 Journal of Biomaterials and Nanobiotechnology, 2012, 3, 499-507 http://dx.doi.org/10.4236/jbnb.2012.324051 Published Online October 2012 (http://www.SciRP.org/journal/jbnb) 499 Effects of Physical Parameters on Bacterial Cell Adsorption onto Pre-Imprinted Sol-Gel Films Jeanna Starosvetsky1, Tally Cohen1, Uta Cheruti1, Dragoljub Bilanović2, Robert Armon1* 1Faculty of Civil and Environmental Engineering Division of Environmental, Water and Agricultural Engineering, Technion, Haifa, Israel; 2Center for Environmental, Earth, and Space Studies, Bemidji State University, Bemidji, USA. Email: *cvrrobi@tx.technion.ac.il Received April 30th, 2012; revised June 15th, 2012; accepted July 16th, 2012 ABSTRACT Organically modified silica (ORMOSILS) thin films produced by sol-gel method were imprinted with two bacterial strains as whole cells in order to develop an easy, fast and specific probe to detect and specifically identify these micro- organisms when present in water samples. An important feature of the imprinting process was the molecular finger- prints left by these microorganisms alongside morphology, into imprinted film cavities. The films also showed high selectivity toward the imprinted template and were able to discriminate between two very close bacterial species (E. coli and S. typhimurium). In addition, several central physical parameters of the experimental water solution were examined (i.e., pH, ionic strength and the organic load exemplified by NaCl and TOC concentration, respectively). The method sensitivity to different bacterial concentrations was studied by confocal microscopy (CLSM) and quartz crystal micro- balance (QCM) tools. Results showed that increased bacterial concentrations favor rapid adsorption onto imprinted sol-gel films with high affinity, while low pH, increased organic load and high ionic concentrations (i.e., seawater) in- terfere with bacteria re-adsorption, reducing detection capability. Under average drinking water chemical composition the method proved to be highly efficient. Keywords: Detection; Imprinting; Sol-Gel; Bacteria; QCM; E. coli O157:H7; S. typhimurium; Sphaerotilus natans; CLSM 1. Introduction The source of 80% of gastrointestinal illnesses in devel- oping countries is caused by consumption of polluted drinking water. As already pointed out, “waterborne dis- ease probably accounts for more than 2.5 million death/ year with a majority of children under 5 years of age” [1]. In view of this fact, rapid, economic and accurate identi- fication and quantification of indicator or pathogenic microorganisms present in these environments is a ne- cessity for public health improvement. There are a number of current methods to identify and quantify bacteria in water in routine use, practiced by most of water laboratories around the globe. For example, MPN (most probable number) is the method that pro- vides a statistic estimate of viable and culturable bacte- rial cell numbers [2]. The technique is applicable to all kinds of samples, however it is most advantageous in turbid and marine waters were direct filtration may be hindered. Variations of this method have been developed for all faecal indicator bacteria in question: E. coli, coli- forms, enterococci, bacteriophages and C. perfringens [3-5]. However, the main drawback of this method is that MPN procedure demands long incubation time and large incubator space due to the numerous experimental tubes required. Another highly used method is membrane fil- tration (MF), an alternative method for identification and quantification of bacteria present in water samples [2]. Following capture of bacteria onto membrane surface, the filter is placed onto various selective media and the detection of specific microorganisms is possible by de- velopment of visible colonies under the restriction of discriminatory substrates. The final result is expressed by colonies growth coupled with color development. The newly developed colonies are counted and identified based on biochemical tests or genotypically by poly- merase chain reaction (PCR) and electrophoresis [3]. The MF method in spite of its global acceptance in water mi- crobiology laboratories still has several drawbacks: rela- tively long incubation times, occasionally false positive results and requirement for relatively clear water sam- ples. Based on specific and different biochemical pathways of bacteria, other methods were developed (i.e., Colilert) but still presenting sometimes false positive/negative *Corresponding author. Copyright © 2012 SciRes. JBNB  Effects of Physical Parameters on Bacterial Cell Adsorption onto Pre-Imprinted Sol-Gel Films 500 results or yet relatively long time [6]. Consequently based on this knowledge, there is a need for a rapid, efficient, relatively simple and inexpensive method for detection of microorganisms present in water. Cohen et al. [7] showed that macromolecular