The location of the pith meristem region in a branch of black walnut.
2% polyvinylpolypyrrolidone, and 2% ß-mecaptoethanol. RNA concentration was measured at 260 nM with a NanoDrop 1000 spectrophotometer (Wilmington, DE).
2.3. Semi-quantitative RT-PCR
To analyze transcript abundance, 10 μg total RNA was incubated with DNase (Ambion, Foster City, CA) at 37˚C for 30 min, followed by the addition of 5 μl DNase inactivation solution (Ambion, Foster City, CA) at room temperature. After 2 min, the mixture was spun for 2 min full-speed in a table-top centrifuge at room temperature, and the supernatant was recovered for reverse transcription (RT). 18S rRNA was used as the internal standard for normalization. RT-PCR primers were designed with the aid of the web-based program Primer3 (http://www.frodo.wi.mit.edu/cgi-bin/primer3/primer3www.cgi), and potential inhibitory secondary structures of primers and the predicted amplicon were checked using the web-based program “mFold” [22]. (http://www.bioweb.pasteur.fr/seqanal/interfaces/mfold-simple.html). Primers were designed to produce amplicons of 150 to 500 bp in size. The primers included: JnCML-like forward, 5’-AGGGCGCTGTCTTCTTACAA-3’, and reverse, 5’-AGTGCCTCTCGCAACT-CATT-3’ (product size around 200 bp); sequences of the 18S rRNA forward and reverse primers were 5’-AGA-GGCCTACAATGGTGGTG-3’ and 5’-CTCCAATGGAT-CCTC-GTTA-3’ (product size round 200 bp), respectively. Firststrand cDNA was synthesized as described by the