imprinting of sol-gel surfaces with specific bacteria provides an in- teresting mean of bacteria detection in liquid samples. Molecular imprinting is a method in which an appro- priate functional monomer is polymerized in the presence of a template forming covalent or non-covalent bonds with the functional monomer [8]. Afterward, following polymerization process, the template is removed that leaves a space or cavity originally occupied by the tem- plate molecule, in its original shape. These cavities due to their steric shape can adopt and bind similar or analo- gous molecule with pronounced selectivity and sensitiv- ity [9]. The next step in this interesting methodology was to imprint much more complex organic structures such as whole cells in sol-gel matrices, termed macromolecular imprinting [7]. The imprinting of whole cells into inorganic sol-gel matrices has been practiced for the past fifteen years, pioneered by the study of Carturan et al., [10] who en- capsulated S. cerevisiae yeast into silica gel and studied its catalytic activity. Barreau et al. [11] and Livage [12] showed that entrapment of parasitic protozoa in sol-gel does not affect cells’ structure. Imprinting of whole cells in sol-gel was reported by Chia [13], and further devel- oped by Dickert [14,15]. These authors manufactured a polyurethane surface imprinted with S. cerevisiae yeasts allowing a selective and highly sensitive enrichment of the microorganisms under flow conditions. As already shown in previous studies, different types of microor- ganisms can also be detected by molecular imprinted polymers [7,15,16]. Detection and quantification of whole yeast cells imprinted on films made of polyure- thane and alkoxide through sol-gel technique was ob- tained by application of QCM [14]. Bacteria and viruses are very small microorganisms (in the range of 1 - 3 μm and 30 to 700 nm, respectively) that behave as colloidal particles in aqueous systems and therefore can be considered as living colloids. Due to their colloidal behavior, many biological aspects of their adsorption on surfaces can be explained in terms of their physicochemical properties. It is not clear whether the primary factors controlling the degree of bacterial asso- ciation with surfaces are related to solid interface capac- ity to support growth, strictly to physicochemical interac- tions or to some permutation of these factors [17]. In a previous study it was shown that imprinted bacteria onto sol-gel surfaces created cavities that allowed for a sec- ondary adsorption of similar bacteria, based on steric similarity [7]. The present research was aimed to reveal the solution physicochemical properties’ effect on the re-adsorption of bacteria onto imprinted sol-gel surface. Among these parameters bacterial cells and NaCl con- centration, pH, TOC were assessed for their impact on the specific adsorption of bacteria to whole-cell-im- printed sol-gel thin films, through live cell imaging (confocal microscopy) and quartz crystal microbalance as evaluating tools. 2. Materials and Methods 2.1. Bacterial Cultures Two members of the family Enterobacteriaceae were used in the present study: E. coli and Salmonella typhi- murium. The first one was selected as a model of indica- tor bacteria (E. coli CN13) and as a known pathogen (E. coli O157:H7) while the second member is a known pathogen with phenotypic and genotypic close similarity to the first one (S. typhimurium). E. coli CN13 (an E. coli C that lacks restriction system against bacteriophages and harbors nalidixic acid resis- tance, isolated in our laboratory was grown in nutrient broth (Difco, USA) for 16 hours at 35˚C ± 1˚C [18]. The grown culture was further centrifuged for 10 minutes at 6000 x g (Sorvall T 6000D). Supernatant medium was discarded and concentrated cells were washed twice with saline (0.85%) and finally resuspended in saline. S. ty- phimurium (ATCC 14028) (G-) was grown on McCon- key agar (Difco, USA) for 16 hour at 35˚C ± 1˚C. Grown colonies were harvested and washed similarly to E. coli CN13 and suspended in saline. E. coli O157:H7 was grown on R2A agar (Acumedia, USA) for 16 hours at 35˚C, harvested and suspended in saline. 2.2. Sol-Gel Thin Films Sol gel solution and thin films were prepared as already described [7,19,20]. Briefly, a tetraethoxysilane (TEOS, 98%, ABCR Co., Germany) precursor was used as bulk sol-gel solution. TEOS (2 ml) was mixed with triple- distilled water (1 ml) and gently stirred for 10 min at room temperature (22˚C ± 1˚C). Next, 0.1 M HCl (0.1 N, 0.25 ml) was added to the solution which was further stirred for 3 - 5 hours until visible homogeneity. The fi- nal solution was aged at 4˚C ± 1˚C for 24 hours and kept for approximately 5 weeks at 4˚C. 2.3. Macromolecular Imprinting Procedure Sol-gel films (SG) were prepared on standard microscope glass slides previously cleaned according to a modified procedure [7]. Briefly, glasses were soaked for 20 - 30 min successively in 1:1:1 acetone-ethanol-chloroform mixture; soap solution; Piranha solution: H2SO4:H2O2 (4:1), subsequently exhaustively rinsed in double-dis- tilled water and finally dried at 100˚C for 2 - 4 hours. Copyright © 2012 SciRes. JBNB  Effects of Physical Parameters on Bacterial Cell Adsorption onto Pre-Imprinted Sol-Gel Films 501 The imprinting procedure was as following: aged sol- gel stock solution (0.5 ml) was added to a bacterial sus- pension (3 ml, 109 - 1010 CFU/ml) and stirred for 8 to 10 minutes. Subsequently a glass slide was drop-coated with 0.25 ml of the experimental mixture. To obtain a thin and uniform film, the glass slide was immediately positioned vertically to drain excess solution. Slides coated with sol-gel films containing live bacteria were placed into a desiccator and dried overnight at room temperature. Re- moval of immobilized bacterial cells from dried sol-gel film was performed by soaking the coated slides for 30 - 40 min in 96% ethanol and washed with sterile dou- ble-distilled water. Slides were further Gram stained (successively 3 min in crystal violet, iodine, 70% ethanol, safranin for 3 minutes each) (Bactolab Diagnostics, Israel) to visually detect cavities formed by entrapped bacteria (Figure 1(A)). It is important to note that in many cases immobilized bacterial cells dropped just after Gram staining procedure short of ethanol treatment. 2.4. Confocal Microscopy (CLSM) In order to monitor the adsorption process with free live bacteria, live imaging was performed on Leica CLSM microscope. Pre-cut and imprinted microscopic slides were glued to Petri dish and let dry for 2 hours. A water dip objective lens (× 63) was used for confocal bright- field scan, in altered solutions, for example: different bacteria concentrations (102 - 107 cells/ml), various pH-s (2.5, 5.3 and 7.5), TOC (1.3, 13 and 60 mg/ml) and NaCl concentrations (1.4, 140 and 300 mM). All experiments were performed with a culture of E. coli CN13 pre-stained with 0.001 gr/ml methylene blue (Spectrum, USA) for 10 minutes, washed twice by cen- trifugation with 0.85% saline to reach a concentration of ~1 - 4 × 107 cells/ml (counted with hemocytometer). In a small petri dish, 1 ml of bacterial suspension was added to 5 ml solution and a scan was made every 30 seconds, for the duration of one hour. Cell adhesion count was performed on each frame. 2.5. Immunofluorescence In order to discriminate between the two enterobacterial strains (E. coli O157:H7 and S. typhimurium) im- munofluorescence was applied. Thin films imprinted with either strain were prepared as mentioned above. Entrapped bacteria were removed, and both types of the imprinted films were exposed to Salmonella typhimurium, E. coli, or a mixed suspension (107 cell/ml mixed culture with identical concentration of both bacterial strains). Attachment of bacteria to cavities formed on the im- printed film surface was detected by immunofluores- cence. Cells were fixed with 4% formaldehyde, washed with cold PBS buffer and blocked with bovine serum albumin (BSA) (1% BSA in PBS) for four hours. Cells were detected by specific antibodies. E. coli O157:H7 was detected with specific antibody (Goat anti E. coli, Zotal, Israel) conjugated with FITC (fluoresceinisothio- cyanate, excitation/emission-488/520 nm). S. typhimu- rium was detected with primary antibody (mouse mono- clonal to S. typhimurium, Zotal, Israel) and a secondary antibody (Goat anti mouse, affinity purified, Zotal, Israel) conjugated with DyLight 594© (excitation/emission-593/ 618). Primary antibodies were incubated at room tem- perature overnight and washed with PBS. Secondary an- tibodies were incubated for two hours at room tempera- ture and washed with PBS. Microscopical observations were performed with Leica Confocal microscope (CLSM) in dual channel state. 2.6. QCM Measurements Procedure Sensitivity assessment of the imprinted sol-gel films for their respective bacterial cells was performed by meas- urements within a flow cell connected to quartz crystal microbalance apparatus (QCM922, Princeton Applied Research). Measurements were made on gold electrode equipped with 9 MHz quartz crystals. Thin film coated crystals were prepared by smear-coating procedure with sol-gel stock solution mixed with bacterial suspension (0.85% saline) following over-night desiccation. Immo- bilized bacterial cells removal from cured sol-gel film was performed by coated crystals immersion in ethanol (96%) for 30 to 40 minutes and additional rinse with ster- ile double distilled water (SDDW). Control crystals were coated with sol-gel films without imprinted bacteria (termed non-imprinted film) and rinsed with suspension medium (0.85% NaCl w/v + 0.2% Tween-80, v/v). The coated crystals (imprinted and non-imprinted) were exposed to UV lamp for 10 minutes, to attain steady measurements. Subsequently, the crystals were inserted in the flow cell and rinsed with SDDW. Once a stable measurement with SDDW was reached, the crystals were washed with the basic solution for additional 20 minutes and finally exposed to experimental bacterial solution accordingly. Measurements were performed by flow of different solutions containing different concentrations of bacteria over the detector (flow cell, 0.5 ml/minute, Cole- Parmer Masterflex L/S pump). 