manufacturer (Invitrogen, Carlsbad, CA). One μl of cDNA derived from 10 μg of total RNA was added to a PCR reaction consisting of 1X reaction buffer, 100 μM MgCl2 100 μM dNTP’s, 10 pmol each primer, 0.25 U of TaqGo polymerase (Promega, Madison, WI) in a total volume of 25 μl. The PCR program consisted of a 2-min denaturation step at 94˚C, followed by 24 cycles of 94˚C (30 s), annealing at 54˚C (30 s), 68˚C (45 s), followed by a 10-min extension at 68˚C. Amplified products were electrophoresed through 1% agarose gels, detected by ethidium bromide staining, and photographed under UV light (UVP, BioDoc-It, Upland, CA).
2.4. Candidate Genes and Real-time PCR
To understand the biology of the TZ, the expression of genes from a partially sequenced and annotated cDNA library made from the TZ of black walnut was evaluated using a Populus microarray [23] and semi-quantitative RT-PCR. The phage titer of this primary cDNA library exceeded 1 × 106 pfu and the cloned cDNA insert sizes (20 clones sized) ranged between 400 to 2000 bp (average size: 800 bp) [unpublished data]. As a preliminary analysis, primers were designed to amplify 80 genes with putative homeodomains or with annotations related to hormone signal transduction or cell death (Table 1).
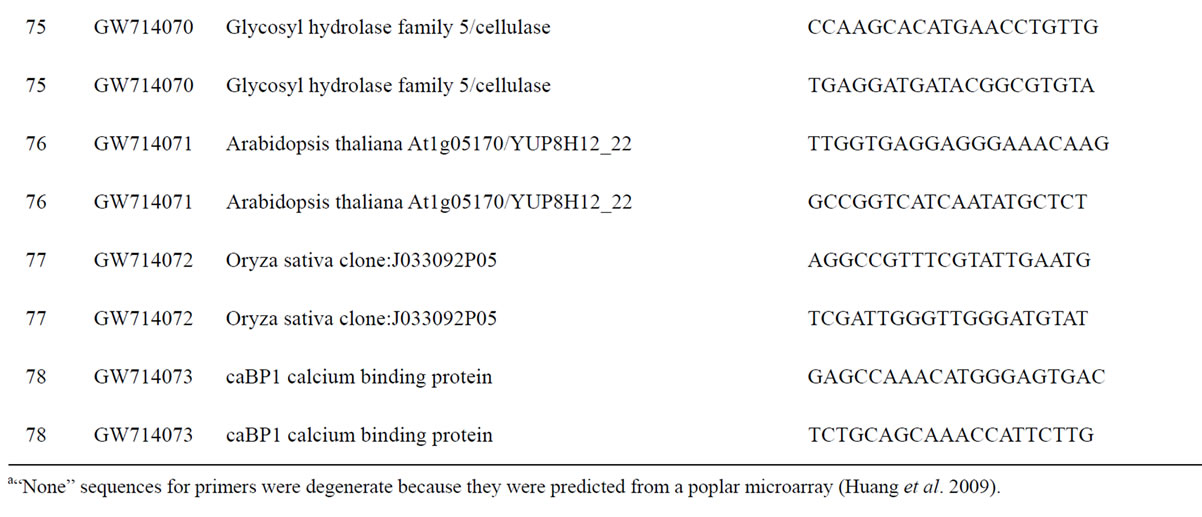

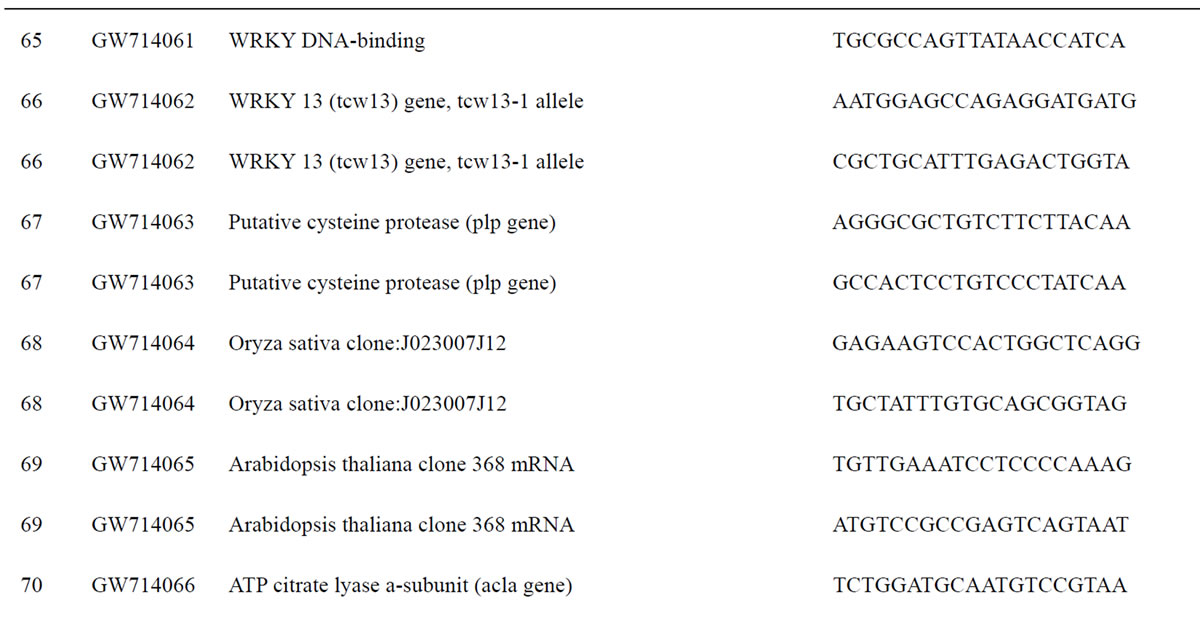
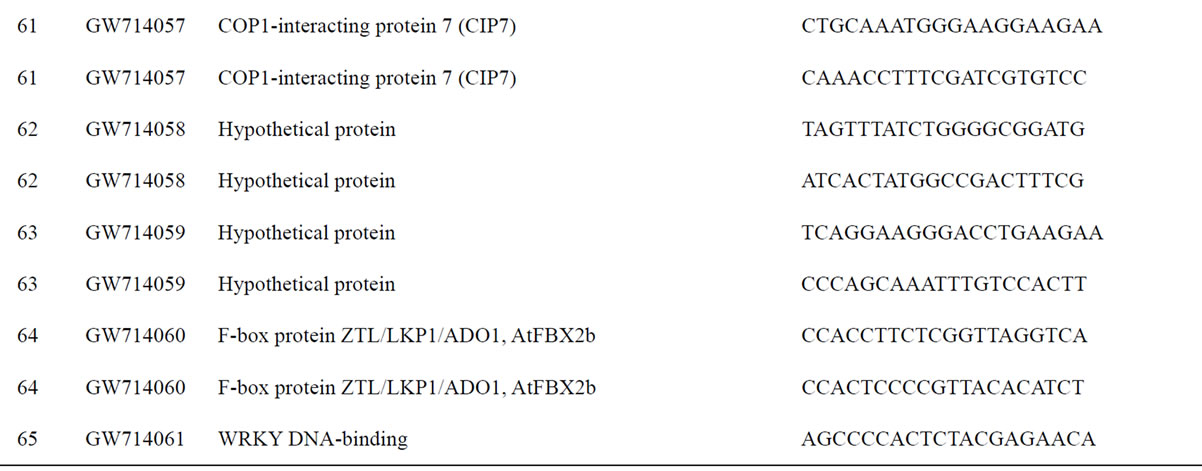
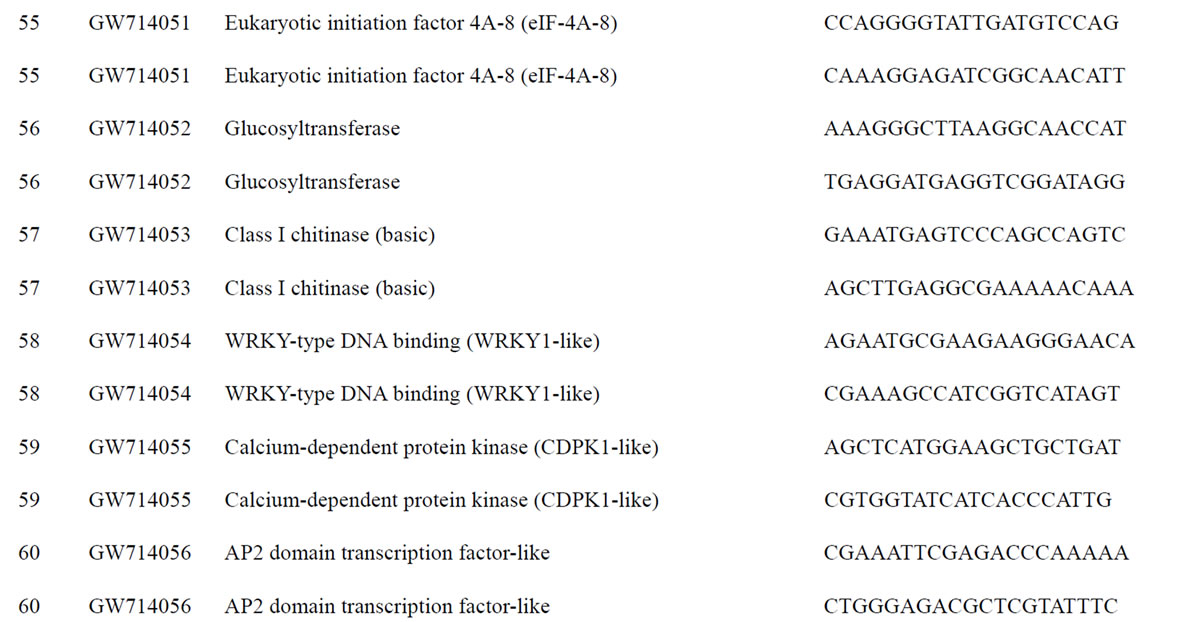
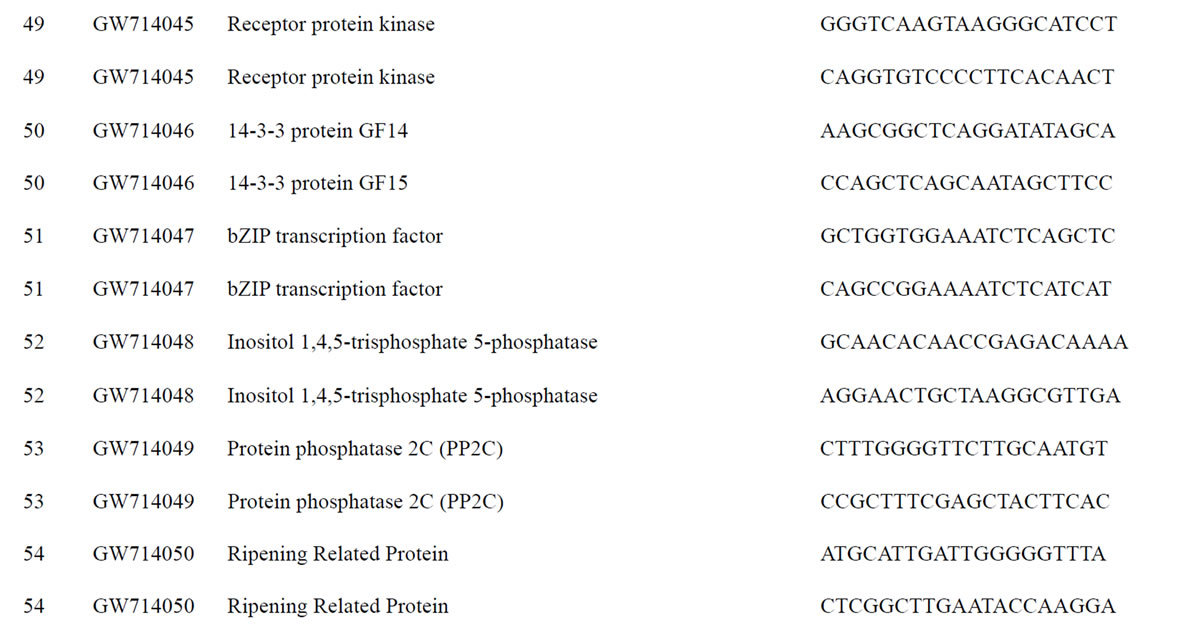