3. Results and Discussion 3.1. Method’s Selectivity In an earlier study it was shown that imprinted sol-gel films differentiate between types of microorganisms based on shape and size variation and only cavities-spe- cific adsorption takes place (Figures 1(A) and (B)) [7]. In the present study different techniques were applied in Copyright © 2012 SciRes. JBNB 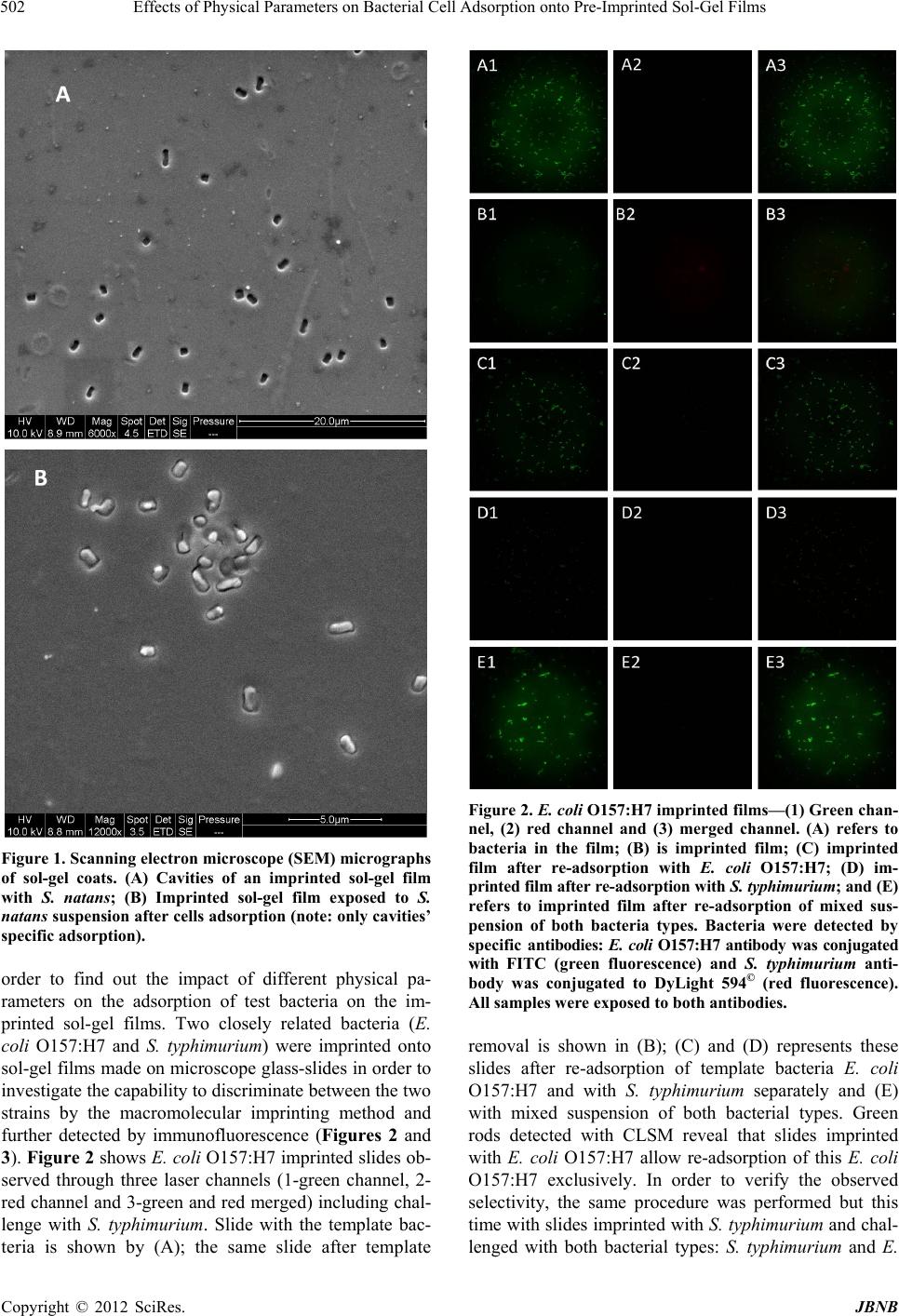 Effects of Physical Parameters on Bacterial Cell Adsorption onto Pre-Imprinted Sol-Gel Films 502 Figure 1. Scanning electron microscope (SEM) micrographs of sol-gel coats. (A) Cavities of an imprinted sol-gel film with S. natans; (B) Imprinted sol-gel film exposed to S. natans suspension after cells adsorption (note: only cavities’ specific adsorption). order to find out the impact of different physical pa- rameters on the adsorption of test bacteria on the im- printed sol-gel films. Two closely related bacteria (E. coli O157:H7 and S. typhimurium) were imprinted onto sol-gel films made on microscope glass-slides in order to investigate the capability to discriminate between the two strains by the macromolecular imprinting method and further detected by immunofluorescence (Figures 2 and 3). Figure 2 shows E. coli O157:H7 imprinted slides ob- served through three laser channels (1-green channel, 2- red channel and 3-green and red merged) including chal- lenge with S. typhimurium. Slide with the template bac- teria is shown by (A); the same slide after template Figure 2. E. col i O157:H7 imprinted films—(1) Green chan- nel, (2) red channel and (3) merged channel. (A) refers to bacteria in the film; (B) is imprinted film; (C) imprinted film after re-adsorption with E. coli O157:H7; (D) im- printed film after re-adsorption with S. typhimurium; and (E) refers to imprinted film after re-adsorption of mixed sus- pension of both bacteria types. Bacteria were detected by specific antibodies: E. coli O157:H7 antibody was conjugated with FITC (green fluorescence) and S. typhimurium anti- body was conjugated to DyLight 594© (red fluorescence). All samples were exposed to both antibodies. removal is shown in (B); (C) and (D) represents these slides after re-adsorption of template bacteria E. coli O157:H7 and with S. typhimurium separately and (E) with mixed suspension of both bacterial types. Green rods detected with CLSM reveal that slides imprinted with E. coli O157:H7 allow re-adsorption of this E. coli O157:H7 exclusively. In order to verify the observed selectivity, the same procedure was performed but this time with slides imprinted with S. typhimurium and chal- lenged with both bacterial types: S. typhimurium and E. Copyright © 2012 SciRes. JBNB 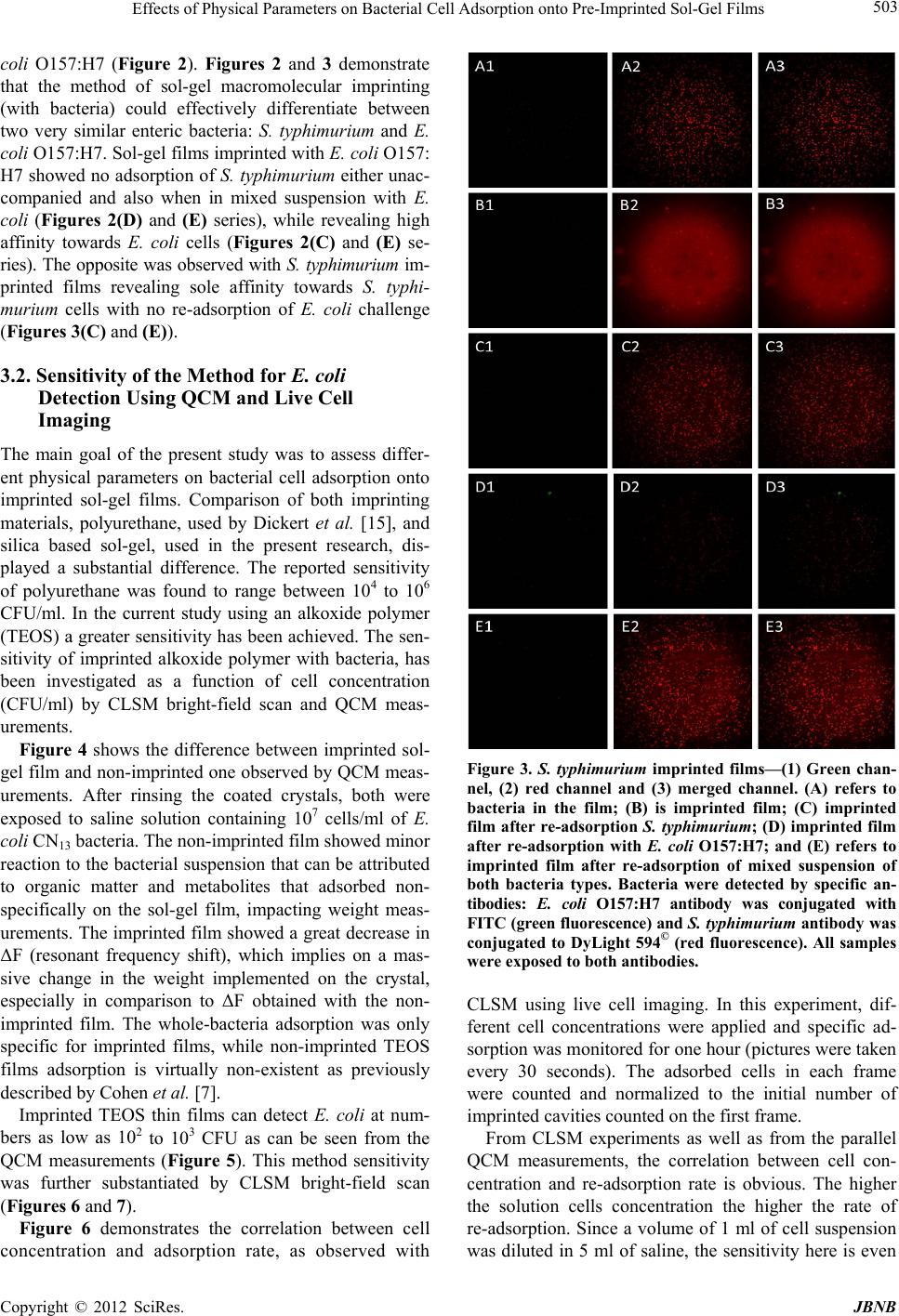 Effects of Physical Parameters on Bacterial Cell Adsorption onto Pre-Imprinted Sol-Gel Films 503 coli O157:H7 (Figure 2). Figures 2 and 3 demonstrate that the method of sol-gel macromolecular imprinting (with bacteria) could effectively differentiate between two very similar enteric bacteria: S. typhimurium and E. coli O157:H7. Sol-gel films imprinted with E. coli O157: H7 showed no adsorption of S. typhimurium either unac- companied and also when in mixed suspension with E. coli (Figures 2(D) and (E) series), while revealing high affinity towards E. coli cells (Figures 2(C) and (E) se- ries). The opposite was observed with S. typhimurium im- printed films revealing sole affinity towards S. typhi- murium cells with no re-adsorption of E. coli challenge (Figures 3(C) and (E)). 3.2. Sensitivity of the Method for E. coli Detection Using QCM and Live Cell Imaging The main goal of the present study was to assess differ- ent physical parameters on bacterial cell adsorption onto imprinted sol-gel films. Comparison of both imprinting materials, polyurethane, used by Dickert et al. [15], and silica based sol-gel, used in the present research, dis- played a substantial difference. The reported sensitivity of polyurethane was found to range between 104 to 106 CFU/ml. In the current study using an alkoxide polymer (TEOS) a greater sensitivity has been achieved. The sen- sitivity of imprinted alkoxide polymer with bacteria, has been investigated as a function of cell concentration (CFU/ml) by CLSM bright-field scan and QCM meas- urements. Figure 4 shows the difference between imprinted sol- gel film and non-imprinted one observed by QCM meas- urements. After rinsing the coated crystals, both were exposed to saline solution containing 107 cells/ml of E. coli CN13 bacteria. The non-imprinted film showed minor reaction to the bacterial suspension that can be attributed to organic matter and metabolites that adsorbed non- specifically on the sol-gel film, impacting weight meas- urements. The imprinted film showed a great decrease in ΔF (resonant frequency shift), which implies on a mas- sive change in the weight implemented on the crystal, especially in comparison to ΔF obtained with the non- imprinted film. The whole-bacteria adsorption was only specific for imprinted films, while non-imprinted TEOS films adsorption is virtually non-existent as previously described by Cohen et al. [7]. Imprinted TEOS thin films can detect E. coli at num- bers as low as 102 to 103 CFU as can be seen from the QCM measurements (Figure 5). This method sensitivity was further substantiated by CLSM bright-field scan (Figures 6 and 7). Figure 6 demonstrates the correlation between cell concentration and adsorption rate, as observed with Figure 3. S. typhimurium imprinted films—(1) Green chan- nel, (2) red channel and (3) merged channel. (A) refers to bacteria in the film; (B) is imprinted film; (C) imprinted film after re-adsorption S. typhimurium; (D) imprinted film after re-adsorption with E. coli O157:H7; and (E) refers to imprinted film after re-adsorption of mixed suspension of both bacteria types. Bacteria were detected by specific an- tibodies: E. coli O157:H7 antibody was conjugated with FITC (green fluorescence) and S. typhimurium antibody was conjugated to DyLight 594© (red fluorescence). All samples were exposed to both antibodies. CLSM using live cell imaging. In this experiment, dif- ferent cell concentrations were applied and specific ad- sorption was monitored for one hour (pictures were taken every 30 seconds). The adsorbed cells in each frame were counted and normalized to the initial number of imprinted cavities counted on the first frame. From CLSM experiments as well as from the parallel QCM measurements, the correlation between cell con- centration and re-adsorption rate is obvious. The higher the solution cells concentration the higher the rate of re-adsorption. Since a volume of 1 ml of cell suspension was diluted in 5 ml of saline, the sensitivity here is even Copyright © 2012 SciRes. JBNB  Effects of Physical Parameters on Bacterial Cell Adsorption onto Pre-Imprinted Sol-Gel Films 504 Figure 4. E. coli specific adsorption onto SG imprinted films as detected by QCM. Black line refers to SG film imprinted with E. coli bacteria and cells elution; gray line refers to control (non-imprinted sol-gel film) exposed to the same E. coli suspension. Figure 5. E. coli specific adsorption at different concentra- tions onto SG imprinted films (with E. coli) detected by QCM: Red line—0 cells/ml; green line—30 cells/ml; purple line—3 × 102 cells/ml; blue line—3 × 103 cells/ml; orange line—3 × 104 cells/ml; gray line—3 ×106 cells/ml. Figure 6. Specific adsorption kinetics of E. coli CN13 cells to imprinted slide, normalized to the initial number of im- printed cavities as observed with CLSM bright-field scan. lower than 102 cells/ml, and should be as low as 80 cells/ ml. 3.3. pH Effect on Bacterial Cell Adsorption onto Imprinted Sol-Gel Film Many ions are associated with normal cellular growth and development. Simple inorganic and organic ions establish both pH and ionic strength of a given system [21]. The overall metabolic function of these ions can be complex, but certain effects are distinct by the ability of such compounds to permeate cell walls, regulate osmotic pressure, facilitate transport and alter surface electrical charges [22]. These effects can be observed microscopi- cally through changes in cell shape and size, or by their interaction with other cells and surfaces. As shown in Figure 7, solution’s pH has great influence on adsorption, most likely as a consequence of changes in the zeta po- tential of the outer membrane [22]. The pH values for optimum adsorption depend on the relative isoelectric points of the microbial cell and the surface. In general, under physiological conditions, the optimal adsorption of bacterial cells onto surfaces occurs at pH range of 3 to 6 [21]. The results presented in Figure 7 reveal better re- adsorption equally in rate and in sheer numbers, at physiological pH of ~7. 