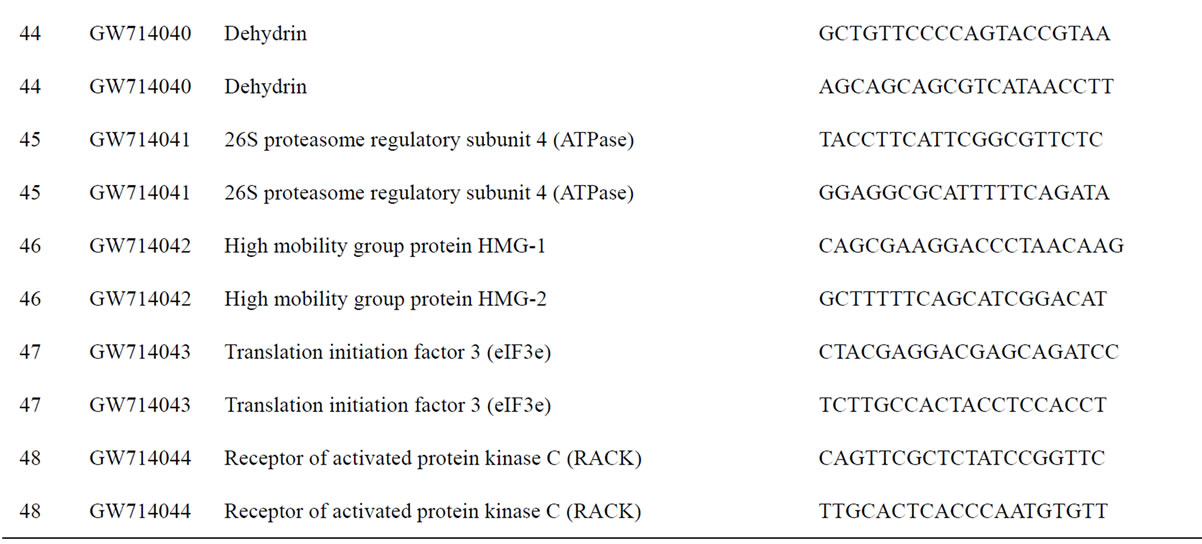
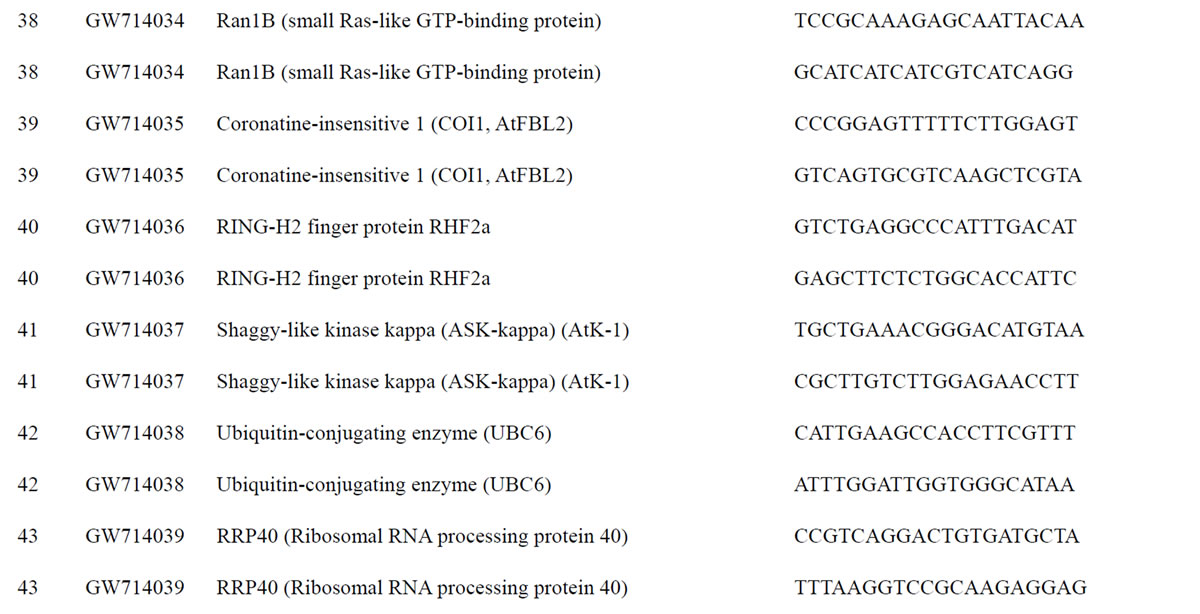



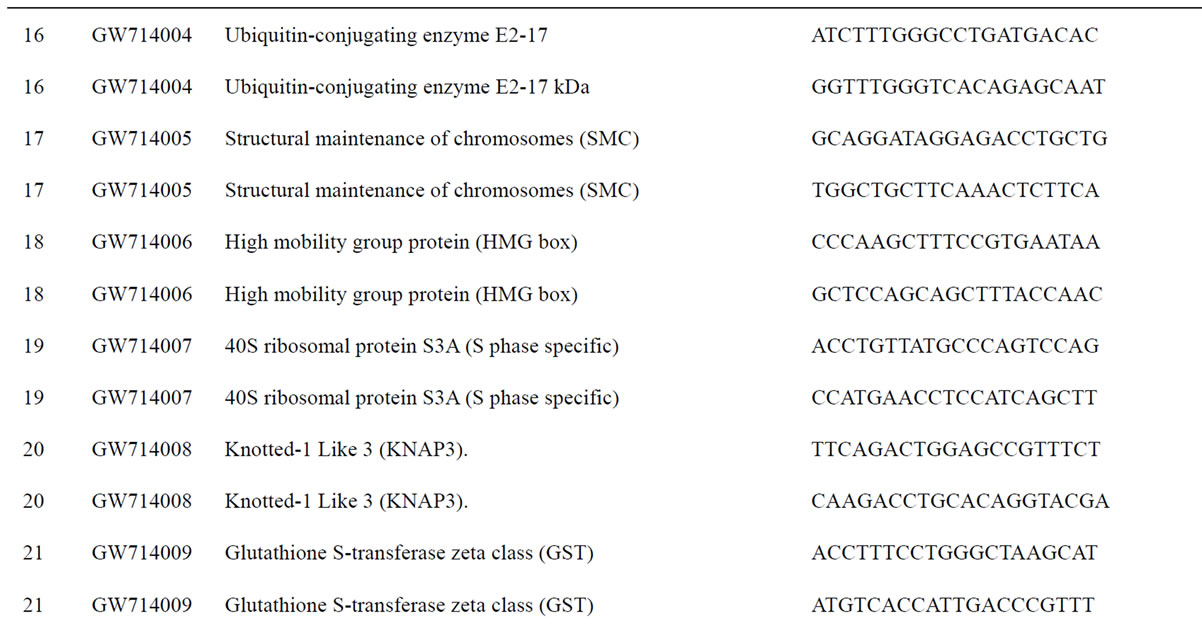
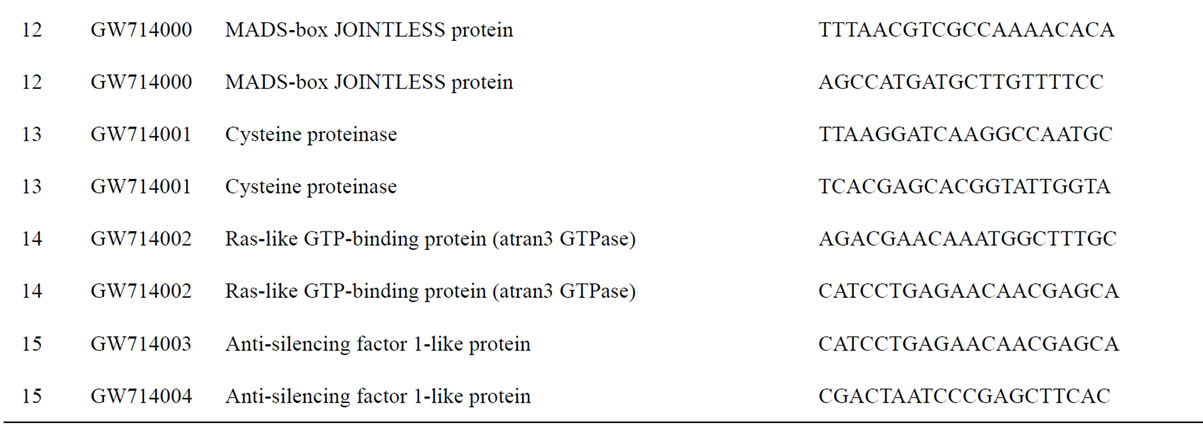


Table 1. Primers for black walnut gene models examined for differential expression in the transition zone.
Our goal was to identify genes expressed at higher levels in the TZ and in the fall than in sapwood and the summer. After identifying candidate genes, we verified expression differences using iQTM SYBR Green Supermix (Bio-Rad, Hercules, CA) and the iQ5 multicolor real-time PCR Detection System (BioRad, Hercules, CA). Control (18S rRNA) and six samples (TZ, interior sapwood, and exterior sapwood, for both summer tree and fall trees) were run as three replicates (distinct RNA pools from distinct trees) and repeated twice (technical replicates). Each 25-μl reaction consisted of 12.5 μl of SYBR Green PCR Master Mix (Bio-Rad), 1 μl of each primer (10 μM, JnCML-like forward and reverse; these primers were also used for semi-quantitative RT-PCR) and 1 μl of cDNA derived from 10 μg of total RNA. The reaction was run at 95˚C for 10 min, followed by 40 cycles at 95˚C for 15 s, 55˚C ~ 58˚C for 30 s, and 72˚C for 30 s, and an extension phase of 81 cycles of melt-curve analysis as described by the manufacturer. BioRad’s iQ5 software was allowed to choose cycle threshold levels and define the log-phase cycle number used for comparing gene-expression levels. The fold change of gene expression relative to the standard (18S rRNA) was defined by the formula 2–ΔΔCT (comparative C T method) (User's Manual, ABI PRISM 7700 Sequence Detection System, Perkin-Elmer Applied Biosystems), where ΔCT = CT (sample) − CT (18S rRNA), ΔΔCT = ΔCT of samples –ΔCT of the TZ harvested from fall. 18S rRNA has been shown to be a robust standard for reverse transcription qPCR [24]. CT values are the number of PCR cycles at which signal significantly rises above background; a consistent CT was applied across all replicates. All samples appearing in a single figure were analyzed simultaneously in a single qPCR run, and then RNA from a separate pool was analyzed as a second replicate, and so on.
2.4.1. Isolation of Full-length cDNA by 5’-and 3’-RACE
The SMART RACE cDNA Amplification kit (Clontech, Mountain View, CA) was used to perform 5’- and 3’-RACE following the manufacturer’s instructions. Samples of 1 μg of total RNA were used for reverse transcription. The gene-specific primers GSP1 (5’- AAGAACAAGCTTCCTCCGCCGCCTCT-3’), designed from the antisense strand was used for 5′-RACE, and the GSP2 (5’-TCGTCGATAATCCCGCTGCCATCTTG-3’), from the sense strand, was used for 3’-RACE. All RACE reactions were performed using the following PCR program: 94˚C for 30 s, 68˚C for 30 s and 72˚C for 3 min for 30 cycles. The PCR product (200 bp ~ 400 bp) was subcloned into pGEM-T vector (Promega, Madison, WI) and recombinant clones sequenced. The contigs were aligned with Sequencer TM 4.1 (Gene Codes, Ann Arbor, MI).