3.4. Organic Matter Effect on Bacterial Cell Adsorption onto Imprinted Sol-Gel Film Stark et al. [23] found that measurable amounts of or- ganic matter accumulate within a few hours on chemi- cally clean glass slides immersed in lake water. They expressed the concept that the accumulation of organic nutrients (conditioning film) favors bacterial growth. Supporting evidence is given by the studies on the rela- tion between food concentration and solid surfaces [24]. Specific structures such as fimbriae [25] and pili [26] may be involved in attachment to specific substrates. However, in aqueous environments adhesion is believed to be mediated by extracellular polymers [27,28]. The extent to which the polysaccharides are involved in the adhesion process is however open to query. Some reports suggest their roles both in the initial, reversible phase of adhesion [29,30] and also in the later, irreversible phase [21,27,30]. Brown et al. [31] presented evidence sug- gesting that excess polymer production may even prevent adhesion, although trace amounts of polysaccharide might be initially required. Although this work does not involve biofilm formation and EPS production by the re-adsorbed bacteria, the role of organic matter in general Figure 7. Effect of solution pH on E. coli CN13 re-adsorption on imprinted TEOS films. Copyright © 2012 SciRes. JBNB  Effects of Physical Parameters on Bacterial Cell Adsorption onto Pre-Imprinted Sol-Gel Films 505 and cell envelope remnants on re-adsorption appears to be of great importance. As described previously, imprints of template cells contain proteins originating from cell membranes [7]. As can be seen in Figure 8, the total organic carbon (TOC) value higher than the one com- monly recorded in drinking water (>4 mg/L) [32] appears to prevent re-adsorption onto imprinted film, while drinking-water levels of TOC (between 0.7 and 4 mg/L) had no significant effect and re-adsorption occurred at the same rate as saline. 3.5. Effect of NaCl Concentration on Cell Re-Adsorption The influence of electrolyte concentration on adsorption processes, is well documented. In any water suspension, a certain potential exists between charged particles and bulk medium, therefore counter-ions are attracted to sur- face to form a diffusible double layer of ions. The thickness of the double layer is a function of the concentration (m-molar) and valency (Z) of the electro- lytes in solution. Double-layer thickness (1/k) is given by the equation k = 3.27 × 107 ZM1/2 [33]. In the present study, k equals 1.79 × 107 for 300, 1.22 × 107 for 140 mM and 1.22 × 106 for 1.4 mM of a NaCl solution. The repul- sive force encountered by a particle as it approaches a sur- face is a function of the double layer thickness. Bacterial attachment is generally reported to increase under electrolytes at low ionic strengths (0 to 0.1 M). At higher ionic strengths, such as those found in estuarine or marine waters, an attachment decline versus increasing salt concentration was generally observed [34]. Studies had also showed that salt solutions will reduce adsorption and increase desorption of some bacteria from ion-ex- change resins also through electrolytes bound to ad- sorption complex [35,36]. Addition of various inorganic salts to a water suspension may prevent adsorption and promote desorption from solid surfaces. Salts such as sodium chloride promote desorption, if adsorption had not became irreversible. Figure 9 demonstrates that sodium chloride concentrations above 140 mM prevent cell ad- sorption onto imprinted sol-gel surface (isotonic and hy- potonic solutions having no effect on cell re-adsorption). Figure 8. Effect of total organic content in the solution on E. coli CN13 re-adsorption on TEOS imprinted films. Finally, in order to assess the feasibility of the present method to detect and identify microorganisms in drinking water in real time, QCM measurements of E. coli CN13 re-adsorption onto imprinted films of TEOS sol-gel were performed. As shown in Figure 10, albeit tap water has a greater background compared to saline, a distinct decline in ∆F was evident following bacteria suspension intro- duction. This ∆F shift indicates that most tap water pa- rameters (e.g. salts, organic matter, trace metals etc.) have little effect on bacterial re-adsorption, therefore through re-adsorption specific bacteria can be specifi- cally and rapidly detected in tap water (when present). 4. Conclusions In the present study different parameters that may influ- ence re-adsorption of bacteria onto previously imprinted sol-gel films were studied. Ormosil (such as TEOS) im- printed with various enteric bacteria revealed a highly specific cell adsorption following exposure to planktonic cells in saline suspensions. The high affinity of imprinted sol-gel films towards planktonic cells was previously found to be determined by cavity morphology (rod, coc- cus or tetrad) and residual components if the microorgan- ism’s outer surface was entrapped into the cavity surface area [7]. These two characteristics were found to be Figure 9. Effect of different NaCl concentrations in solution on E. coli CN13 re-adsorption on TEOS imprinted films. Figure 10. Re-adsorption of E. coli CN13 (107 cells/ml) on imprinted TEOS thin films in tap water, measured by QCM. Copyright © 2012 SciRes. JBNB  Effects of Physical Parameters on Bacterial Cell Adsorption onto Pre-Imprinted Sol-Gel Films 506 specific enough to discriminate between the various