2.5. Multiple Alignment and Neighbor-joining Tree Construction
Putative orthologs of JnCML-like in a range of plant and animal species, including Pisum sativum, Zea mays, Oryza sativa, Arabidopsis thaliana, Populus trichocarpa, Ricinus communis, and Oncorhynchus mykiss, were selected based on results from Basic Local Alignment Search Tool (BLAST). The predicted protein sequences from the putative orthologs were aligned using Clustal W [25] with the purpose of discerning the EF-hand motif domain and identifying other regions of similarity. A Neighbor-joining tree was constructed with COBALT software [26].
2.5.1. Crystal Structure Prediction
The tertiary structure of JnCML-like was modeled by a combination of threading and homology modeling methods. First, known protein tertiary structures that are compatible with the amino acid sequence of JnCML-like are searched by the SP4 threading method [27]. SP4 considers sequence similarity, predicted secondary structure, and amino acid propensity of the solvent accessibility in searching known protein structures. Once the threading method identified a model structure, we employed MODELLER, a homology modeling program [28] to construct the tertiary structure model of JnCMLlike, using the model structure as the template.
3. RESULTS
We identified and analyzed a small number of candidate genes that were expressed at higher levels in the TZ than in sapwood, and at higher levels in the fall than summer (Table 1, Figure 2). Preliminary results showed that a cDNA clone containing an EF-hand motif, designated JnCML-like, was differentially expressed in the TZ of a tree entering dormancy (Figure 2). The full-length coding sequence of JnCML-like (FJ665980) was isolated by 5’- and 3’-RACE. The coding sequence of this transcript was 618 bp in length. It encoded a predicted protein of 206 amino acids with a molecular weight of 23 kD (ACN39566). Analysis via the Conserved Domain Database and Search Service, v2.13 NCBI [29] showed that JnCML-like contains two EF-hand domains (Figure 3). BLAST searches in Swiss-Prot and other databases indicated that JnCML-like showed significant homology to members of the Penta-EF-hand (PEF) group of Ca2+ binding proteins, including calpain, peflin and grancalcin. Sequence alignment of the predicted proteins showed that JnCML-like is identical to grancalcin in the EF-Hand containing region and the C-terminus, and overall shares 57% similarity to a calcium-binding EF hand family protein in Arabidopsis that may be related

Figure 2. Examples of semi-quantitative PCR to screen candidate genes for differential expression in the transition zone and in summer versus fall. See Table 1 for a complete list of evaluated genes. Lanes: (1) Water as template; (2) TZ from summer tree; (3) Interior sapwood from summer tree; (4) Exterior sapwood from summer tree; (5) TZ from fall tree; (6) Interior sapwood from fall tree; (7) Exterior sapwood from fall tree

 (a)
(a) 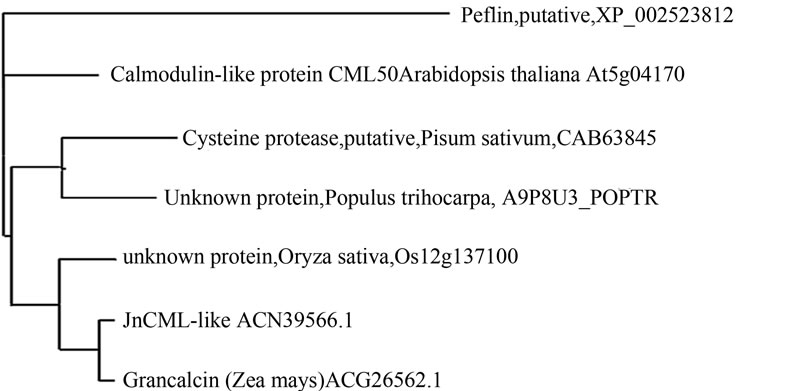 (b)
(b)
Figure 3. (a) Deduced amino acid sequence for JnCML-like aligned with proteins of other species. Invariant residues are indicated as *. Conservation of a strong group is indicated as “:”, conservation of a weak group is indicated as “.”, see Larkin et al., 2007, for details. The consensus sequence derived from the alignment is underlined and invariant residues are indicated as * in the consensus. The underscored amino acids represent an EF-hand motif from amino acids 29 to 68, a second EF-hand motif stretches from amino acids 101 to 131; (b) Neighbor-joining tree of JnCML-like and similar proteins produced using the software COBALT. The distance along the horizontal axis is proportional to the difference between sequences, whereas the distance along the vertical axis has no significance.
to grancalcin [30], 63% similarity with Os12g0137100 (which is also similar to grancalcin, e–30), 64% with an uncharacterized EF-hand family protein in Populus trichocarpa, and 84% identity with Zea mays grancalcin (Figure 3).
3.1.1. Structure of JnCML-like
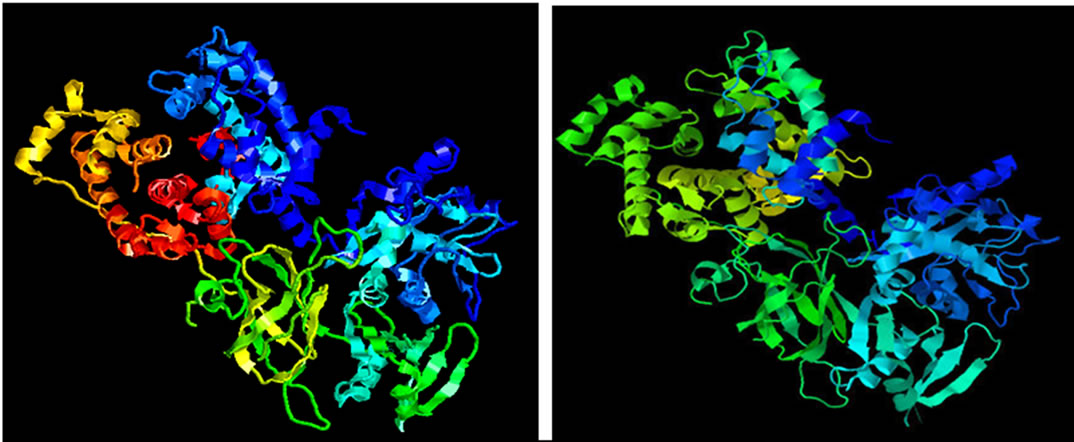
The model of JnCML-like protein made by threading and homology modeling showed that JnCML-like was predicted to be similar to the crystal structure of M-calpain (PDB ID: 1df0A) with a Z-score of 16.52. The available structural models of grancalcin do not include the N-terminal region suitable for comparison to the N-terminal of JnCML-like (Figure 4); the C-terminal regions of m-Calpain, grancalcin and JnCML-like were nearly identical.