bac- teria tested without interference between the different species when mixed together. Therefore the bacterial type is of great importance and even bacteria strains that have many similar properties (i.e. enterobacteriaceae) do not re-adsorb to cavities other than their own imprints. The method is consequently specific enough to differen- tiate between different bacteria. Application of QCM and CLSM bright-field scans revealed improved sensitivity of this recognition method compared with previous re- sults published by Dickert et al. [14,15]. Concentrations as low as 102 CFU/ml were detectable on QCM, and an even lower concentration (80 CFU/ml) was detectable with CLSM. The present study results have showed that water various parameters may influence re-adsorption onto imprinted sol-gel films (i.e. ionic strength, pH and total organic carbon (TOC)). Among these parameters, TOC showed a significant effect on the extent of bacte- rial cells re-adsorption. Application of this method for drinking water must consider these parameters for its successful implementation. However under normal con- ditions, such as potable water the method revealed high sensitivity and fast results. For further development of macromolecular imprinting method as a scientific tool for rapid examination of water samples to rapidly detect indicator and pathogenic bacte- ria, additional research is necessary related to commer- cial aspects of these imprinted films [37]. For instance the shelf-life of the imprinted SG films that is currently unknown. In the present research, SG films were used not late than two weeks after preparation, therefore es- tablishing their shelf life is very much required. 5. Acknowledgements The authors would like to thank the “Phyllis and Joseph Gurwin Fund for Scientific Advancement at the Tech- nion” for its financial support of the present study and to Dr. Moshe Herzberg (Ben-Gurion University) for his help on QCM application. REFERENCES [1] C. J. Woodall, “Waterborne Disease—What Are the Pri- mary Killers?” Desalination, Vol. 248, No. 1-3, 2009, pp. 616-621. doi:10.1016/j.desal.2008.05.110 [2] American Public Health Association, American Water Works Association and Water Pollution Control Feder- ation, “Standard Methods for the Examination of Water and Wastewater,” 21st Edition, American Public Health Association, Washington, 2005. [3] V. C. A. Junqueira, R. C. Neto, N. da Silva and J. H. Terra, “Comparison of Methods for the Enumeration of Clostridium Perfringens Perfringens Spores in Water,” Water Science and Technology, Vol. 65, No. 2, 2012, pp. 227-232. doi:10.2166/wst.2012.758 [4] W. J. Dore, K. Henshilwood and D. N. Lees, “Evaluation of F-Specific RNA Bacteriophage as a Candidate Human Enteric Virus Indicator for Bivalve Molluscan Shellfish,” Applied and Environmental Microbiology, Vol. 66, No. 4, 2000, pp. 1280-1285. doi:10.1128/AEM.66.4.1280-1285.2000 [5] Y. Kott and E. F. Glona, “Correlating Coliform Bacteria with Escherichia coli Bacteriophages in Shellfish,” Water & Sewage Works, Vol. 112, No. 11, 1965, pp. 424-426. [6] J. M. Pisciotta, D. F. Rath, P. A. Stanek, D. M. Flanery and V. J. Harwood, “Marine Bacteria Cause False-Positive Results in the Colilert-18 Rapid Identification Test for Escherichia coli in Florida Waters,” Applied and Envi- ronmental Microbiology, Vol. 68, No. 2, 2002, pp. 539- 544. doi:10.1128/AEM.68.2.539-544.2002 [7] T. Cohen, J. Starosvetsky, U. Cheruti and R. Armon, “Whole Cell Imprinting in Sol-Gel Thin Films for Bac- terial Recognition in Liquids: Macromolecular Finger- printing,” International Journal of Molecular Sciences, Vol. 11, No. 4, 2010, pp. 1236-1252. doi:10.3390/ijms11041236 [8] K. Mosbach, “Molecular Imprinting,” Trends in Bioche- mical Sciences, Vol.19, No. 1, 1994, pp. 9-14. doi:10.1016/0968-0004(94)90166-X [9] K. Mosbach and O. Ramström, “The Emerging Tech- nique of Molecular Imprinting and Its Future Impact on Biotechnology,” Nature Biotechnology, Vol. 14, No. 2, 1996, pp. 163-170. doi:10.1038/nbt0296-163 [10] G. Carturan, R. Campostrini, S. Diré, V. Scardi and E. De Alteriis “Inorganic Gels for Immobilization of Biocata- lysts: Inclusion of Invertase-Active Whole Cells of Yeast (Saccharomyces Cerevisiae) into Thin Layers of SiO2 Gel Deposited on Glass Sheets,” Journal of Molecular Ca- talysis, Vol. 57, No. 1, 1989, pp. L13-L16. doi:10.1016/0304-5102(89)80121-X [11] J. Y. Barreau, J. M. Da Costa, I. Desportes, J. Livage, L. Monjour and M. Gentilini, “Fixation and Immunological Reactivity of Parasitic Protozoa in Sol-Gel Matrices,” Proceedings of the French Academy of Science, Series III, Life Sciences, Vol. 317, No. 7, 1994, pp. 653-657. [12] J. Livage, C. Roux, J. M. Da Costa, I. Desportes and J.F. Quinson, “Immunoassays in Sol-Gel Matrices,” Journal of Sol-Gel Science and Technology, Vol. 7, No. 1-2, 1996, pp. 45-51. doi:10.1007/BF00401882 [13] J. U. Chia, F. Tamanoi and