3.1.2. Expression of JnCML-like
Real-time PCR (Figure 5) revealed that JnCML-like was most abundant in the TZ and interior sapwood, and present only at low levels in exterior sapwood. The transcript JnCML-like was five times more abundant in the TZ of trees harvested in the fall as compared to the TZ of the trees harvested in summer (Figure 5), and was greater than five times more abundant in the TZ in fall versus other xylem tissues, including interior and exterior sapwood harvested in summer, and exterior sapwood of trees harvested in fall.
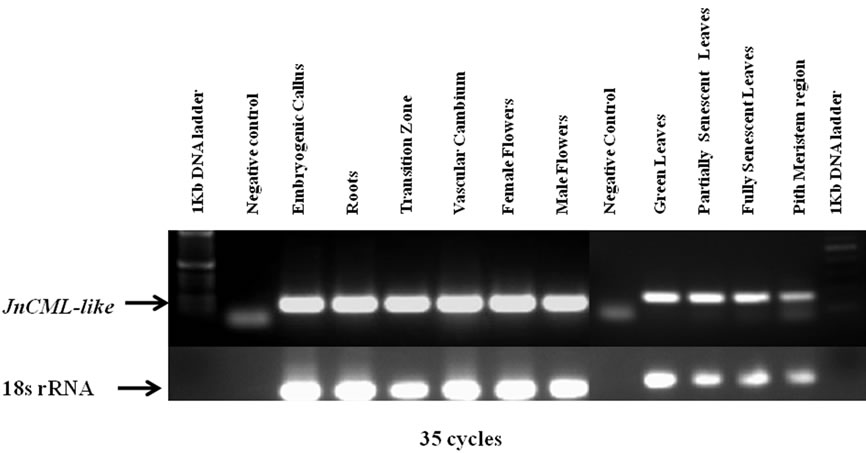
To further understand the role of JnCML-like in tree growth and development, and to determine if it is associated with differentiation and PCD in tissues other than the TZ, transcript abundance was quantified in vascular cambium, embryogenic callus, roots, female and male flowers, green leaves, and partially and fully senescent leaves. Quantitative real-time PCR showed that in these tissues, JnCML-like was expressed most abundantly in male flowers (76-fold greater than the vascular cambium zone tissue sample), followed by fully senescent leaves (six-fold greater), and female flowers (three-fold greater). JnCML-like was weakly expressed in other tissues including embryogenic calli, roots, green leaves, and partially senescent leaves, as compared to vascular cambium (Figure 6). Expression in these tissues (Figure 6) was not tested against those from the xylem tissues (Figure 5), so expression of JnCML-like in the TZ and in male flowers was not directly compared.
4. DISCUSSION
Analysis of sequence data indicated that JnCML-like is most likely a member of the PEF sub-family of calcium signaling proteins (Figure 3). Other members of the PEF family have roles in cell growth, cell death, and exocytosis [5]. JnCML-like was highly similar to m-calpain and grancalcin, proteins that are actively researched in animal systems [31-32], but their roles in plants are poorly understood. Calpains uniquely combine thiol protease activity and calmodulin-like EF-hands [33] (Figure 4), and calpains and caspases engage in cross-talk in the regulation of cell death in animals [34]. Aside from the previously mentioned DEK1, other putative calpain-like cysteine proteases from plants include Q6SSJ2 (Nictotiana benthamiana), OS02g47970 (Oryza sativa), and
 JnCML-like, m-Calpain, Grancalcin
JnCML-like, m-Calpain, Grancalcin
Figure 4. The predicted structures of JnCML-like, m-Calpain and grancalcin. The available structure of grancalcin is missing the N-terminus, as a consequence, the grancalcin model matches only the upper half of the JnCML-like model.
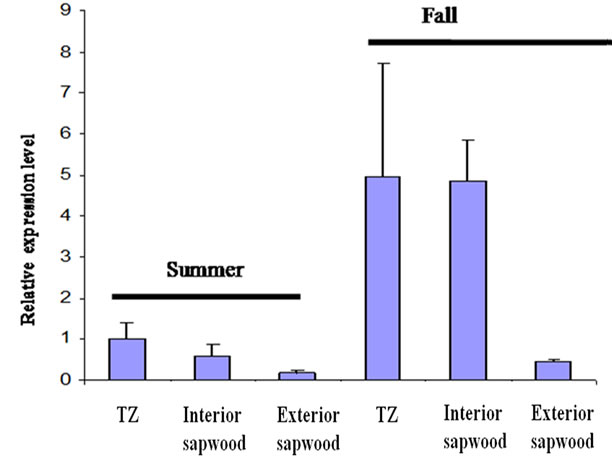
Figure 5. Transcript abundance of JnCML-like in the TZ, interior sapwood, and exterior sapwood in summer and fall. The fold changes were quantified by real-time PCR and were analyzed by the comparative CT method by comparison with the JnCML-like transcript level in TZ of the summer tree. Values are the means ± SD for three biological replicates. 18S rRNA was used as a standard.
 (a)
(a) (b)
(b)
Figure 6. Quantification of JnCML-like expression in black walnut tissues using real-time PCR. 18S rRNA was used as a standard. The fold changes were quantified and analyzed by the comparative CT method, expression in the vascular cambium zone was set to unity for comparison.
Vv024818001 (Vitis vinifera). The high degree of similarity between grancalcin and JnCML-like indicates that the two proteins probably have similar functions, but it may be premature to assign JnCML-like to a protein sub-family until additional gene models and roles for PEF proteins are described in plants.
The details of cell death in or near the TZ of trees and its regulation are emerging slowly. Heartwood formation probably occurs in the autumn, and there is evidence that this is true in particular for black walnut [11,15-16]. If so, then the observed increase in expression of JnCML-like in the TZ in the fall may broadly reflect a role for calcium in regulating heartwood formation. Alternatively, it may simply reflect the adjustment of the xylem to stresses associated with leaf senescence, cold temperatures, and dormancy [35], but this would not explain why JnCML-like is expressed at higher levels in the TZ than in the sapwood. The abundance of JnCML-like transcript in xylem tissues harvested in both summer and fall displayed a radial gradient increasing toward the heartwood. This pattern was consistent with the observed distribution of phospholipase C, and sucrose synthase in the trunkwood of R. pseudoacacia [19, 36], and starch in four tropical tree species Anacardium excelsum, Luehea seemannii, Cecropia longipes, and Urera caracasana [37]. Magel [38] proposed sucrose synthase as a biomarker for heartwood formation because it reflects the metabolic demand associated with the biosynthesis of phenolic extractives deposited in heartwood. We found that JnCML-like expression was not limited to xylem, although JnCML-like transcript abundance is low in healthy vegetative tissues. By examining the expression of JnCML-like throughout the course of tree development, we hoped to correlate its expression with specific physiological processes. For example, we reasoned that pith formation and leaf senescence, developmental processes that require PCD [39-40], might be more convenient models for understanding the function of JnCML-like. In black walnut, the pith is prominent and distinctive, and its dark color is similar to heartwood (Figure 1(b)). Similarly, leaf sensecence is a tractable phenotype in an accessible tissue. We were surprised to find that JnCML-like was expressed at relatively low levels in the region of the stem where pith forms and at the onset of leaf senescence, although JnCML-like expression seemed to increase in abundance as senescence progressed (Figure 6). The high level of expression of JnCML-like in male flowers may indicate that it plays an important role in pollen formation, which also requires cell death [39].
5. CONCLUSIONS
Sequence alignment and similarity of predicted structure support the view that JnCML-like isolated from black walnut is related to intracellular calcium perception and or response. Its possible orthologs include several members of the PEF family of calcium-dependent signal transduction elements. PEF proteins in mammals play a role in regulating cell growth, death, and exocytosis, but their role in plants is still under investigation. Real-time PCR and semi-quantitative RT-PCR revealed that JnCML-like was expressed at low levels in healthy, vegetative tissues. Interestingly, JnCML-like was expressed abundantly in male flowers, indicating that this gene might play an important role in male development through calcium physiology. Finally, our results show that JnCML-like transcript is more abundant in the fall than the summer and more highly expressed in TZ than all other xylem tissues, suggesting a possible role for this gene in the development of heartwood.
6. ACKNOWLEDGEMENTS
We thank Drs. Chung-Jui Tsai and Maureen McCann for useful suggestions, and Charles Michler for providing embryogenic calli. Funds for this research were provided by the United States Department of Agriculture Forest Service and the Department of Forestry and Natural Resources at Purdue University. The use of trade names is for the information and convenience of the reader and does not imply official endorsement or approval by the United States Department of Agriculture or the Forest Service of any product to the exclusion of others that may be suitable.
REFERENCES
- Henikoff, S., Greene, E.A., Pietrokovski, S., Bork, P., Attwood, T.K. and Hood, L. (1997) Gene families: The taxonomy of protein paralogs and chimeras. Science, 278, 609-637. doi:10.1126/science.278.5338.609
- Nakayama, S., Kawasaki, H. and Krestinger, R. (2000) Evolution of EF-hand proteins. In: Carafoli, E. and Krebs, J., Eds., Calcium Homeostasis, Springer, New York.
- Kretsinger, R.H. and Nockolds, C.E. (1973) Carp muscle calcium-binding protein II, Structure determination and general description. Journal of Biological Chemistry, 248, 3313-3326.
- Day, I., Reddy, V.S., Ali, G.S. and Reddy, A.S.N. (2002) Analysis of EF-hand-containing proteins in Arabidopsis. Genome Biology, 3, 1-24. doi:10.1186/gb-2002-3-10-research0056
- Nagata, T., Iizumi, S., Satoh, K., Ooka, H., Kawai, J., Carninci, P., Hayashizaki, Y., Otomo, Y., Murakami, K., Matsubara, K. and Kikuchi, S. (2004) Comparative analysis of plant and animal calcium signal transduction element using plant full-length cDNA data. Molecular Biology and Evolution, 21, 1855-1870. doi:10.1093/molbev/msh197
- Maki, M., Kitaura, Y., Satoh, H., Ohkouchi, S. and Shibata, H. (2002) Structures, functions and molecular evolution of the penta-EF-hand Ca2+ binding proteins. Biochim Biophysica Acta, 1600, 51-60.
- Johnson, K.L., Faulkner, C., Jeffree, C.E. and Ingram, G.C. (2008) The phytocalpain Defective Kernel 1 is a novel arabidopsis growth regulator whose activity is regulated by proteolytic processing. Plant Cell, 20, 2619-2630. doi:10.1105/tpc.108.059964
- Spicer, R. (2005) Senescence in secondary xylem: Heartwood formation as an active developmental program. In: Holbrook, N.M. and Zwieniecki, M.A., Eds., Vascular Transport in Plants, Elsevier Academic Press, Amsterdam, 457-475. doi:10.1016/B978-012088457-5/50024-1
- Andrews, J.A. and Siccama, T.G. (1999) Restranslocation of calcium and magnesium at the heartwood sapwood boundary of Atlantic white cedar. Ecology, 76, 659-663. doi:10.2307/1941225
- Berthier, S., Kokutse, A.D., Stokes, A. and Fourcaud, T. (2001) Irregular heartwood formation in Maritime pine (Pinus pinaster Ait.): Consequences for biomechanical and hydraulic tree functioning. Annals of Botany, 87, 19-25. doi:10.1006/anbo.2000.1290
- Dehon, L., Macheix, J.J. and Durand, M. (2002) Involvement of peroxidases in the formation of the brown coloration of heartwood in Juglans nigra. Journal of Experimental Botany, 53, 303-311. doi:10.1093/jexbot/53.367.303
- Yang, J., Kamdem, D.P., Keathley, D.E. and Han, K.H. (2004) Seasonal changes in gene expression at the sapwood-heartwood transition zone of black locust (Robinia pseudoacacia) revealed by cDNA microarray analysis. Tree Physiology, 24, 461-474.