B. J. I. Z. Dunn, “Patterned Hexagonal Arrays of Living Cells in Sol-Gel Silica Films,” Journal of the American Chemical Society, Vol. 122, No. 27, 2000, pp. 6488-6489. doi:10.1021/ja0011515 [14] F. L. Dickert and G. Hayden, “Bioimprinting of Polymers and Sol-Gel Phases. Selective Detection of Yeasts with Imprinted Polymers,” Analytical Chemistry, Vol. 74, No. 6, 2002, pp. 1302-1306. doi:10.1021/ac010642k [15] F. L. Dickert, P. Lieberzeit and O. Hayden, “Sensor Stra- tegies for Microorganism Detection—From Physical Principles to Imprinting Procedures,” Analytical and Bio- analytical Chemistry, Vol. 377, No. 3, 2003, pp. 540-549. Copyright © 2012 SciRes. JBNB  Effects of Physical Parameters on Bacterial Cell Adsorption onto Pre-Imprinted Sol-Gel Films Copyright © 2012 SciRes. JBNB 507 doi:10.1007/s00216-003-2060-5 [16] O. Hayden, C. Haderspock and F. L. Dickert, “Microor- ganism Detection with Selective Bioimprinted Polymers and Sol-Gels,” International Journal of Bio-Chroma- tography, Vol. 6, No. 4, 2001, pp. 303-311. [17] S. G. Andrews and F. J. Millero, “Electrolyte Effects on Attachment of an Estuarine Bacterium,” Applied and En- vironmental Microbiology, Vol. 47, No. 3, 1984, pp. 495- 499. [18] R. Armon, M. Arella and P. Payment, “A Highly Effi- cient Second-Step Concentration Technique for Bacte- riophages and Enteric Viruses Using Ammonium Sulfate and Tween 80,” Canadian Journal of Microbiology, Vol. 34, No. 5, 1988, pp. 651-655. [19] D. Avnir, “Organic Chemistry within Ceramic Materials: Doped Sol-Gel Materials,” Accounts of Chemical Re- search, Vol. 28, No. 8, 1995, pp. 328-334. doi:10.1021/ar00056a002 [20] R. Zusman, C. Rotman, M. Ottolenghi and D. Avnir, “Doped Sol-Gel Glasses as Chemical Sensors,” Journal of Non- Crystalline Solids, Vol. 122, No. 1, 1990, pp. 107-109. doi:10.1016/0022-3093(90)90232-B [21] G. Bitton and K. C. Marshall, “Adsorption of Microor- ganisms to Surfaces,” Wiley-Interscience, Weinheim, 1980. [22] C. E. Zobell, “The Effect of Solid Surfaces upon Bacterial Activity,” Journal of Bacteriology, Vol. 46, No. 1, 1943, pp. 39-56. [23] W. H. Stark, J. Stadler and E. McCoy, “Some Factors Affecting the Bacterial Population of Freshwater Lakes,” Journal of Bacteriology, Vol. 36, No. 1, 1938, pp. 653- 654. [24] H. Heukelekian and A. Heller, “Relation between Food Concentration and Surface for Bacterial Growth,” Journal of Bacteriology, Vol. 40, No. 4, 1940, pp. 547-558. [25] M. Rosenberg, E. A. Bayer, J. Delarea and E. Rosenberg, “Role of Thin Fimbriae in Adherence and Growth of Acinetobacter Calcoaceticus RAG-1 on Hexadecane,” Applied and Environmental Microbiology, Vol. 44, No. 4, 1982, pp. 929-937. [26] R. L. Weiss, “Attachment of Bacteria to Sulphur in Ex- treme Environments,” Journal of General Microbiology, Vol. 77, No. 2, 1973, pp. 501-507. [27] M. Fletcher and G. D. Floodgate, “An Electron-Micro- scope Demonstration of an Acidic Polysaccharide In- volved in the Adhesion of a Marine Bacterium to Solid Surfaces,” Journal of General Microbiology, Vol. 74, No. 2, 1973, pp. 325-334. [28] G. G. Geesey, W. T. Richardson, H. G. Yeomans, R. T. Irvin and J. W. Costerton, “Microscopic Examination of Natural Sessile Bacterial Populations from an Alpine Stream,” Canadian Journal of Microbiology, Vol. 23, No. 2, 1977, pp. 1733-1736. [29] W. A. Corpe, “Attachment of Marine Bacteria to Solid Surfaces,” In: R. S. Manley, Ed., Adhesion in Biological Systems, Academic Press, New York, 1970, pp. 73-87. [30] M. Fletcher, “The Attachment of Bacteria to Surfaces in Aqueous Environments,” In: D. C. Ellwood, J. Melling and P. Rutter, Eds., Adhesion of Microorganisms to Sur- faces, Academic Press, London, 1979, pp. 88-108. [31] C. M. Brown, D. C. Ellwood and J. R. Hunter, “Growth of Bacteria at Surfaces: Influence of Nutrient Limitation,” FEMS Microbiology Letters, Vol. 1, No. 3, 1977, pp. 163-166. [32] I. T. Miettinen, T. Vartiainen and P. J. Martikainen, “Phos- phorus and Bacterial Growth in Drinking Water,” Applied and Environmental Microbiology, Vol. 63, No. 8, 1997, pp. 3242-3245. [33] S. G. Andrews and F. J. Millero, “Electrolyte Effects on Attachment of an Estuarine Bacterium,” Applied and En- vironmental Microbiology, Vol. 47, No. 3, 1984, pp. 495-499. [34] T. Kaneko and R. R. Colwell, “Adsorption of Vibrio Parahaemolyticus onto Chitin and Copepods,” Applied Microbiology, Vol. 29, No. 2, 1975, pp. 269-274. [35] S. L. Daniels, “The Adsorption of Microorganisms onto Solid Surfaces: A Review,” Developments in Industrial Microbiology Series, Vol. 13, 1972, pp. 211-253. [36] J. Wood, “Interaction of Micro-Organisms with Ion Ex- change Resins,” In: R. C. W. Berkeley, J. M. Lynch, J. Melling, P. R. Rutter and B. Vincent, Eds., Microbial Ad- hesion to Surfaces, Ellis-Horwood, West Sussex, 1980, pp. 163-186. [37] T. Takeuchi, T. Mukawa and H. Asanuma, “Molecular Imprinting from Fundamentals to Applications,” Wiley- VCH, Weinheim, 2003.
|