- Spicer, R. and Holbrook, N.M. (2007) Parenchyma cell respiration and survival in secondary xylem: Does metabolic activity decline with age? Plant and Cell Environment, 30, 934-943. doi:10.1111/j.1365-3040.2007.01677.x
- Nakaba, S., Kubo, T. and Funada, R. (2008) Differences in patterns of cell death between ray parenchyma cells and ray tracheids in the conifers Pinus densiflora and Pinus rigida. Trees, 22, 623-630. doi:10.1007/s00468-008-0220-0
- Hillis, W.E. (1987) Heartwood and tree exudates. Springer-Verlag, Berlin, New York.
- Nelson, N.D. (1978) Xylem ethylene, phenol-oxidizing enzymes and nitrogen and heartwood formation in walnut and cherry. Canadian Journal of Botany, 56, 626-634. doi:10.1139/b78-070
- Frey-Wyssling, A. and Bosshard, H.H. (1959) Cytology of the ray cells in sapwood and heartwood. Holzforschung, 13, 129-137. doi:10.1515/hfsg.1959.13.5.129
- Bosshard, H.H. (1965) Aspects of the ageing process in cambium and xylem. Holzforschung, 19, 65-69. doi:10.1515/hfsg.1965.19.3.65
- Hauch, S. and Magel, E. (1998) Extractable activities and protein content of sucrose phosphate synthase, sucrose synthase and neutral invertase in trunk tissues of Robinia pseudoacacia L. are related to cambial wood production and heartwood formation. Planta, 207, 266-274. doi:10.1007/s004250050482
- Chapple, C.S., Vogt, T., Ellisyba, B.E. and Somerville, C.R. (1992) An Arabidopsis mutant defective in the general phenylpropanoid pathway. Plant Cell, 4, 1413-1424.
- Kolosova, N., Miller, B., Ralph, S., Ellis, B.E., Douglas, C., Ritland, K. and Bohlmann, J. (2004) Isolation of high-quality RNA from gymnosperm and angiosperm trees. Biotechnology, 36, 821-824.
- Zuker, M. (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Research, 31, 3406-3415. doi:10.1093/nar/gkg595
- Huang, Z., Tsai, C.J., Harding, S., Meilan, R. and Woeste, K.E. (2009) A Cross-species transcriptional profile analysis of heartwood formation in black walnut. Plant Molecular Biology Reporter, 28, 222-230. doi:10.1007/s11105-009-0144-x
- Kim, B-R., Nam, H-Y., Kim, S-I. and Chang, Y. J. (2004) Normalization of reverse transcription quantitative-PCR with housekeeping genes in rice. Biotech Letters, 25, 1869-1872. doi:10.1023/A:1026298032009
- Larkin, M.A., Blackshields, G., Brown, N.P., Chenna, R., McGettigan, P.A., McWilliam, H., Valentin, F., Wallace, I.M., Wilm, A., Lopez, R., Thompson, J.D., Gibson, T.J. and Higgins, D.G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics, 23, 2947-2948. doi:10.1093/bioinformatics/btm404
- Papadopoulos, J.S. and Agarwala, R. (2007) COBALT: Constraint-based alignment tool for multiple protein sequences. Bioinformatics, 23, 1073-1079. doi:10.1093/bioinformatics/btm076
- Liu, S., Zhang, C., Liang, S. and Zhou, Y. (2007) Fold recognition by concurrent use of solvent accessibility and residue depth. Proteins, 68, 636-645. doi:10.1002/prot.21459
- Eswar, N., Eramian, D., Webb, B., Shen, M.Y. and Sali, A. (2008) Protein structure modeling with MODELLER. Methods in Molecular Biology, 426,145-159. doi:10.1007/978-1-60327-058-8_8
- Jackson, D., Veit, B. and Hake, S. (1994) Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development, 120, 405-413.
- Schwacke, R., Schneider, A., Van Der Graaff, E., Fischer, K., Catoni, E., Desimone, M., Frommer, W.B., Flugge, U.I. and Kunze, R. (2003) ARAMEMNON, a novel database for arabidopsis integral membrane proteins. Plant Physiology, 131, 16-26. doi:10.1104/pp.011577
- Xu, P., Roes, J., Segal, A.W. and Radulovic, M. (2006) The role of grancalcin in adhesion of neutrophils. Cellular Immunology, 240, 116-121. doi:10.1016/j.cellimm.2006.07.004
- Shao, H., Chou, J., Baty, C.J., Burke, N.A., Watkins, S.C., Stolz, D.B. and Wells, A. (2006) Spatial localization of m-Calpain to the plasma membrane by phosphoinositide bisphosphate binding during epidermal growth factor receptor-mediated activation. Molecular Cell Biology, 26, 5481-5496. doi:10.1128/MCB.02243-05
- Hosfield, C.M., Elce, J.S., Davies, P.L. and Jia, Z. (1999) Crytal structure of calpain reveals the structural basis for Ca2+-dependent protease activity and a novel mode of enzyme activation. EMBO Journal, 18, 6880-6889. doi:10.1093/emboj/18.24.6880
- Orrenius, S., Zhivotovsky, B. and Nicotera, P. (2003) Regulation of cell death: The calcium apoptosis link. Nature Reviews Molecular Cell Biology, 4, 552-565. doi:10.1038/nrm1150
- Lautner, S. and Fromm, J. (2009) Calcium-dependent physiological processes in trees. Plant Biology, 12, 268- 274. doi:10.1111/j.1438-8677.2009.00281.x
- Hillinger, C., Holl, W. and Ziegler, H. (1996) Lipids and lipolytic enzymes in the trunkwood of Robinia pseudoacacia L. during heartwood formation. Tree, 10, 376-381.
- Newell, E. A., Mulkey, S.S. and Wright, S.J. (2002) Seasonal patterns of carbohydrate storage in four tropical tree species. Oecologia, 131, 333-342. doi:10.1007/s00442-002-0888-6
- Magel, E. (2000) Biochemistry and physiology of heartwood formation. In: Savidge, R., Barnett, J. and Napier, R., Eds., Molecular and Cell Biology of Wood Formation. BIOS Scientific Publishers, Oxford.
- van Doorn, W.G. and Woltering, E.J. (2005) Many ways to exit? Cell death categories in plants. Trends in Plant Science, 10, 117-122. doi:10.1016/j.tplants.2005.01.006
- Lim, P.O., Kim, H.J. and Nam, H.G. (2007) Leaf senescence. Annual Review Plant Physiology, 58, 115-136.
Abbreviations
EST Expressed sequence tag
RACE Rapid amplification of cDNA ends
RT-PCR Reverse transcription polymerase chain reaction

